Construction of a High-Density Genetic Linkage Map Based on Bin Markers and Mapping of QTLs Associated with Fruit Size in Jujube (Ziziphus jujuba Mill.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Test Materials and Phenotypic Determination
2.2. Extraction of Genomic DNA, Library Construction, and Sequencing
2.3. SNP Marker Screening and Genotyping
2.4. Construction of Genetic Linkage Map
2.5. Gene Mapping and Nomenclature of QTLs
2.6. Screening and Annotation of Candidate Genes
2.7. Data Processing and Analysis
3. Results
3.1. Identification of SNP Markers Based on Whole-Genome Resequencing Data
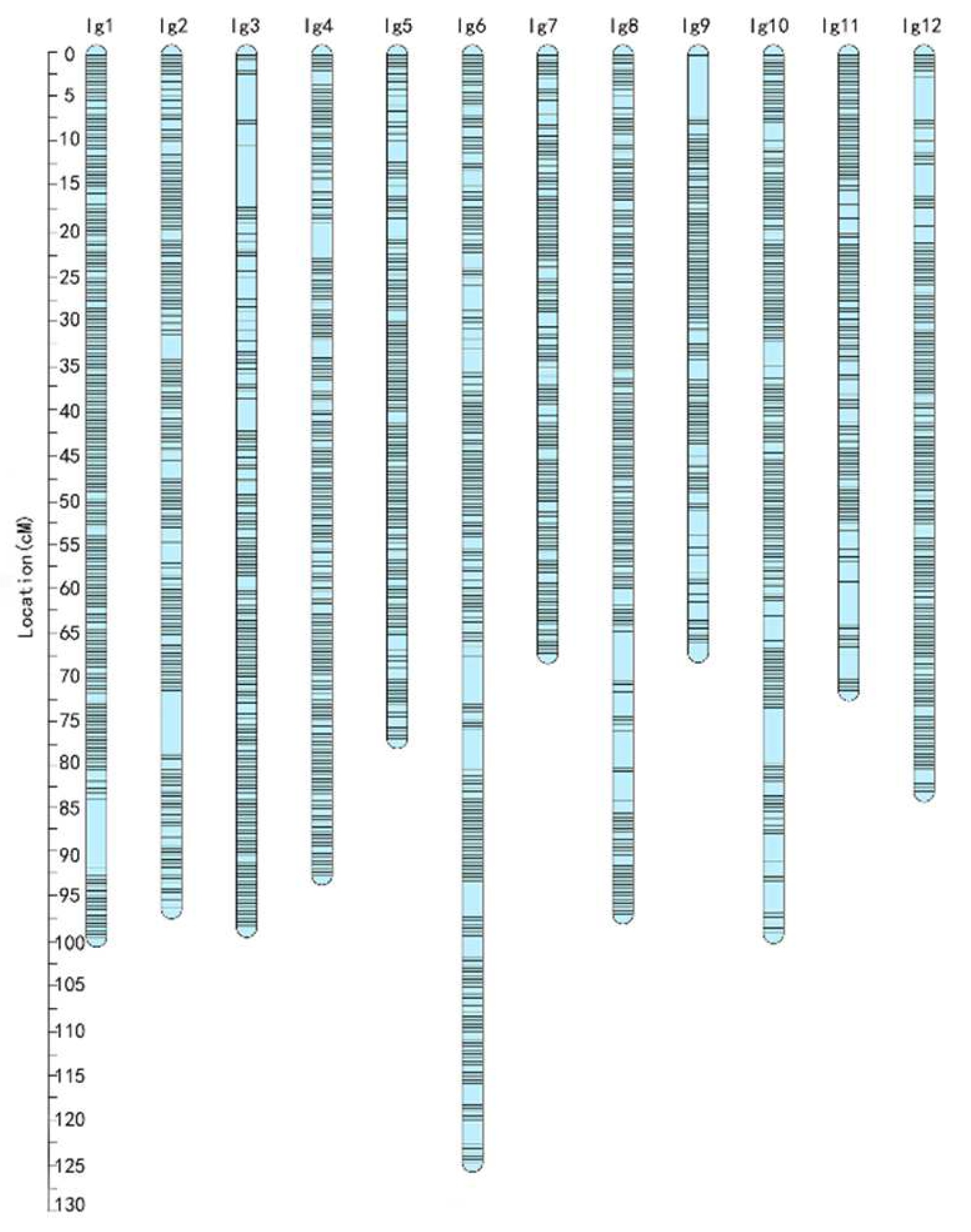
3.2. Construction of a High-Density Genetic Map Using the F1 Segregating Population Obtained from “JMS2” × “Xing16”
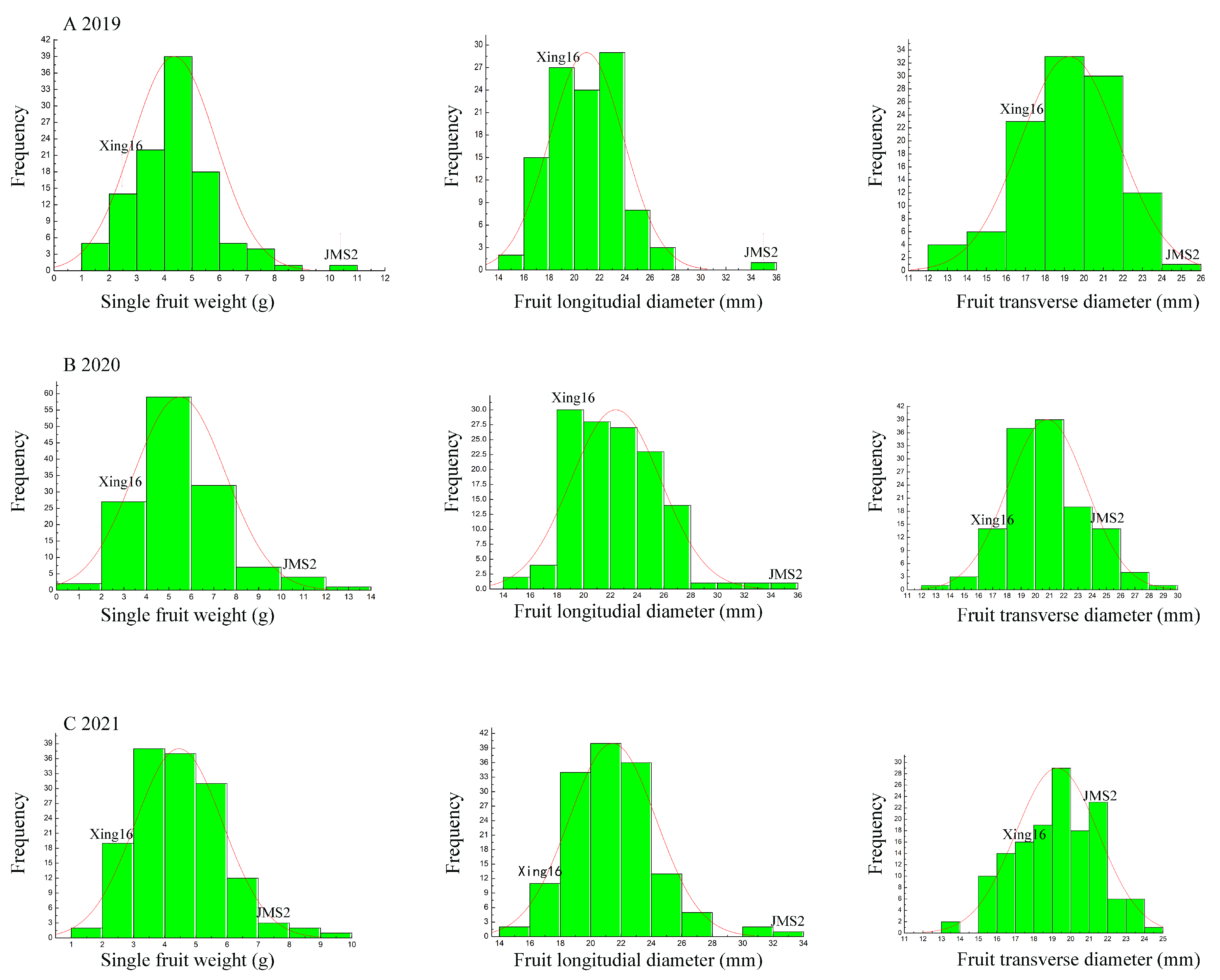
3.3. Phenotypic Analysis of Traits Relating to Fruit Size in the F1 Segregating Population
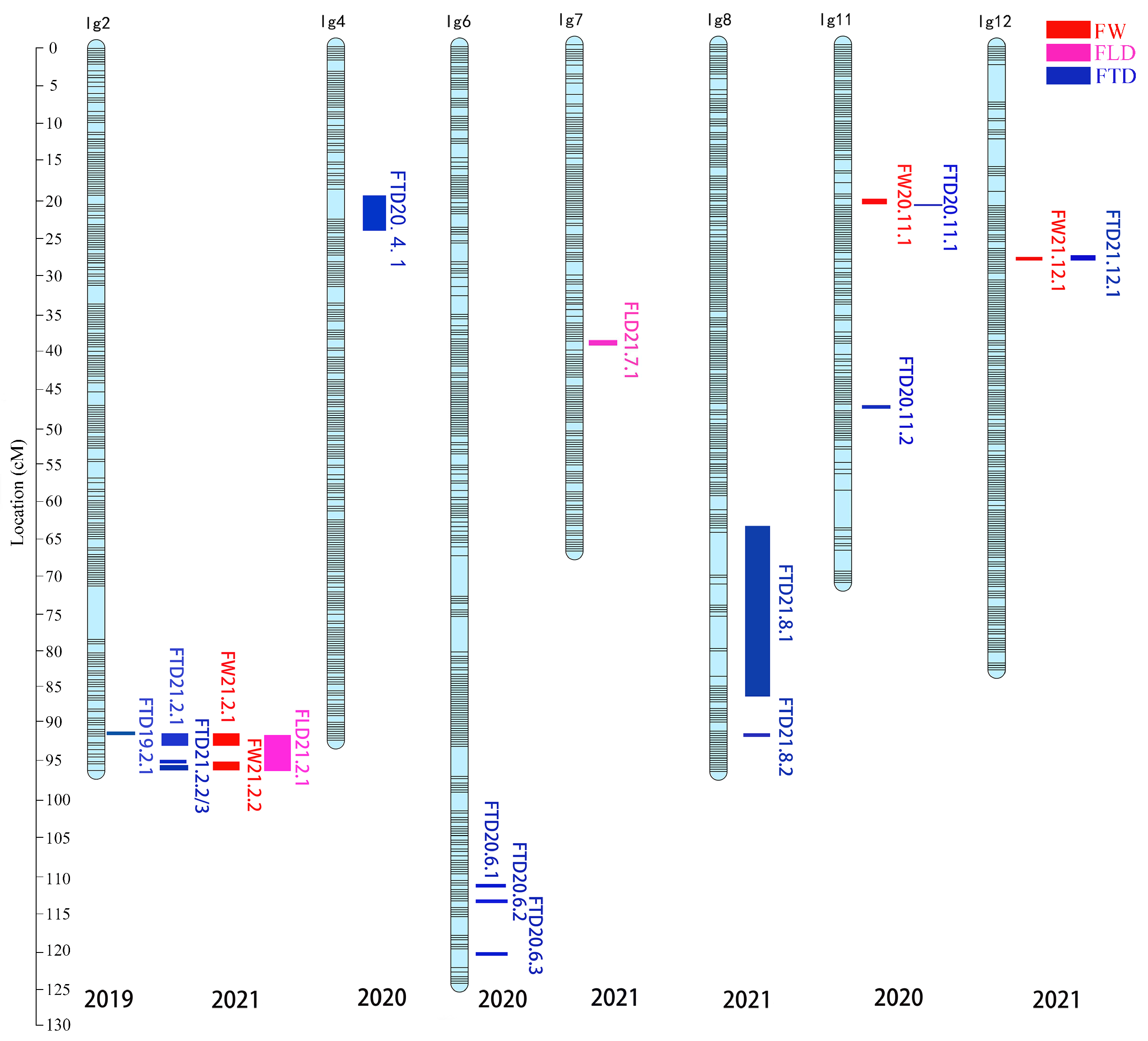
3.4. QTL Analysis of Traits Relating to Fruit Size in the F1 Segregating Population Derived from “JMS2” × “Xing16”
3.5. Prediction of the Putative Functions of Candidate Genes
4. Discussion
4.1. Advantages of Constructing a High-Density Genetic Linkage Map Based on Bin Markers
4.2. Mapping of QTLs Associated with Traits Relating to Fruit Size in Jujube
4.3. In Silico Prediction of the Putative Functions of the Candidate Genes Determining
Fruit Size
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liu, M.; Wang, J.; Wang, L.; Liu, P.; Zhao, J.; Zhao, Z.; Yao, S.; Stănică, F.; Liu, Z.; Wang, L. The historical and current research progress on jujube—A superfruit for the future. Hortic. Res. 2020, 7, 119. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Wang, J. Fruit scientific research in New China in the past 70 years: Chinese jujube. J. Fruit Sci. 2019, 36, 1369–1381. [Google Scholar]
- Meng-jun, L.; Jiu-rui, W.; Ping, L.; Jin, Z.; Zhi-hui, Z.; Li, D.; Xian-Song, L.; Zhi-guo, L. Historical achievements and frontier advances in the production and research of Chinese jujube (Ziziphus jujuba) in China. Acta Hortic. Sin. 2015, 42, 1683. [Google Scholar]
- Richardson, J.E.; Fay, M.F.; Cronk, Q.C.; Bowman, D.; Chase, M.W. A phylogenetic analysis of Rhamnaceae using rbcL and trnL-F plastid DNA sequences. Am. J. Bot. 2000, 87, 1309–1324. [Google Scholar] [CrossRef]
- Qu, Z.-Z.; Wang, Y.-H. Chinese Fruit Trees Record-Chinese Jujube; The Forestry Publishing House of China: Beijing, China, 1993. [Google Scholar]
- Liu, M.J.; Wang, M. Germplasm Resources of Chinese Jujube; China Forestry Pubilishing House: Beijing, China, 2009. [Google Scholar]
- Yang, H.; Zou, Y.; Li, X.; Zhang, M.; Zhu, Z.; Xu, R.; Xu, J.; Deng, X.; Cheng, Y. QTL analysis reveals the effect of CER1-1 and CER1-3 to reduce fruit water loss by increasing cuticular wax alkanes in citrus fruit. Postharvest Biol. Technol. 2022, 185, 111771. [Google Scholar] [CrossRef]
- Calle, A.; Wünsch, A. Multiple-population QTL mapping of maturity and fruit-quality traits reveals LG4 region as a breeding target in sweet cherry (Prunus avium L.). Hortic. Res. 2020, 7, 127. [Google Scholar] [CrossRef]
- Wu, Y.; Popovsky-Sarid, S.; Tikunov, Y.; Borovsky, Y.; Baruch, K.; Visser, R.G.; Paran, I.; Bovy, A. CaMYB12-like underlies a major QTL for flavonoid content in pepper (Capsicum annuum) fruit. New Phytol. 2023, 237, 2255–2267. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.Y. Study on Identifieation of Hybrids and Hereditary Variation of Natural Pollinated Chinese Jujube Seedlings; Agricultural University of Hebei: Baoding, China, 2003. [Google Scholar]
- Xu, L.S. QTL Mapping of Fruit Traits and Superior Genotypes Selecting in Chinese Jujube (Ziziphus jujuba Mill.). Ph.D. Thesis, Agricultural University of Hebei, Baoding, China, 2012. [Google Scholar]
- Qi, J.; Dong, Z.; Mao, Y.M.; Shen, L.Y.; Zhang, Y.X.; Liu, J.; Wang, X.L. Construction of a Dense Genetic Linkage Map and QTL Analysis of Trunk Diameter in Chinese Jujube. Sci. Silvar Sin. 2009, 45, 44–49. [Google Scholar]
- Shen, L.Y. Construction of Genetic Linkage Map and Mapping QTLs for Some Traits in Chinese Jujube (Ziziphus jujuba Mill.). Ph.D. Thesis, Agricultural University of Hebei, Baoding, China, 2005. [Google Scholar]
- Liu, M.-J.; Zhao, J.; Cai, Q.-L.; Liu, G.-C.; Wang, J.-R.; Zhao, Z.-H.; Liu, P.; Dai, L.; Yan, G.; Wang, W.-J. The complex jujube genome provides insights into fruit tree biology. Nat. Commun. 2014, 5, 5315. [Google Scholar] [CrossRef] [PubMed]
- Yan, F.; Luo, Y.; Bao, J.; Pan, Y.; Wang, J.; Wu, C.; Liu, M. Construction of a highly saturated genetic map and identification of quantitative trait loci for leaf traits in jujube. Front. Plant Sci. 2022, 13, 1001850. [Google Scholar] [CrossRef]
- Zhang, Z.; Wei, T.; Zhong, Y.; Li, X.; Huang, J. Construction of a high-density genetic map of Ziziphus jujuba Mill. using genotyping by sequencing technology. Tree Genet. Genomes 2016, 12, 76. [Google Scholar] [CrossRef]
- Hou, L.; Chen, W.; Zhang, Z.; Pang, X.; Li, Y. Genome-wide association studies of fruit quality traits in jujube germplasm collections using genotyping-by-sequencing. Plant Genome 2020, 13, e20036. [Google Scholar] [CrossRef] [PubMed]
- Qin, M.-F.; Li, L.-T.; Singh, J.; Sun, M.-Y.; Bai, B.; Li, S.-W.; Ni, J.-P.; Zhang, J.-Y.; Zhang, X.; Wei, W.-L. Construction of a high-density bin-map and identification of fruit quality-related quantitative trait loci and functional genes in pear. Hortic. Res. 2022, 9, uhac141. [Google Scholar] [CrossRef] [PubMed]
- Vision, T.J.; Brown, D.G.; Shmoys, D.B.; Durrett, R.T.; Tanksley, S.D. Selective mapping: A strategy for optimizing the construction of high-density linkage maps. Genetics 2000, 155, 407–420. [Google Scholar] [CrossRef]
- Peng, Z.; Zhao, C.; Li, S.; Guo, Y.; Xu, H.; Hu, G.; Liu, Z.; Chen, X.; Chen, J.; Lin, S.; et al. Integration of genomics, transcriptomics and metabolomics identifies candidate loci underlying fruit weight in loquat. Hortic. Res. 2022, 9, uhac037. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Aizezi, S.; Li, Y.L.; Sun, F.; Wu, G.H. Inheritance trend of the fruit traits of ‘huozhouheiyu’ grape. Acta Bot. Boreal.-Occident. Sin. 2015, 35, 275–281. [Google Scholar]
- Bu, H.; Sun, X.; Yue, P.; Qiao, J.; Sun, J.; Wang, A.; Yuan, H.; Yu, W. The MdAux/IAA2 Transcription Repressor Regulates Cell and Fruit Size in Apple Fruit. Int. J. Mol. Sci. 2022, 23, 9454. [Google Scholar] [CrossRef]
- Che, G.; Song, W.; Zhang, X. Gene network associates with CsCRC regulating fruit elongation in cucumber. Veg. Res. 2023, 3, 7. [Google Scholar] [CrossRef]
- Ma, B.; Zhao, S.; Wu, B.; Wang, D.; Peng, Q.; Owiti, A.; Fang, T.; Liao, L.; Ogutu, C.; Korban, S.S.; et al. Construction of a high density linkage map and its application in the identification of QTLs for soluble sugar and organic acid components in apple. Tree Genet. Genomes 2015, 12, 1. [Google Scholar] [CrossRef]
- Wang, L.; Li, X.; Wang, L.; Xue, H.; Wu, J.; Yin, H.; Zhang, S. Construction of a high-density genetic linkage map in pear (Pyrus communis × Pyrus pyrifolia nakai) using SSRs and SNPs developed by SLAF-seq. Sci. Hortic. 2017, 218, 198–204. [Google Scholar] [CrossRef]
- Jiang, J.; Fan, X.; Zhang, Y.; Tang, X.; Li, X.; Liu, C.; Zhang, Z. Construction of a High-Density Genetic Map and Mapping of Firmness in Grapes (Vitis vinifera L.) Based on Whole-Genome Resequencing. Int. J. Mol. Sci. 2020, 21, 797. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Liu, J.; Liu, W.; Liu, N.; Zhang, Y.; Xu, M.; Liu, S.; Ma, X.; Zhang, Y. Construction of a High-Density Genetic Map and Identification of Quantitative Trait Loci Linked to Fruit Quality Traits in Apricots Using Specific-Locus Amplified Fragment Sequencing. Front Plant Sci 2022, 13, 798700. [Google Scholar] [CrossRef] [PubMed]
- Paterson, A.H.; Brubaker, C.L.; Wendel, J.F. A rapid method for extraction of cotton (Gossypium spp.) genomic DNA suitable for RFLP or PCR analysis. Plant Mol. Biol. Report. 1993, 11, 122–127. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Cingolani, P.; Platts, A.; Wang, L.L.; Coon, M.; Nguyen, T.; Wang, L.; Land, S.J.; Lu, X.; Ruden, D.M. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. fly 2012, 6, 80–92. [Google Scholar] [CrossRef]
- Liu, D.; Ma, C.; Hong, W.; Huang, L.; Liu, M.; Liu, H.; Zeng, H.; Deng, D.; Xin, H.; Song, J. Construction and analysis of high-density linkage map using high-throughput sequencing data. PLoS ONE 2014, 9, e98855. [Google Scholar] [CrossRef]
- Yandell, B.S.; Mehta, T.; Banerjee, S.; Shriner, D.; Venkataraman, R.; Moon, J.Y.; Neely, W.W.; Wu, H.; Von Smith, R.; Yi, N. R/qtlbim: QTL with Bayesian interval mapping in experimental crosses. Bioinformatics 2007, 23, 641–643. [Google Scholar] [CrossRef]
- Zhao, J.; Xu, Y.; Li, H.; Yin, Y.; An, W.; Li, Y.; Wang, Y.; Fan, Y.; Wan, R.; Guo, X. A SNP-based high-density genetic map of leaf and fruit related quantitative trait loci in wolfberry (Lycium Linn.). Front. Plant Sci. 2019, 10, 977. [Google Scholar] [CrossRef]
- Pereira, L.; Ruggieri, V.; Pérez, S.; Alexiou, K.G.; Fernández, M.; Jahrmann, T.; Pujol, M.; Garcia-Mas, J. QTL mapping of melon fruit quality traits using a high-density GBS-based genetic map. BMC Plant Biol. 2018, 18, 324. [Google Scholar] [CrossRef] [PubMed]
- Huang, X.; Feng, Q.; Qian, Q.; Zhao, Q.; Wang, L.; Wang, A.; Guan, J.; Fan, D.; Weng, Q.; Huang, T. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009, 19, 1068–1076. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Koonin, E.V.; Lipman, D.J. A Genomic Perspective on Protein Families. Science 1997, 278, 631–637. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; Xu, L.; Wang, Y.; Dong, J.; Chen, Y.; Tang, M.; Fan, L.; Zhu, Y.; Liu, L. An ultra-high-density genetic map provides insights into genome synteny, recombination landscape and taproot skin colour in radish (Raphanus sativus L.). Plant Biotechnol. J. 2020, 18, 274–286. [Google Scholar] [CrossRef] [PubMed]
- Dong, Z.; Alam, M.K.; Xie, M.; Yang, L.; Liu, J.; Helal, M.; Huang, J.; Cheng, X.; Liu, Y.; Tong, C. Mapping of a major QTL controlling plant height using a high-density genetic map and QTL-seq methods based on whole-genome resequencing in Brassica napus. G3 2021, 11, jkab118. [Google Scholar] [CrossRef] [PubMed]
- Lian, Q.; Fu, Q.; Xu, Y.; Hu, Z.; Zheng, J.; Zhang, A.; He, Y.; Wang, C.; Xu, C.; Chen, B. QTLs and candidate genes analyses for fruit size under domestication and differentiation in melon (Cucumis melo L.) based on high resolution maps. BMC Plant Biol. 2021, 21, 126. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhao, Y.; Guo, Y.; Su, K.; Shi, X.; Liu, D.; Zhang, J. High-density genetic linkage-map construction of hawthorn and QTL mapping for important fruit traits. PLoS ONE 2020, 15, e0229020. [Google Scholar] [CrossRef]
- Shi, G.; Sun, D.; Wang, Z.; Liu, X.; Guo, J.; Zhang, S.; Zhao, Y.; Ai, J. Construction of a resequencing-based high-density genetic map for grape using an interspecific population (Vitis amurensis × Vitis vinifera). Hortic. Environ. Biotechnol. 2022, 63, 489–497. [Google Scholar] [CrossRef]
- Bao, J.K. Construction of High-Density Genetic Map and QTL Mapping of Fruit Size and Sugar-Acid Traits on ‘JMS2′ × ‘Jiaocheng 5′ Jujuba Hybrid Progeny. Master’s Thesis, Tarim University, Alar, China, 2022. [Google Scholar]
- Wang, Z. High-density Genetic Map Construction and QTL Mapping of Agronomic Traits of Ziziphus jujuba Mill. Ph.D. Thesis, College of Forestry Northwest A&F University, Yangling, China, 2020. [Google Scholar]
- Zhang, H.; Tan, J.; Zhang, M.; Huang, S.; Chen, X. Comparative transcriptomic analysis of two bottle gourd accessions differing in fruit size. Genes 2020, 11, 359. [Google Scholar] [CrossRef]
- Liu, X. Studies on Mechanism of Fruit Growth and Development of Different Ripening-Season of Pears. Ph.D. Thesis, Sichuan Agriculture University, Ya’an, China, 2008. [Google Scholar]
- Bashline, L.; Lei, L.; Li, S.; Gu, Y. Cell wall, cytoskeleton, and cell expansion in higher plants. Mol. Plant 2014, 7, 586–600. [Google Scholar] [CrossRef]
- Guo, M.; Zhang, Z.; Cheng, Y.; Li, S.; Shao, P.; Yu, Q.; Wang, J.; Xu, G.; Zhang, X.; Liu, J. Comparative population genomics dissects the genetic basis of seven domestication traits in jujube. Hortic. Res. 2020, 7, 89. [Google Scholar] [CrossRef] [PubMed]
- Mahjoub, A.; Hernould, M.; Joubès, J.; Decendit, A.; Mars, M.; Barrieu, F.; Hamdi, S.; Delrot, S. Overexpression of a grapevine R2R3-MYB factor in tomato affects vegetative development, flower morphology and flavonoid and terpenoid metabolism. Plant Physiol. Biochem. 2009, 47, 551–561. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Liu, X.; Sun, C.; Song, X.; Li, X.; Cui, H.; Guo, J.; Liu, L.; Ying, A.; Zhang, Z.; et al. CsTRM5 regulates fruit shape via mediating cell division direction and cell expansion in cucumber. Hortic. Res. 2023, 10, uhad007. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Song, X.; Li, H.; Zhang, Z.; Zhang, J. Transcriptome and hormones analysis provide insights into elongated fruit somaclonal mutant in ‘Akihime’ strawberry. Sci. Hortic. 2023, 309, 111608. [Google Scholar] [CrossRef]
- Yang, L.; Zhou, H.i.; Bo, W.h.; Li, Y.y.; Pang, X.m. Identification of genes related with jujube fruit size based on selective sweep analysis. J. Beijing For. Univ. 2019, 41, 30–36. [Google Scholar]
| Sample | Total Clean Bases (Gb) | Q30 Proportion (%) | GC Proportion (%) | Average Sequencing Depth (×) | Mapped (%) | Coverage Ratio (%) |
|---|---|---|---|---|---|---|
| Female parent | 13.93 | 93.19 | 33.71 | 28× | 97.91 | 87.31 |
| Male parent | 9.92 | 93.61 | 33.74 | 19× | 97.84 | 86.24 |
| Offspring | 362.65 | 92.82 | 33.96 | 4.14× | 97.04 | 76.33 |
| Parents | SNP Number | Transition Number | Transverison Number | Ti/Tv Ratio | Heterozygous SNP Number | Homozygous SNP Number |
|---|---|---|---|---|---|---|
| Female parent | 1,569,033 | 998,634 | 570,399 | 1.75 | 615,822 | 953,211 |
| Male parent | 1,569,033 | 998,634 | 570,399 | 1.75 | 713,815 | 855,218 |
| Linkage Group | Number of Bin Markers | Number of SNP Markers | Genetic Length (cM) | Average Distance (cM) | Max Gap (cM) | Gap < 5 cM (%) |
|---|---|---|---|---|---|---|
| LG1 | 263 | 16,203 | 99.45 | 0.38 | 7.76 | 99.62 |
| LG2 | 211 | 10,638 | 96.26 | 0.46 | 7.06 | 99.52 |
| LG3 | 188 | 9802 | 98.36 | 0.52 | 7.06 | 99.47 |
| LG4 | 228 | 10,716 | 92.54 | 0.41 | 4.05 | 100.00 |
| LG5 | 182 | 11,390 | 77.14 | 0.42 | 2.46 | 100.00 |
| LG6 | 249 | 9032 | 124.86 | 0.50 | 5.37 | 99.60 |
| LG7 | 165 | 9878 | 67.46 | 0.41 | 1.83 | 100.00 |
| LG8 | 221 | 8205 | 96.89 | 0.44 | 5.70 | 99.55 |
| LG9 | 135 | 9366 | 67.43 | 0.50 | 7.06 | 99.25 |
| LG10 | 206 | 7228 | 99.12 | 0.48 | 6.38 | 99.51 |
| LG11 | 151 | 6948 | 71.71 | 0.47 | 5.04 | 99.33 |
| LG12 | 199 | 6906 | 83.11 | 0.42 | 5.04 | 99.49 |
| Totals | 2398 | 116,312 | 1074.33 | - | - | - |
| Overall average | - | - | - | 0.45 | - | 99.61 |
| Max | - | - | - | - | 7.76 | - |
| Traits | Year | Parents | F1 population | |||||
|---|---|---|---|---|---|---|---|---|
| JMS2 | Xing16 | Range | Mean ± SD | Ta % | Skewness | Kurtosis | ||
| Single fruit weight (g) | 2019 | 11.00 | 2.76 | 1.02–8.45 | 4.30 ± 1.38 | 62.52 | 0.200 | 0.287 |
| 2020 | 10.50 | 2.81 | 1.46–13.41 | 5.47 ± 1.99 | 82.22 | 0.989 | 1.817 | |
| 2021 | 7.82 | 2.52 | 1.67–9.01 | 4.45 ± 1.38 | 86.09 | 0.574 | 0.557 | |
| Fruit longitudinal diameter (mm) | 2019 | 34.47 | 17.76 | 15.04–27.19 | 20.87 ± 2.64 | 79.92 | 0.020 | −0.509 |
| 2020 | 35.04 | 18.49 | 14.32–32.49 | 22.33 ± 3.18 | 83.44 | 0.200 | 0.108 | |
| 2021 | 32.58 | 17.45 | 14.88–31.34 | 21.40 ± 2.71 | 85.54 | 0.566 | 1.205 | |
| Fruit transverse diameter (mm) | 2019 | 25.58 | 16.16 | 12.27–23.57 | 19.22 ± 2.44 | 92.10 | −0.512 | 0.111 |
| 2020 | 24.88 | 16.56 | 13.60–28.20 | 20.83 ± 2.74 | 100.53 | 0.146 | 0.067 | |
| 2021 | 21.70 | 16.44 | 13.71–24.49 | 19.27 ± 2.24 | 101.04 | 0.566 | 1.205 | |
| Year | FW vs. FLD | FW vs. FTD | FLD vs. FTD |
|---|---|---|---|
| 2019 | 0.8360 ** | 0.9280 ** | 0.7512 ** |
| 2020 | 0.8771 ** | 0.9424 ** | 0.7972 ** |
| 2021 | 0.8291 ** | 0.9370 ** | 0.6832 ** |
| Trait | QTLs | Intervals on Maps (cM) | LOD | Peak Marker Position (cM) | PVE (%) | Containing Markers |
|---|---|---|---|---|---|---|
| Single fruit weight | FW20.11.1 | 20.475–21.382 | 3.15 | 20.475 | 10.7 | 2 |
| FW21.2.1 | 91.738–92.944 | 3.44 | 92.644 | 10.8 | 3 | |
| FW21.2.2 | 94.45–96.259 | 3.63 | 94.45 | 11.3 | 4 | |
| FW21.12.1 | 28.383 | 3.02 | 28.383 | 9.5 | 1 | |
| Fruit transverse diameter | FTD19.2.1 | 91.437–91.738 | 3.08 | 91.437 | 13.3 | 2 |
| FTD20.4.1 | 18.968–23.921 | 3.27 | 23.02 | 11.1 | 5 | |
| FTD20.6.1 | 111.226 | 3.02 | 111.226 | 10.3 | 1 | |
| FTD20.6.2 | 113.029–113.93 | 3.15 | 113.33 | 10.7 | 4 | |
| FTD20.6.3 | 122.75 | 3.2 | 122.75 | 10.9 | 1 | |
| FTD20.11.1 | 21.382 | 3.04 | 21.382 | 10.3 | 1 | |
| FTD20.11.2 | 47.29–47.891 | 3.04 | 47.891 | 10.4 | 3 | |
| FTD21.2.1 | 91.738–92.944 | 3.35 | 92.644 | 10.5 | 3 | |
| FTD21.2.2 | 94.45 | 3.07 | 94.45 | 9.7 | 1 | |
| FTD21.2.3 | 95.352–96.259 | 3.15 | 95.352 | 9.9 | 2 | |
| FTD21.8.1 | 64.38–86.663 | 4.51 | 71.891 | 13.9 | 18 | |
| FTD21.8.2 | 92.085 | 3.00 | 92.085 | 9.5 | 1 | |
| FTD21.12.1 | 27.783–28.683 | 3.47 | 28.383 | 10.9 | 4 | |
| Fruit longitudinal diameter | FLD21.2.1 | 91.137–96.259 | 3.91 | 94.45 | 12.1 | 11 |
| FLD21.7.1 | 38.567–39.468 | 3.10 | 38.867 | 9.7 | 4 |
| Traits | Corresponding QTL | Coincidence Interval (cM) | Marker Number | LG |
|---|---|---|---|---|
| Fruit transverse diameter, Single fruit weight, Fruit longitudinal diameter | FTD21.1, FW21.1, FLD21.1 | 91.738–92.944 | 3 | 2 |
| Single fruit weight, Fruit transverse diameter, Fruit longitudinal diameter | FW21.2, FTD21.2, FLD21.1 | 94.450–96.259 | 4 | 2 |
| Fruit transverse diameter, Single fruit weight | FTD21.1, FW21.1 | 28.383 | 1 | 12 |
| Fruit transverse diameter, Single fruit weight | FTD20.1, FW20.1 | 21.382 | 1 | 11 |
| LG | Coincidence Interval (cM) | QTL Lloci | Gene Number | COG Anno | GO Anno | KEGG Anno | Swissprot Anno | Nr Anno |
|---|---|---|---|---|---|---|---|---|
| 2 | 91.738 | FTD19.1 | 9 | 3 | 4 | 5 | 9 | 9 |
| FTD21.1 | 19 | 0 | 10 | 10 | 14 | 19 | ||
| 2 | 91.738–92.944 | FTD21.1 | 19 | 0 | 10 | 10 | 14 | 19 |
| FW21.1 | 19 | 0 | 10 | 10 | 14 | 19 | ||
| FLD21.1 | 99 | 33 | 60 | 40 | 71 | 99 | ||
| 2 | 94.450–96.259 | FW21.2 | 61 | 21 | 36 | 21 | 47 | 61 |
| FTD21.2 | 41 | 12 | 24 | 16 | 32 | 41 | ||
| FLD21.1 | 99 | 33 | 60 | 40 | 71 | 99 | ||
| 12 | 28.383 | FTD21.1 | 30 | 12 | 25 | 14 | 28 | 30 |
| FW21.1 | 3 | 2 | 3 | 2 | 2 | 3 | ||
| 11 | 21.382 | FTD20.1 | 0 | 0 | 0 | 0 | 0 | 0 |
| FW20.1 | 1 | 0 | 1 | 0 | 1 | 1 | ||
| 2 | 94.45–96.259 | FW21.2 | 61 | 21 | 36 | 21 | 47 | 61 |
| 2 | 91.137–96.259 | FLD21.1 | 99 | 33 | 60 | 40 | 71 | 99 |
| 8 | 64.38–86.663 | FTD21.1 | 167 | 55 | 111 | 68 | 125 | 167 |
| Total | - | - | 727 | 225 | 450 | 297 | 546 | 727 |
| Range (cM) | QTL Name | Candidate Genes | Candidate Gene ID | Gene Annotation |
|---|---|---|---|---|
| 91.738 | FTD19.2.1 | - | - | - |
| FTD21.2.1 | LOC107410242 | rna-XM_025070976.1 | anatomical structure morphogenesis;tissue development; regulation of gene expression; cellular process; developmental process; regulation of cellular process | |
| LOC107409642 | rna-XM_016017070.2 | regulation of cellular process | ||
| 91.738–92.944 | FW21.2.1 | LOC107410242 | rna-XM_025070976.1 | anatomical structure morphogenesis; tissue development; regulation of gene expression; cellular process; developmental process; regulation of cellular process |
| LOC107409642 | rna-XM_016017070.2 | regulation of cellular process | ||
| FLD21.2.1 | LOC107409704 | rna-XM_016017148.2 | Cell wall | |
| LOC107409953 | rna-XM_016017375.2 | LRR receptor-like serine/threonine-protein At3g47570 | ||
| LOC112490770 | rna-XM_025071274.1 | LRR receptor-like serine/threonine-protein EFR | ||
| LOC107410143 | rna-XM_016017551.2 | Leucine-rich repeat-containing protein | ||
| LOC107410070 | rna-XM_016017481.1 | regulation of mitotic cell cycle | ||
| LOC107409704 | rna-XM_016017134.2 | Cell wall | ||
| LOC107409998 | rna-XM_016017417.2 | LRR receptor-like serine/threonine-protein EFR | ||
| LOC107410070 | rna-XM_025070726.1 | regulation of mitotic cell cycle | ||
| LOC107410070 | rna-XM_016017502.2 | regulation of mitotic cell cycle | ||
| LOC107410242 | rna-XM_025070976.1 | anatomical structure morphogenesis; tissue development; regulation of gene expression; cellular process; developmental process; regulation of cellular process | ||
| LOC107409704 | rna-XM_016017140.2 | Cell wall | ||
| LOC107409426 | rna-XM_016016863.2 | Zinc finger protein | ||
| LOC107410070 | rna-XM_016017488.1 | regulation of mitotic cell cycle | ||
| LOC107409919 | rna-XM_016017343.2 | LRR receptor-like serine/threonine-protein At3g47570 | ||
| LOC107409987 | rna-XM_016017405.2 | NAC domain-containing protein | ||
| LOC107410070 | rna-XM_025070727.1 | regulation of mitotic cell cycle | ||
| LOC107410143 | rna-XM_016017558.2 | Leucine-rich repeat-containing protein | ||
| LOC107409642 | rna-XM_016017070.2 | regulation of cellular process | ||
| 94.450–96.259 | FW21.2.2 | LOC107409953 | rna-XM_016017375.2 | LRR receptor-like serine/threonine-protein At3g47570 |
| LOC107409987 | rna-XM_016017405.2 | NAC domain-containing protein | ||
| LOC112490770 | rna-XM_025071274.1 | LRR receptor-like serine/threonine-protein EFR | ||
| LOC107409919 | rna-XM_016017343.2 | LRR receptor-like serine/threonine-protein At3g47570 | ||
| LOC107409426 | rna-XM_016016863.2 | Zinc finger protein | ||
| LOC107410143 | rna-XM_016017551.2 | Leucine-rich repeat-containing protein | ||
| LOC107409998 | rna-XM_016017417.2 | LRR receptor-like serine/threonine-protein EFR | ||
| LOC107410143 | rna-XM_016017558.2 | Leucine-rich repeat-containing protein | ||
| FTD21.2.2 | LOC107409426 | rna-XM_016016863.2 | Zinc finger protein | |
| LOC107410143 | rna-XM_016017551.2 | Leucine-rich repeat-containing protein | ||
| LOC107409953 | rna-XM_016017375.2 | LRR receptor-like serine/threonine-protein At3g47570 | ||
| LOC107410143 | rna-XM_016017558.2 | Leucine-rich repeat-containing protein | ||
| FLD21.2.1 | LOC107409704 | rna-XM_016017148.2 | Cell wall | |
| LOC107409953 | rna-XM_016017375.2 | LRR receptor-like serine/threonine-protein At3g47570 | ||
| LOC112490770 | rna-XM_025071274.1 | LRR receptor-like serine/threonine-protein EFR | ||
| LOC107410143 | rna-XM_016017551.2 | Leucine-rich repeat-containing protein | ||
| LOC107410070 | rna-XM_016017481.1 | regulation of mitotic cell cycle | ||
| LOC107409704 | rna-XM_016017134.2 | Cell wall | ||
| LOC107409998 | rna-XM_016017417.2 | LRR receptor-like serine/threonine-protein EFR | ||
| LOC107410070 | rna-XM_025070726.1 | regulation of mitotic cell cycle | ||
| LOC107410070 | rna-XM_016017502.2 | regulation of mitotic cell cycle | ||
| LOC107410242 | rna-XM_025070976.1 | anatomical structure morphogenesis; tissue development; regulation of gene expression; cellular process; developmental process; regulation of cellular process | ||
| LOC107409704 | rna-XM_016017140.2 | Cell wall | ||
| LOC107409426 | rna-XM_016016863.2 | Zinc finger protein | ||
| LOC107410070 | rna-XM_016017488.1 | regulation of mitotic cell cycle | ||
| LOC107409919 | rna-XM_016017343.2 | LRR receptor-like serine/threonine-protein At3g47570 | ||
| LOC107409987 | rna-XM_016017405.2 | NAC domain-containing protein | ||
| LOC107410070 | rna-XM_025070727.1 | regulation of mitotic cell cycle | ||
| LOC107410143 | rna-XM_016017558.2 | Leucine-rich repeat-containing protein | ||
| LOC107409642 | rna-XM_016017070.2 | regulation of cellular process | ||
| 28.383 | FTD21.12.1 | LOC107431773 | rna-XM_016042763.2 | E3 ubiquitin-protein ligase |
| LOC107431775 | rna-XM_025079541.1 | Cindole-3-acetic acid amido synthetase activity | ||
| LOC107431773 | rna-XM_016042765.2 | E3 ubiquitin-protein ligase | ||
| LOC107431773 | rna-XM_025066560.1 | E3 ubiquitin-protein ligase | ||
| LOC107431775 | rna-XM_016042766.2 | indole-3-acetic acid amido synthetase activity | ||
| LOC107431773 | rna-XM_025066561.1 | E3 ubiquitin-protein ligase | ||
| LOC107431772 | rna-XM_016042762.2 | plant-type secondary cell wall biogenesis | ||
| LOC107431773 | rna-XM_016042764.2 | E3 ubiquitin-protein ligase | ||
| FW21.12.1 | - | - | - | |
| 94.450–96.259 | FW21.2.2 | LOC107409953 | rna-XM_016017375.2 | LRR receptor-like serine/threonine-protein At3g47570 |
| LOC112490770 | rna-XM_025071274.1 | LRR receptor-like serine/threonine-protein EFR | ||
| LOC107409919 | rna-XM_016017343.2 | LRR receptor-like serine/threonine-protein At3g47570 | ||
| LOC107410143 | rna-XM_016017551.2 | Leucine-rich repeat-containing protein | ||
| LOC107409998 | rna-XM_016017417.2 | LRR receptor-like serine/threonine-protein EFR | ||
| LOC107410143 | rna-XM_016017558.2 | Leucine-rich repeat-containing protein | ||
| 91.137–96.259 | FLD21.2.1 | LOC107409704 | rna-XM_016017148.2 | Cell wall |
| LOC107409953 | rna-XM_016017375.2 | LRR receptor-like serine/threonine-protein At3g47570 | ||
| LOC112490770 | rna-XM_025071274.1 | LRR receptor-like serine/threonine-protein EFR | ||
| LOC107410143 | rna-XM_016017551.2 | Leucine-rich repeat-containing protein | ||
| LOC107410070 | rna-XM_016017481.1 | regulation of mitotic cell cycle | ||
| LOC107409704 | rna-XM_016017134.2 | Cell wall | ||
| LOC107409998 | rna-XM_016017417.2 | LRR receptor-like serine/threonine-protein EFR | ||
| LOC107410070 | rna-XM_025070726.1 | regulation of mitotic cell cycle | ||
| LOC107410070 | rna-XM_016017502.2 | regulation of mitotic cell cycle | ||
| LOC107410242 | rna-XM_025070976.1 | anatomical structure morphogenesis; tissue development; regulation of gene expression; cellular process; developmental process; regulation of cellular process | ||
| LOC107409704 | rna-XM_016017140.2 | Cell wall | ||
| LOC107409426 | rna-XM_016016863.2 | Zinc finger protein | ||
| LOC107410070 | rna-XM_016017488.1 | regulation of mitotic cell cycle | ||
| LOC107409919 | rna-XM_016017343.2 | LRR receptor-like serine/threonine-protein At3g47570 | ||
| LOC107410070 | rna-XM_025070727.1 | regulation of mitotic cell cycle | ||
| LOC107410143 | rna-XM_016017558.2 | Leucine-rich repeat-containing protein | ||
| 64.380–86.663 | FTD21.8.1 | LOC107423900 | rna-XM_016033552.2 | cell wall; auxin-activated signaling pathway |
| LOC107423928 | rna-XM_025076890.1 | cytokinin metabolic process | ||
| LOC107423913 | rna-XM_016033565.2 | indoleacetic acid biosynthetic process; response to auxin | ||
| LOC107423813 | rna-XM_016033450.2 | E3 ubiquitin-protein ligase | ||
| LOC107423814 | rna-XM_016033451.1 | cell wall; cell wall modification | ||
| LOC107423928 | rna-XM_025076888.1 | cytokinin metabolic process | ||
| LOC107423861 | rna-XM_016033508.2 | protein serine/threonine kinase activity | ||
| LOC107423912 | rna-XM_016033564.2 | indoleacetic acid biosynthetic process; response to auxin | ||
| LOC107423811 | rna-XM_016033446.2 | Transcription factor bHLH | ||
| LOC107423821 | rna-XM_025076730.1 | mitotic cell cycle; regulation of cell cycle | ||
| LOC107423857 | rna-XM_016033503.2 | cell wall | ||
| LOC107423801 | rna-XM_016033440.1 | E3 ubiquitin-protein ligase | ||
| LOC107423922 | rna-XM_016033576.2 | plant-type cell wall biogenesis; regulation of meristem growth | ||
| LOC107423928 | rna-XM_016033582.2 | cytokinin metabolic process | ||
| LOC107423848 | rna-XM_016033491.2 | cell wall; Pectin methylesterase | ||
| LOC107423928 | rna-XM_025076887.1 | cytokinin metabolic process | ||
| LOC107423928 | rna-XM_025076889.1 | cytokinin metabolic process | ||
| LOC107423846 | rna-XM_016033490.2 | cell wall; pectinesterase activity; cell wall modification; pectin catabolic process | ||
| LOC107423858 | rna-XM_025076143.1 | cell wall | ||
| LOC107423925 | rna-XM_016033579.2 | cell wall | ||
| LOC107423923 | rna-XM_016033577.2 | protein serine/threonine kinase activity | ||
| LOC107423815 | rna-XM_025076224.1 | putative pectinesterase/pectinesterase inhibitor 45-like | ||
| LOC107423897 | rna-XM_016033547.2 | Leucine-rich repeat receptor-like protein kinase | ||
| LOC107423905 | rna-XM_016033556.2 | response to auxin | ||
| LOC107423856 | rna-XM_025076290.1 | cell wall | ||
| LOC107423846 | rna-XM_016033489.2 | cell wall; Pectinesterase PPE8B | ||
| LOC107423903 | rna-XM_016033555.1 | cell wall | ||
| LOC107423821 | rna-XM_016033457.2 | mitotic cell cycle; meiotic cell cycle; regulation of cell cycle | ||
| LOC107423816 | rna-XM_016033453.2 | E3 ubiquitin-protein ligase | ||
| LOC107423881 | rna-XM_016033528.2 | Cell wall | ||
| LOC107423820 | rna-XM_016033456.2 | Pectinesterase 54 | ||
| 21.382 | FTD20.11.1 | - | - | - |
| FW20.11.1 | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, T.; Qiu, Q.; Yan, F.; Wang, Z.; Bao, J.; Yang, Z.; Xia, Y.; Wang, J.; Wu, C.; Liu, M. Construction of a High-Density Genetic Linkage Map Based on Bin Markers and Mapping of QTLs Associated with Fruit Size in Jujube (Ziziphus jujuba Mill.). Horticulturae 2023, 9, 836. https://doi.org/10.3390/horticulturae9070836
Guo T, Qiu Q, Yan F, Wang Z, Bao J, Yang Z, Xia Y, Wang J, Wu C, Liu M. Construction of a High-Density Genetic Linkage Map Based on Bin Markers and Mapping of QTLs Associated with Fruit Size in Jujube (Ziziphus jujuba Mill.). Horticulturae. 2023; 9(7):836. https://doi.org/10.3390/horticulturae9070836
Chicago/Turabian StyleGuo, Tianfa, Qianqian Qiu, Fenfen Yan, Zhongtang Wang, Jingkai Bao, Zhi Yang, Yilei Xia, Jiurui Wang, Cuiyun Wu, and Mengjun Liu. 2023. "Construction of a High-Density Genetic Linkage Map Based on Bin Markers and Mapping of QTLs Associated with Fruit Size in Jujube (Ziziphus jujuba Mill.)" Horticulturae 9, no. 7: 836. https://doi.org/10.3390/horticulturae9070836
APA StyleGuo, T., Qiu, Q., Yan, F., Wang, Z., Bao, J., Yang, Z., Xia, Y., Wang, J., Wu, C., & Liu, M. (2023). Construction of a High-Density Genetic Linkage Map Based on Bin Markers and Mapping of QTLs Associated with Fruit Size in Jujube (Ziziphus jujuba Mill.). Horticulturae, 9(7), 836. https://doi.org/10.3390/horticulturae9070836





