Abstract
Camellia oleifera Abel., which produces fruits of high comprehensive utilization value, is an important woody oil tree in China. ZEITLUPE (ZTL) is a blue light receptor and clock component protein that is involved in various physiological and biochemical processes. However, the expression pattern and function of C. oleifera ZTL (CoZTL) remain unclear. In this study, the coding sequence of the CoZTL gene was isolated and the protein function was explored using bioinformatics and expression analyses and heterologous expression techniques. The results showed that the CoZTL protein was highly conserved during evolution and was on the same branch of the evolutionary tree as the ZTL proteins from Ipomoea nil and Nicotiana attenuata. CoZTL was mainly expressed in the fruit shells and stems of C. oleifera, and its expression level fluctuated greatly during flower bud development. Transgenic CoZTL-overexpressing Arabidopsis plants showed delayed flowering under long-day conditions as well as light-dependent promotion of hypocotyl elongation. Furthermore, yeast two-hybrid library screening revealed that seven C. oleifera proteins (CoAAT, Coβ-GAL, CoLAT52-like, CoCAR4-like, CoAO, CoUQCC1, and CoADF 2) interacted with CoZTL. Our results indicate that CoZTL plays an important role in C. oleifera flowering and hypocotyl growth.
1. Introduction
Camellia oleifera Abel. is a woody oil tree of the family Theaceae [1]. The seeds of this plant can be pressed to release tea oil, which has a long history of consumption in China and is well received by consumers. Studies have shown that tea oil contains up to 90% unsaturated fatty acids and a variety of bioactive substances, such as flavonoids, tocopherols, squalene, and triterpenoids, all of which have both medicinal and edible value [2,3,4]. For a long time, the phenomenon of more flowers and fewer fruits was common in the production of C. oleifera, mainly for two reasons. First, the flowering period occurs in autumn and winter, when the temperature is low and the weather is changeable, which is not conducive to flowering and pollination [5,6]. Second, the self-incompatibility of C. oleifera leads to a low fruit-set rate [7]. At present, cross-pollination is used to increase the fruit-set rate to raise the fruit yield of C. oleifera [8]. However, bad weather during the flowering period still has an uncontrollable influence on fruit setting and the early development of young fruits. Therefore, it would be beneficial to increase the yield of C. oleifera by selecting a variety with a more suitable flowering period.
Light is the main environmental indicator used by plants to detect seasonal and diurnal changes [9]. Various plant photoreceptors can sensitively perceive light signals and transmit them to the core oscillators of the biological clock, which integrates various external environmental signals for adaptive regulation, the primary way in which plant growth and development are regulated [10,11,12]. Blue light, which has a wavelength range of 400–500 nm, is involved in the regulation of plant photomorphogenesis, photosynthesis, and the synthesis of active substances [13,14,15]. Blue light receptors have been shown to be involved not only in these above-mentioned processes but also in the regulation of the plant photoperiod, phototropism, stress response, and organ development [16,17].
Zeitlupe (ZTL) is a blue light receptor and clock component protein that plays a specific role in many biological processes [18]. The protein is mainly made up of light–oxygen–voltage (LOV) and F-box domains. The LOV domain senses blue light and interacts physically with other proteins [19], whereas the F-box domain serves as a ubiquitin ligase-binding site for formation of the Skp–Cullin–F-box-protein (SCF)ZTL complex, which degrades proteins bound to the LOV domain [20]. Studies have shown that ZTL affects the period of the core oscillator of the circadian clock by degrading timing of CAB expression 1 (TOC1) and pseudo-response regulator (PRR) proteins [21]. Overexpression of the ZTL gene in Arabidopsis thaliana (AtZTL) can lead to circadian rhythm disorder, whereas its deletion results in circadian rhythm shortening. ZTL can indirectly regulate hypocotyl elongation, the shading response, and thermomorphogenesis by degrading TOC1 and PRRs and interacting with phytochromes A/B to maintain an abundance of phytochrome-interacting factors (PIFs) [22]. The transcription level of the CONSTANS (CO) protein and its regulation of FLOWERING LOCUS T (FT) gene expression are key to the photoperiodic flowering response [23]. AtZTL has also been found to maintain the abundance of CO protein by interacting with the GIGANTEA (GI) protein [24], thus promoting expression of the CO/FT module and further affecting the flowering time of A. thaliana. Additionally, researchers found that ZTL-overexpressing plants were more sensitive to abscisic acid (ABA) [25]. PRR5, a known target of ZTL, interacts with the Open Stomata 1 (OST1) protein to regulate the ABA response in Arabidopsis and Poplar [26], thus linking the circadian clock to ABA regulation. Given that ZTL is a key protein connecting the light, circadian clock, and ABA pathways, exploring its function and interaction network would have reference value in aiding our understanding of the mechanisms behind the coordinated regulation of plant growth and development through different pathways.
In this study, we found that the ZTL protein of C. oleifera (CoZTL) delayed the flowering time and promoted hypocotyl elongation in transgenic Arabidopsis lines. The expression pattern of CoZTL in C. oleifera revealed that the gene was relatively highly expressed in fruit shells and stems simultaneously, whereas its expression was significantly changed at different flower bud stages. Yeast two-hybrid library screening identified seven C. oleifera proteins that interacted with CoZTL; namely, aspartate aminotransferase_cytoplasmic (CoAAT), beta-glactosidase (Coβ-GAL), CoLAT52-like, C2-DOMAIN ABA-RELATED 4-like (CoCAR4-like), L-ascorbate oxidase (CoAO), ubiquinol-cytochrome-c reductase complex assembly factor 1 (CoUQCC1), and actin-depolymerizing factor 2 (CoADF2). These results provide a reference for understanding the biological role of ZTL in C. oleifera as well as the flowering mechanism of the plant.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
The 5-year-old C. oleifera ‘Huashuo’ trees were planted in Central South University of Forestry and Technology (112.994587° E, 28.131841° N), Arabidopsis plants were planted in the greenhouse (light 16 h, dark 8 h, 22 °C), seeds of transgenic Arabidopsis needed to be disinfected and sown on 1/2 MS (50 mg/L Kanamycin) solid medium plates for screening, after vernalizing in the dark at 4 °C for 2 d, the plates were transferred to the greenhouse for two weeks, and then the seedlings were transplanted to perlite/nutrient soil mixed substrate for further cultivation.
2.2. RNA, DNA Extraction and cDNA Synthesis
The plant materials were ground into powder by liquid nitrogen, and the total RNA of C. oleifera and Arabidopsis was extracted by using EZ-10 DNA Away RNA mini-preps kit (BBI, Shanghai, China), and the genomic DNA of Arabidopsis was extracted by Super Plant Genomic DNA Kit (Polychardes & Polychromatics-Rich) (Tiangen, Beijing, China) and cDNA synthesis used HISCRIPT II 1st strand cDNA Synthesis Kit (+GDNA Wiper) (Vazyme, Nanjing, China).
2.3. CDS Isolation and Vector Construction of CoZTL
The CDS sequence of CoZTL was isolated from the cDNA of C. oleifera leaves and cloned into the pCAMBIA2300 overexpression vector to produce the recombinant plasmid p2300-35S-CoZTL. The CoZTL specific primers (Table A1) were designed in Primer Premier 5, and the PCR amplification reaction was performed using Phantamax Super-Fidelity DNA Polymerase (Vazyme, Nanjing, China), the reaction condition is shown in Table A2, then PCR product was detected with 1% agarose. It was ligated to a pCAMBIA2300 vector which was linearized by FastDigest BamHI (Thermo Fisher, Shanghai, China), and the recombinant plasmid was transformed into E. coli DH5α and sequenced. (Tsingke, Beijing, China).
2.4. Bioinformatics Analysis of CoZTL
The sequencing results were compared with the CDS sequence of CoZTL using DNAMAN software and translated nucleic acid sequences into protein sequences on the online website (https://www.lynnon.com/, accessed on 8 November 2022). The Expasy (https://www.expasy.org/, accessed on 8 November 2022) and DeepTMHMM (https://dtu.biolib.com/DeepTMHMM, accessed on 8 November 2022) were used to analyze the physicochemical properties of CoZTL protein, the conserved domains and motifs search were acquired from NCBI (https://www.ncbi.nlm.nih.gov/, accessed on 20 February 2023) and MEME (https://meme-suite.org/meme/index.html, accessed on 21 March 2023), the NCBI was also used for blastp search of ZTL protein sequences, which were subjected to multiple alignments and phylogenetic tree construction in DNAMAN and MEGA 11, the phylogenetic tree was constructed by Maximum Likelihood [27].
2.5. Expression Analysis of CoZTL in C. oleifera
The flower buds of C. oleifera were collected every 15 d from 31 July until 30 October [28]. The roots, stems, leaves, flowers, and fruits of C. oleifera were collected at same time in mid-September. These materials were put into liquid nitrogen and stored at −80 °C for extracting total RNA and cDNA synthesis, the expression level of CoZTL was determined by qRT-PCR experiments using ChamQ Universal SYBR qPCRMaster MixRT-qPCR (Vazyme, Nanjing, China), the internal reference gene was CoGAPDH [29]. The qRT-PCR primers are shown in Table A1, and the reaction conditions are shown in Table A3. The qRT-PCRs were performed on the CFX96 real-time PCR system (Bio-Rad, California, USA). Each reaction was performed with three technical replicates. The calculation method for qRT-PCR was 2−∆∆CT.
2.6. Transformation and Identification of Arabidopsis
The recombinant plasmid was transformed into Agrobacterium tumefaciens AGL0 by electroporation, then transformed Col-0 plants using the floral dip method [30]. The transformed T0 seeds were harvested and sowed on 1/2 MS (50 mg/L Kana) solid medium for screening. After two weeks of cultivation, the T0 seeding with four leaves and long roots on the plate were selected, then transferred them to pots for cultivation. When the plants had about 10 leaves, we collected the leaves of each Col-0 and T0 generation Arabidopsis plants to extract RNA and DNA. The transgenic Arabidopsis lines were identified by PCR and qRT-PCR, the internal reference gene was AtACTIN2 [31], the primers are shown in Table A1.
2.7. Phenotypic Observation of Transgenic Arabidopsis
The transgenic Arabidopsis plants were repeatedly screened for the T2 generation, and the T2 seeds were used for phenotypic observation. We analyzed the phenotypic data using SPSS’s ANOVA and Tukey’s HSD (Honest Significant Difference) test tool.
2.7.1. Flowering Observation
After the seeds of Arabidopsis were disinfected, col-0 was sprinkled on the solid medium, the seeds of Col-0 and ztl (SALK_035701) were sown on 1/2 MS solid medium plates, the T2 generation transgenic Arabidopsis seeds were sown on 1/2 MS (50 mg/L Kana) solid medium plates, after vernalization at 4 °C for 2 d, the plates were transferred to the greenhouse at 22 °C for 16 h/8 h (light/dark) for two weeks, then transplanted 15 seedlings of each Arabidopsis line into pots and cultivated at 22 °C, 16 h/8 h (light/dark), we recorded the bolting time and number of rosette leaves at the time of bolting.
2.7.2. Hypocotyl Observation
After disinfecting of Col-0, ztl and T2 generation transgenic Arabidopsis seeds, they were sown on the same 1/2 MS solid medium plate, which needed two plates, the plates were vernalized at 4 °C for 4 d. Then placed in a 5000 xl incubator at 22 °C, 16 h/8 h (light/dark) for 1 d; after wrapping one of the plates in tinfoil, the culture was continued for 6 d. Finally, each Arabidopsis strain was measured the hypocotyl length of 20 seedlings.
2.8. The Autoactivation Activity Test of CoZTL
The CoZTL fragment was cloned into the pGBKT7 vector (the primers are shown in Table A1), and the recombinant plasmid was named CoZTL-BD. the experimental group CoZTL-BD+pGADT7, the positive control group pGBKT7-53+pGADT7-T, the negative control group pGBKT7-Lam+pGADT7-T and the blank control group pGBKT7+pGADT7 were transformed into yeast AH109. the yeast colony of each group was selected and resuspended with 50 μL sterile water, then transferred 2.5 μL to SD/-Trp/-Leu/-His (TDO) solid plates with 3-AT gradient settings of 0, 2.5, 5, 10, 15, 20, 30, 40, 50, and 60 mM/L. After the plates were dried, sealed and cultured upside down for 4 d, the lowest 3-AT concentration corresponding to the plate without colony growth was selected for subsequent library screening.
2.9. Screen of Yeast Two-Hybrid Library
The mating method was used for yeast two-hybrid screening, protein-protein interactions could be screened with different selected media [32,33]. First, CoZTL-BD was transformed into the yeast AH109, and the yeast colony was picked out into 50 mL of SD/-Trp liquid culture medium and cultured at 30 °C, 200 rpm until the OD600 was about 0.8. Then, the yeast cells were collected at the speed of 3000 rpm for 5 min, and the precipitated yeasts were resuspended with 5 mL of SD/-Trp liquid medium to make the cell concentration greater than 1 × 108/mL, and mixed with 1 mL Y187 library yeast. The mixture and 45 mL of 2 YPDA liquid medium were added into a sterile 2 L triangular flask and cultured at 30 °C, 40 rpm for about 16 to 20 h until the appearance of heterozygotes in the shape of “clover” under a microscope. Finally, the yeast cells were collected again at 3000 rpm for 10 min, and resuspended with 5 mL sterile water. The resuspended solution was coated on TDO solid plates with 3-AT about 200 μL/plate, and cultured for 4–6 days. We selected all yeast colonies resuspended in 20 μL sterile water and transferred 2.5 μL of the resuspension to TDO and SD/-Trp/-Leu-His-Ade (QDO) + X-α-Gal plates with 3-AT. Blue yeast colonies were selected and cultured overnight in 5 mL SD/-Trp-leu medium at 30 °C and 200 rpm. Rapid yeast plasmid was used the yeast plasmid was extracted using Quick Yeast Plasmid Preps Kit and transformed into E. coli DH5α to sequence. the yeast plasmid was extracted using Quick Yeast Plasmid Preps Kit and transformed into E. coli DH5α to sequence.
After the sequencing results were translated into protein sequences, they were analyzed using NCBI blastp tool, and the results were collated. After the plasmids corresponding to each fragment were extracted from E. coli DH5α, they were transformed into yeast AH109 with CoZTL-BD and verified on TDO and QDO+X-α-gal plates with 3-AT.
3. Results
3.1. Cloning of the CoZTL Coding Sequence and Analysis of the Protein
We isolated the CDS of ZTL from the cDNA of C. oleifera ‘Huashuo’ (Figure 1a). The sequence was 1827 bp in length and encoded 609 amino acids. The CoZTL protein, the molecular formula of which was predicted to be C2947H4616N826O872S27, had a relative molecular weight of 66.44 kDa, a theoretical pI of 5.78, and an instability index of 41.02 (indicating it is an unstable protein). The aliphatic index of the protein was 88.31, and the average hydrophilicity was −0.104, which was consistent with the ProtScale analysis. The ProtParam tool predicted CoZTL to be an intracellular protein, as it did not contain a transmembrane region. The conserved domain search identified two domains: Per–ARNT–Sim (PAS) and F-box.
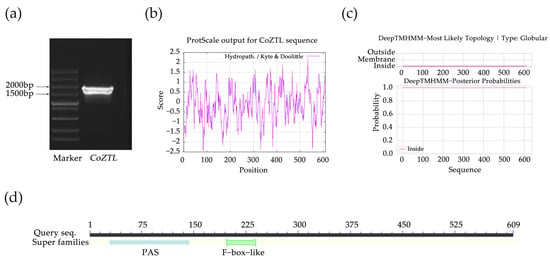
Figure 1.
CDS fragment detection and bioinformatics analysis of CoZTL protein Notes: (a) CDS fragment detection of CoZTL gene. (b) Hydrophobicity analysis of CoZTL protein, negative number is hydrophilic, positive number is hydrophobic. (c) Transmembrane analysis of CoZTL protein. (d) Conserved domains of CoZTL protein.
3.2. Phylogenetic Analysis of CoZTL
We compared the CoZTL protein sequence with the ZTL protein sequences of A. thaliana, Picea abies, Triticum aestivum, Dimocarpus longan, and Glycine max. As shown in Figure 2a, the lengths of the sequences from the different species were similar. The degree of sequence composition similarity between C. oleifera and the five species was 82.98%, 79.39%, 71.82%, 89.82%, and 88.46%, respectively, with the differences being mainly from the N-terminal sequence to the PAS domain and from the PAS domain to the F-box domain. The other regions were highly conserved, which was consistent with the motif analysis results. The motifs of CoZTL and 24 ZTL family members were analyzed, and a phylogenetic tree was constructed (Figure 2b). The tree revealed that CoZTL belonged in the same branch as its counterparts from Ipomoea nil (InZTL) and Nicotiana attenuata (NaZTL). It was also in the same branch as LOV kelch protein 2 (LKP2) of Brassica rapa and A. thaliana, indicating that the CoZTL protein might be related to LKP2. Additionally, motif analysis revealed that the ZTL family of proteins shared more conserved regions. We hypothesize that CoZTL has been highly conserved during evolution.
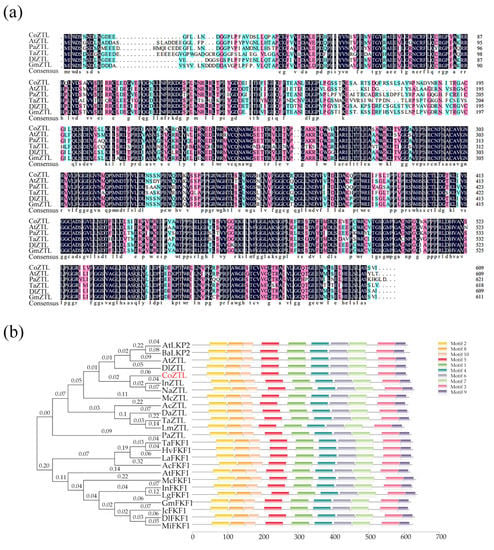
Figure 2.
Multiple sequence alignment and phylogenetic tree. Notes: (a) Multiple sequence alignment, the ZTL protein sequences are from A. thaliana, Picea abies, Triticum aestivum, Dimocarpus longan, and Glycine max. Highlight homology level:  = 100%,
= 100%,  ≥ 75%,
≥ 75%,  ≥ 50%. (b) Phylogenetic tree and motifs of ZEITLUPE protein family from C. oleifera and other species. The information of 24 protein sequences is shown in Table A4.
≥ 50%. (b) Phylogenetic tree and motifs of ZEITLUPE protein family from C. oleifera and other species. The information of 24 protein sequences is shown in Table A4.
3.3. Pattern of CoZTL Expression in C. oleifera
C. oleifera is a type of tree that bears flowers and fruits at the same time. Therefore, the roots, stems, leaves, flower buds, and fruits can be collected at one time, and the fruit shells and seeds can be separated to detect their CoZTL expression levels. As indicated in Figure 3a, the gene expression levels were the highest in the stems and fruit shells and the lowest in the roots. Additionally, flower buds from seven stages were collected every 15 d for CoZTL expression analysis. The results (Figure 3b) showed that the level of CoZTL expression changed significantly during development of the flower buds and peaked in mid-September.
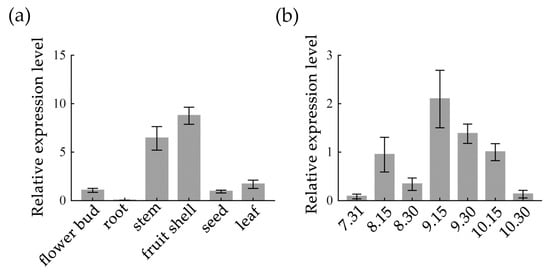
Figure 3.
Relative expression level of CoZTL in C. oleifera. Notes: (a) Relative expression level of CoZTL in different tissues; (b) Relative expression level of CoZTL in different stage flower buds. Data are means ± SE.
3.4. Heterologous Overexpression of CoZTL in Arabidopsis
We identified transgenic Arabidopsis plants using PCR and qRT-PCR and selected three transgenic lines with different CoZTL expression levels (designated 35S-CoZTL-1, 35S-CoZTL-2, and 35S-CoZTL-3) for subsequent phenotype observation (Figure 4a,b). We statistically analyzed the bolting time and number of rosette leaves during bolting of the Col-0 strain, the ztl mutant strain, and the three transgenic Arabidopsis lines (Figure 4c–e). Compared with Col-0, the three transgenic lines showed delayed bolting and had a significantly higher number of rosette leaves. By contrast, there was no difference in number of rosette leaves between the ztl mutant and Col-0 strains. Therefore, we suggest that CoZTL overexpression delays flowering in A. thaliana.
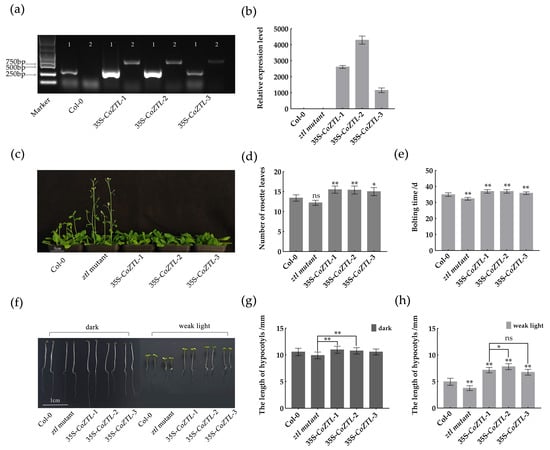
Figure 4.
Identification and phenotypic observation of transgenic Arabidopsis. Notes: (a) Identification of transgenic Arabidopsis plants by PCR, the 1 was AtACTIN2 and 2 was CoZTL in picture. (b) CoZTL gene relative expression levels in Arabidopsis plants. Data are means±SE. (c) Flowering observation. (d) Leaves number at bolting time. (e) Bolting time. (f) Hypocotyl observation. (g) Hypocotyl length under dark. (h) Hypocotyl length under weak light. the data are means ± SD. p-values were determined using Tukey’s HSD test. ns, no statistically significant difference; * p < 0.05; ** p < 0.01.
After vernalization, the seeds of Col-0, the ztl mutant, and the three transgenic Arabidopsis lines were exposed to light for 1 d to ensure seed germination. After 1 d, the plates were placed in the dark and under weak light conditions. After 6 d, the hypocotyl lengths of each line under the two conditions were measured (Figure 4f–h). Under the dark condition, no significant differences were observed among the groups except for the ztl mutant, 35S-CoZTL-1, and 35S-CoZTL-2. However, under the weak light condition, the hypocotyl length of the three transgenic lines was significantly longer than that of Col-0, whereas that of the ztl mutant was significantly shorter. Additionally, the hypocotyl length of 35S-CoZTL-2 was significantly longer than that of 35S-CoZTL-1. Therefore, we suggest that CoZTL overexpression promotes hypocotyl growth in transgenic Arabidopsis plants, an effect that requires light conditions.
3.5. Autoactivation Activity Test and Yeast Two-Hybrid Screening of CoZTL
CoZTL displayed autoactivation activity (Figure 5a), which could be inhibited by the addition of 20 mM 3-AT. Through yeast two-hybrid library screening, we obtained 64 yeast strains for sequencing, which yielded 19 gene fragments. We translated these 19 fragments and searched them using the BLASTP tool. Then, we extracted 10 plasmids corresponding to 10 known protein sequences from E. coli DH5α and transferred them into yeast strain AH109 with CoZTL-BD. Only seven protein sequences interacted with CoZTL (Figure 5b). The protein information is shown in Table 1.

Figure 5.
Autoactivation activity test and yeast two-hybrid screening of CoZTL. Notes: (a) Detection of autoactivation activity of CoZTL protein (b) Validation of interaction between seven proteins and CoZTL.

Table 1.
7 List of proteins that interact with CoZTL protein.
4. Discussion
In this study, we isolated the CDS of CoZTL and performed translation and bioinformatics analyses of its encoded protein. We found that CoZTL was an intracellular protein, and its primary structure contained PAS and F-box domains. The CoZTL protein sequence was highly conserved, exhibiting more than 70% similarity with ZTLs from other species. These results are consistent with the ZTL protein characteristics reported for other species [34,35].
Additionally, the phylogenetic tree revealed that CoZTL belonged to the same branch as InZTL and NaZTL, which are more closely related to each other. These findings provide a reference for studying the function of CoZTL during the development of C. oleifera. Although previous studies have shown that ZTL is a blue light receptor, its expression is not affected by light. The expression of InZTL in Ipomoea nil was higher in the cotyledons and sepals and lowest in the roots [36], whereas the expression of AtZTL in A. thaliana was also lowest in the roots. These results were different from ours, where CoZTL expression was highest in the fruit shells and lowest in the roots of C. oleifera. We also found that the CoZTL expression level changed dynamically during the development of flower buds, which was consistent with the results of rice and soybean studies in which the expression patterns of OsZTL and GmZTL also changed at different plant development stages [37,38,39]. Researchers also found that GmZTL3 mRNA in soybean was accumulated in various leaves and concentrated in mature seeds before flowering [40], which was similar to our finding of a relatively high level of CoZTL expression in the fruits of C. oleifera before flowering. At present, not many studies that have reported relative expression levels have described the developmental stages of the plants, and the common points of ZTL expression patterns are not clear. Therefore, we speculate that the reason for the changes in the CoZTL expression pattern may be related to the degree of organ development at different periods.
Changes in the circadian rhythm often lead to abnormal hypocotyl growth and flowering time [41]. The abundance of ZTL—a clock protein—is controlled by the biological clock, and its expression depends on protease degradation. It is generally assumed that ZTL overexpression regulates the level of CO gene transcription and reduces the inhibition of phytochrome-interacting factors by phytochromes A/B, which leads to delayed flowering and promotes hypocotyl elongation [42,43]. These observations are consistent with the results of this study, in which transgenic CoZTL-overexpressing Arabidopsis plants showed delayed flowering under long-day conditions as well as light-dependent promotion of hypocotyl elongation.
As a blue light receptor protein, ZTL may be involved in blue light-regulated photomorphology and biosynthetic pathways, such as hypocotyl elongation, phototropic movement, shade avoidance syndrome, and primary metabolite synthesis. However, the functions and regulatory control mechanisms of ZTL in these pathways remain unclear. In this study, yeast two-hybrid library screening identified seven protein sequences that interacted with CoZTL. One of those proteins, CAR4-like, binds to and recruits ABA receptors in response to high concentrations of Ca2+ on the cell membrane [44,45,46]. CAR1-overexpressing A. thaliana plants were demonstrated to be more sensitive to ABA-mediated seedling and bud growth inhibition than their wild-type counterparts, whereas car3 mutant seedlings were less sensitive to the ABA-mediated inhibition of seedling establishment and root growth [45,46], which was opposite to the sensitivity to ABA shown by our CoZTL-overexpressing and ztl mutant lines of A. thaliana. These results provide new insights into the correlation between ZTL and ABA signaling pathways.
CoADF2, the major function of which is to cut off and depolymerize filamentous actin, is directly or indirectly involved in cell division, elongation, and signal transduction [47]. The knockout of AtADF1 significantly increased the lengths of the hypocotyl, roots, and stem, whereas its overexpression significantly shortened those lengths. Moreover, the flowering time was significantly delayed in plants with downregulated AtADF1 expression, whereas it was similar between AtADF1-overexpressing and wild-type plants. These findings are opposite to the effects of ZTL on hypocotyl elongation and flowering in our study [48,49]. Huang conducted a comprehensive analysis of the promoter structure and expression patterns of 11 ADFs of Oryza sativa and found that there were cis-acting elements related to stress in the 1 kb promoter region, including ABA response elements, dehydration response elements, and low-temperature response elements [50], but no direct association was found between ADF and ZTL.
CoAAT and Coβ-GAL are enzymes that synthesize amino acids and hydrolyze d-galactosyl residues from polymers, oligosaccharides, or secondary metabolites. They are involved in plant amino acid and sugar metabolism, which is consistent with the role of blue light in regulating the content of free amino acids and soluble sugars in plants [51,52]. AO, which converts ascorbate to monodehydroascorbate and is located in the cell wall, is related to plant growth and development, stress resistance, and senescence [53,54]. The AO expression levels in flowers and fruits are high, which is conducive to the induction of flowering and fruit ripening. Although the expression level of CoZTL is also high in fruits, its overexpression could delay flowering, and there may be a negative feedback relationship involved in this process. With regard to the anther-specific proteins CoLAT52-like and CoUQCC1, their correlation with ZTL could not be surmised owing to the lack of reports about their functions.
5. Conclusions
In this study, we conducted a comprehensive analysis of the ZTL gene in C. oleifera. We observed a close relationship between the ZTL protein of C. oleifera and the ZTL protein of Ipomoea nil by constructing a phylogenetic tree. Meanwhile, we also found that the expression of ZTL gene showed significant changes during the organ development of C. oleifera, rice and soybean. In addition, transgenic CoZTL-overexpressing Arabidopsis plants showed delayed flowering under long-day conditions as well as light-dependent promotion of hypocotyl elongation. Moreover, the yeast two-hybrid experiment results showed that CoZTL protein physically interacted with ABA receptor protein CAR4-like and two proteins (ADF2, AO) which were involved in plant growth and development. Overall, our results enhance our understanding of the ZTL gene in C. oleifera, while also providing potential directions for future investigations into the function of the ZTL gene in plant growth and development.
Author Contributions
Conceptualization, J.Y. and J.L.; methodology, J.Y. and S.R.; software, S.R.; validation, S.R. and J.Y.; formal analysis, S.R., L.J., J.H. and Q.L.; investigation, S.R., L.J., J.H. and Q.L.; resources, J.Y. and J.L.; data curation, S.R., L.J., J.H. and Q.L.; writing—original draft preparation, S.R.; writing—review and editing, S.R., Q.L. and J.Y.; visualization, J.Y.; supervision, J.Y. and J.L.; project administration, J.Y. and J.L.; funding acquisition, J.Y. and J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by China Postdoctoral Science Foundation, grant number 2021M703653, https://jj.chinapostdoctor.org.cn/website/zhengshuchayan.html (accessed on 19 July 2023), Natural Science Foundation of Hunan Province, grant number 2023JJ41041, https://kjt.hunan.gov.cn/kjt/xmxx/xmlx/202305/t20230516_29339472.html (accessed on 19 July 2023), Changsha Natural Science Foundation, grant number kq2202281, http://kjj.changsha.gov.cn/zfxxgk/tzgg_27202/202202/t20220221_10477123.html (accessed on 19 July 2023), and Hunan Forestry Science and Technology Innovation Project, grant number XLK202101-2.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
Appendix A

Table A1.
The list of Primer.
Table A1.
The list of Primer.
| Primer Name | Primer Sequence (5′ to 3′) |
|---|---|
| CoZTL-2300-F | GGTACCCGGGGATCCATGGAGTGGGACAGCAATTC |
| CoZTL-2300-R | CCTCTAGAGGATCCGATAACAGAACTTGCCAAAGATAG |
| CoZTL-trans-F | GAAGATGCCTCTGCCGACA |
| CoZTL-trans-R | TCTTCAAGGCATCTTCTTATTTCTG |
| CoZTL-qPCR-F | ATTCTGATCTGAGCGGCGAC |
| CoZTL-qPCR-R | TTCCGCCCAAGAACCTCTTC |
| GAPDH-F | CTACTGGAGTTTTCACCGA |
| GAPDH-R | TAAGACCCTCAACAATGCC |
| ACTIN-F | CACTGTGCCAATCTACGAGGGT |
| ACTIN-R | CACAAACGAGGGCTGGAACAAG |
| CoZTL-BD-F | AATTCCCGGGGATCCATGGAGTGGGACAGCAATTC |
| CoZTL-BD-R | CAGGTCGACGGATCCGATAACAGAACTTGCCAAAGATAG |

Table A2.
The reaction condition of PCR.
Table A2.
The reaction condition of PCR.
| Step | Temperature/°C | Time | Cycles |
|---|---|---|---|
| Predenaturation | 95 °C | 3 min | 35 |
| Denaturation | 95 °C | 15 s | |
| Annealing | 58 °C | 15 s | |
| Extension | 72 °C | 2 min | |
| Complete extension | 72 °C | 5 min | |
| Finish | 22 °C | forever |

Table A3.
The reaction condition of qRT-PCR.
Table A3.
The reaction condition of qRT-PCR.
| Step | Temperature/°C | Time | Cycles |
|---|---|---|---|
| Predenaturation | 95 °C | 5 min | 45 |
| Denaturation | 95 °C | 30 s | |
| Annealing | 58 °C | 30 s | |
| Extension | 72 °C | 30 s |

Table A4.
The information of 24 proteins.
Table A4.
The information of 24 proteins.
| Protein | Accession | Species | Protein | Accession | Species |
|---|---|---|---|---|---|
| AtLKP2 | NP_849983.1 | Arabidopsis thaliana | AcZTL | ACT22763.1 | Allium cepa |
| AtFKF1 | AAF32298.2 | AcFKF1 | ACT22762.1 | ||
| AtZTL | OAO90691.1 | TaZTL | ABR14627.1 | Triticum aestivum | |
| DlZTL | AHZ89710.1 | Dimocarpus longan | TaFKF1 | ABL11478.1 | |
| DlFKF1 | AHZ89704.1 | GmFKF1 | NP_001235886.2 | Glycine max | |
| InZTL | ABC25060.2 | Ipomoea nil | MiFKF1 | UDP61404.1 | Mangifera indica |
| InFKF1 | AIZ66163.1 | JcFKF1 | AXF53797.1 | Jatropha curcas | |
| NaZTL | AFA35963.1 | Nicotiana attenuata | HvFKF1 | KAE8795993.1 | Hordeum vulgare |
| McZTL | AAQ73527.1 | Mesembryanthemum crystallinum | PaZTL | AGH20050.1 | Picea abies |
| McFKF1 | AAQ73528.1 | LgFKF1 | UDM54773.1 | Luculia gratissima | |
| BaLKP2 | AIC37536.1 | Brassica rapa | LmZTL | BDI21198.1 | Lemna minor |
| LaFKF1 | QTZ25449.1 | Lolium arundinaceum | DaZTL | KAH7671732.1 | Dioscorea alata |
References
- Chen, T.; Liu, L.; Zhou, Y.L.; Zheng, Q.; Luo, S.Y.; Xiang, T.T.; Zhou, L.J.; Feng, S.L.; Yang, H.Y.; Ding, C.B. Characterization and comprehensive evaluation of phenotypic characters in wild Camellia oleifera germplasm for conservation and breeding. Front. Plant Sci. 2023, 14, 1052890. [Google Scholar] [CrossRef]
- Ma, J.L.; Ye, H.; Rui, Y.K.; Chen, G.C.; Zhang, N.Y. Fatty acid composition of Camellia oleifera oil. J. Verbr. Lebensm. 2011, 6, 9–12. [Google Scholar] [CrossRef]
- Quan, W.X.; Wang, A.P.; Gao, C.; Li, C.C. Applications of Chinese Camellia oleifera and its By-Products: A Review. Front. Chem. 2022, 10, 921246. [Google Scholar] [CrossRef]
- Luan, F.; Zeng, J.S.; Yang, Y.; He, X.R.; Wang, B.J.; Gao, Y.B.; Zeng, N. Recent advances in Camellia oleifera Abel: A review of nutritional constituents, biofunctional properties, and potential industrial applications. J. Funct. Foods 2020, 75, 104242. [Google Scholar] [CrossRef]
- Wu, L.L.; Li, J.A.; Gu, Y.Y.; Zhang, F.H.; Gu, L.; Tan, X.F.; Shi, M.W. Effect of Chilling Temperature on Chlorophyll Florescence, Leaf Anatomical Structure, and Physiological and Biochemical Characteristics of Two Camellia oleifera Cultivars. Int. J. Agric. Biol. 2020, 23, 777–785. [Google Scholar]
- Zhang, Y.Q.; Guo, Q.Q.; Luo, S.Q.; Pan, J.W.; Yao, S.; Gao, C.; Guo, Y.Y.; Wang, G. Light Regimes Regulate Leaf and Twigs Traits of Camellia oleifera (Abel.) in Pinus massoniana Plantation Understory. Forests 2022, 13, 918. [Google Scholar] [CrossRef]
- He, Y.F.; Song, Q.Q.; Chen, S.P.; Wu, Y.F.; Zheng, G.H.; Feng, J.L.; Yang, Z.J.; Lin, W.J.; Li, Y.; Chen, H. Transcriptome analysis of self- and cross-pollinated pistils revealing candidate unigenes of self-incompatibility in Camellia oleifera. J. Hortic. Sci. Biotechnol. 2020, 95, 19–31. [Google Scholar] [CrossRef]
- Hu, G.X.; Gao, C.; Fan, X.M.; Gong, W.F.; Yuan, D.Y. Pollination Compatibility and Xenia in Camellia oleifera. Hortscience 2020, 55, 898–905. [Google Scholar] [CrossRef]
- Panchy, N.; von Arnim, A.G.; Hong, T. Early Detection of Daylengths with a Feedforward Circuit Coregulated by Circadian and Diurnal Cycles. Biophys. J. 2020, 119, 1878–1895. [Google Scholar] [CrossRef]
- Kong, S.G.; Okajima, K. Diverse photoreceptors and light responses in plants. J. Plant Res. 2016, 129, 111–114. [Google Scholar] [CrossRef]
- Mawphlang, O.I.L.; Kharshiing, E.V. Photoreceptor Mediated Plant Growth Responses: Implications for Photoreceptor Engineering toward Improved Performance in Crops. Front. Plant Sci. 2017, 8, 1181. [Google Scholar] [CrossRef]
- Voitsekhovskaja, O.V. Phytochromes and Other (Photo)Receptors of Information in Plants. Russ. J. Plant Physiol. 2019, 66, 351–364. [Google Scholar] [CrossRef]
- Abidi, F.; Girault, T.; Douillet, O.; Guillemain, G.; Sintes, G.; Laffaire, M.; Ben Ahmed, H.; Smiti, S.; Huche-Thelier, L.; Leduc, N. Blue light effects on rose photosynthesis and photomorphogenesis. Plant Biol. 2013, 15, 67–74. [Google Scholar] [CrossRef]
- Fraszczak, B. The Effect of Different Doses of Blue Light on the Biometric Traits and Photosynthesis of Dill Plants. Not. Bot. Horti Agrobot. Cluj-Napoca 2016, 44, 34–40. [Google Scholar] [CrossRef]
- Liang, Y.; Kang, C.Q.; Kaiser, E.; Kuang, Y.; Yang, Q.C.; Li, T. Red/blue light ratios induce morphology and physiology alterations differently in cucumber and tomato. Sci. Hortic. 2021, 281, 109995. [Google Scholar] [CrossRef]
- Christie, J.M.; Blackwood, L.; Petersen, J.; Sullivan, S. Plant Flavoprotein Photoreceptors. Plant Cell Physiol. 2015, 56, 401–413. [Google Scholar] [CrossRef]
- Lehmann, P.; Nothen, J.; von Braun, S.S.; Bohnsack, M.T.; Mirus, O.; Schleiff, E. Transitions of gene expression induced by short-term blue light. Plant Biol. 2011, 13, 349–361. [Google Scholar] [CrossRef]
- Pudasaini, A.; Zoltowski, B.D. Zeitlupe Senses Blue-Light Fluence to Mediate Circadian Timing in Arabidopsis thaliana. Biochemistry 2013, 52, 7150–7158. [Google Scholar] [CrossRef]
- Demarsy, E.; Fankhauser, C. Higher plants use LOV to perceive blue light. Curr. Opin. Plant Biol. 2009, 12, 69–74. [Google Scholar] [CrossRef]
- Ito, S.; Song, Y.H.; Imaizumi, T. LOV Domain-Containing F-Box Proteins: Light-Dependent Protein Degradation Modules in Arabidopsis. Mol. Plant 2012, 5, 573–582. [Google Scholar] [CrossRef]
- Seo, D.; Park, J.; Park, J.; Hwang, G.; Seo, P.J.; Oh, E. ZTL regulates thermomorphogenesis through TOC1 and PRR5. Plant Cell Environ. 2023, 46, 1442–1452. [Google Scholar] [CrossRef] [PubMed]
- Zou, Y.; Li, R.; Baldwin, I.T. ZEITLUPE is required for shade avoidance in the wild tobacco Nicotiana attenuata. J. Integr. Plant Biol. 2020, 62, 1341–1351. [Google Scholar] [CrossRef] [PubMed]
- Takase, T.; Nishiyama, Y.; Tanihigashi, H.; Ogura, Y.; Miyazaki, Y.; Yamada, Y.; Kiyosue, T. LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowering under non-inductive conditions, dependent on FLAVIN-BINDING KELCH REPEAT F-BOX1. Plant J. 2011, 67, 608–621. [Google Scholar] [CrossRef]
- Hwang, D.Y.; Park, S.; Lee, S.; Lee, S.S.; Imaizumi, T.; Song, Y.H. GIGANTEA Regulates the Timing Stabilization of CONSTANS by Altering the Interaction between FKF1 and ZEITLUPE. Mol. Cells 2019, 42, 693–701. [Google Scholar] [CrossRef]
- Yu, Y.T.; Portoles, S.; Ren, Y.; Sun, G.Y.; Wang, X.F.; Zhang, H.H.; Guo, S.G. The key clock component ZEITLUPE (ZTL) negatively regulates ABA signaling by degradation of CHLH in Arabidopsis. Front. Plant Sci. 2022, 1, 9959073. [Google Scholar] [CrossRef]
- Jurca, M.; Sjoelander, J.; Ibanez, C.; Matrosova, A.; Johansson, M.; Kozarewa, I.; Takata, N.; Bako, L.; Webb, A.A.R.; Israelsson-Nordstroem, M.; et al. ZEITLUPE Promotes ABA-Induced Stomatal Closure in Arabidopsis and Populus. Front. Plant Sci. 2022, 13, 829121. [Google Scholar] [CrossRef]
- Chor, B.; Hendy, M.D.; Holland, B.R.; Penny, D. Multiple Maxima of Likelihood in Phylogenetic Trees: An Analytic Approach. Mol. Biol. Evol. 2000, 17, 1529–1541. [Google Scholar] [CrossRef]
- Wang, X.N. Research on Phenology and Blossom Biology of Oil-Tea Camellia. Master’s Thesis, Central South University of Forestry and Technology, Changsha, China, 2011. [Google Scholar]
- Tai, Y.; Wei, C.; Yang, H.; Zhang, L.; Chen, Q.; Deng, W.; Wei, S.; Zhang, J.; Fang, C.; Ho, C.; et al. Transcriptomic and phytochemical analysis of the biosynthesis of characteristic constituents in tea (Camellia sinensis) compared with oil tea (Camellia oleifera). BMC Plant Biol. 2015, 15, 190. [Google Scholar] [CrossRef]
- Wiktorek-Smagur, A.; Hnatuszko-Konka, K.; Kononowicz, A.K. Flower bud dipping or vacuum infiltration-two methods of Arabidopsis thaliana transformation. Russ. J. Plant Physiol. 2009, 56, 560–568. [Google Scholar] [CrossRef]
- Chang, M.M.; Li, A.; Feissner, R.; Ahmad, T. RT-qPCR demonstrates light-dependent AtRBCS1A and AtRBCS3B mRNA expressions in Arabidopsis thaliana leaves. Biochem. Mol. Biol. Educ. 2016, 44, 405–411. [Google Scholar] [CrossRef]
- Makuch, L. Yeast Two-Hybrid Screen. Methods Enzymol. 2014, 539, 31–51. [Google Scholar] [CrossRef]
- Soellick, T.R.; Uhrig, J.F. Development of an optimized interaction-mating protocol for large-scale yeast two-hybrid analyses. Genome Biol. 2001, 2, research0052.0051. [Google Scholar] [CrossRef]
- Baudry, A.; Ito, S.; Song, Y.H.; Strait, A.A.; Kiba, T.; Lu, S.; Henriques, R.; Pruneda-Paz, J.L.; Chua, N.H.; Tobin, E.M.; et al. F-Box Proteins FKF1 and LKP2 Act in Concert with ZEITLUPE to Control Arabidopsis Clock Progression. Plant Cell 2010, 22, 606–622. [Google Scholar] [CrossRef]
- Zoltowski, B.D.; Imaizumi, T. Structure and Function of the ZTL/FKF1/LKP2 Group Proteins in Arabidopsis. Enzymes 2014, 35, 213–239. [Google Scholar] [CrossRef] [PubMed]
- Zienkiewicz, A.; Smoliński, D.J.; Zienkiewicz, K.; Glazińska, P.; Wojciechowski, W.; Kopcewicz, J. Molecular and cytological characterization of ZTL in Ipomoea nil. Biol. Plant 2009, 53, 435–443. [Google Scholar] [CrossRef]
- Xue, Z.G. Cloning and Functional Analysis of GmZTL3 and GmZTL4 in Soybean. Master’s Thesis, Henan Agricultural University, Zhengzhou, China, 2011. [Google Scholar]
- Yang, W.Q. Studies on the Functions of OsZTL1 and OsZTL2 in Rice. Ph.D. Thesis, Shenyang Agricultural University, Liaoning, China, 2019. [Google Scholar] [CrossRef]
- Zhao, F. Functional Analysis of Soybean GmZTL Gene. Master’s Thesis, Guizhou University, Guiyang, China, 2008. [Google Scholar]
- Xue, Z.G.; Zhang, X.M.; Lei, C.F.; Chen, X.J.; Fu, Y.F. Molecular cloning and functional analysis of one ZEITLUPE homolog GmZT L3 in soybean. Mol. Biol. Rep. 2012, 39, 1411–1418. [Google Scholar] [CrossRef] [PubMed]
- Niwa, Y.; Yamashino, T.; Mizuno, T. The circadian clock regulates the photoperiodic response of hypocotyl elongation through a coincidence mechanism in Arabidopsis thaliana. Plant Cell Physiol. 2009, 50, 838–854. [Google Scholar] [CrossRef] [PubMed]
- Fujiwara, S. Novel blue light receptors with an F-box: Their direct control of the circadian clock and the flowering timing in Arabidopsis. Plant Biotechnol. 2008, 25, 123–129. [Google Scholar] [CrossRef]
- Song, Y.H.; Estrada, D.A.; Johnson, R.S.; Kim, S.K.; Lee, S.Y.; MacCoss, M.J.; Imaizumi, T. Distinct roles of FKF1, Gigantea, and Zeitlupe proteins in the regulat ion of Constans stability in Arabidopsis photoperiodic flowering. Proc. Natl. Acad. Sci. USA 2014, 111, 17672–17677. [Google Scholar] [CrossRef]
- Lockhart, J. Membrane bound: C2-domain abscisic acid-related proteins help abscisic acid receptors get where they need to go. Plant Cell 2014, 26, 4566. [Google Scholar] [CrossRef]
- Xiong, T.; Tan, Q.; Li, S.; Mazars, C.; Galaud, J.P.; Zhu, X. Interactions between calcium and ABA signaling pathways in the regulat ion of fruit ripening. J. Plant Physiol. 2021, 256, 153309. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.; Sanchez-Barrena, M.J.; Gonzalez-Rubio, J.M.; Rodriguez, L.; Fernandez, D.; Antoni, R.; Yunta, C.; Belda-Palazon, B.; Gonzalez-Guzman, M.; Peirats-Llobet, M.; et al. Calcium-dependent oligomerization of CAR proteins at cell membrane mod ulates ABA signaling. Proc. Natl. Acad. Sci. USA 2016, 113, E396–E405. [Google Scholar] [CrossRef] [PubMed]
- Sengupta, S.; Rajasekaran, K.; Baisakh, N. Natural and targeted isovariants of the rice actin depolymerizing factor 2 can alter its functional and regulatory binding properties. Biochem. Biophys. Res. Commun. 2018, 503, 1516–1523. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Qiu, T.Q.; Yue, J.R.; Guo, N.N.; He, Y.J.; Han, X.P.; Wang, Q.Y.; Jia, P.F.; Wang, H.D.; Li, M.Z.; et al. Arabidopsis ADF1 is Regulated by MYB73 and is Involved in Response to Salt Stress Affecting Actin Filament Organization. Plant Cell Physiol. 2021, 62, 1387–1395. [Google Scholar] [CrossRef]
- Qian, D.; Zhang, Z.; He, J.X.; Zhang, P.; Ou, X.B.; Li, T.; Niu, L.P.; Nan, Q.; Niu, Y.; He, W.L.; et al. Arabidopsis ADF5 promotes stomatal closure by regulating actin cytoskeleton remodeling in response to ABA and drought stress. J. Exp. Bot. 2019, 70, 435–446. [Google Scholar] [CrossRef]
- Huang, S.J.; Qu, X.L.; Zhang, R.H. Plant villins: Versatile actin regulatory proteins. J. Integr. Plant Biol. 2015, 57, 40–49. [Google Scholar] [CrossRef]
- Alcântara, P.; Martim, L.; Silva, C.; Dietrich, S.; Buckeridge, M. Purification of a β-galactosidase from cotyledons of Hymenaea courbaril L. (Leguminosae). Enzyme properties and biological function. Plant Physiol. Biochem. 2006, 44, 619–627. [Google Scholar] [CrossRef]
- McAllister, C.H.; Wolansky, M.; Good, A.G. The impact on nitrogen-efficient phenotypes when aspartate aminotransferase is expressed tissue-specifically in Brassica napus. New Negat. Plant Sci. 2016, 3–4, 1–9. [Google Scholar] [CrossRef]
- Garchery, C.; Gest, N.; Do, P.T.; Alhagdow, M.; Baldet, P.; Menard, G.; Rothan, C.; Massot, C.; Gautier, H.; Aarrouf, J.; et al. A diminution in ascorbate oxidase activity affects carbon allocation and improves yield in tomato under water deficit. Plant Cell Environ. 2013, 36, 159–175. [Google Scholar] [CrossRef]
- Stevens, R.; Truffault, V.; Baldet, P.; Gautier, H. Ascorbate Oxidase in Plant Growth, Development, and Stress Tolerance. In Ascorbic Acid in Plant Growth, Development and Stress Tolerance; Hossain, M.A., Munné-Bosch, S., Burritt, D.J., Diaz-Vivancos, P., Fujita, M., Lorence, A., Eds.; Springer International Publishing: Cham, Switzerland, 2017; pp. 273–295. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).