An Analysis of the Potential Regulatory Mechanisms of Sophora Flower Development and Nutritional Component Formation Using RNA Sequencing
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Samples Collection
2.2. RNA Extraction
2.3. Sequencing and Transcriptome Assembly
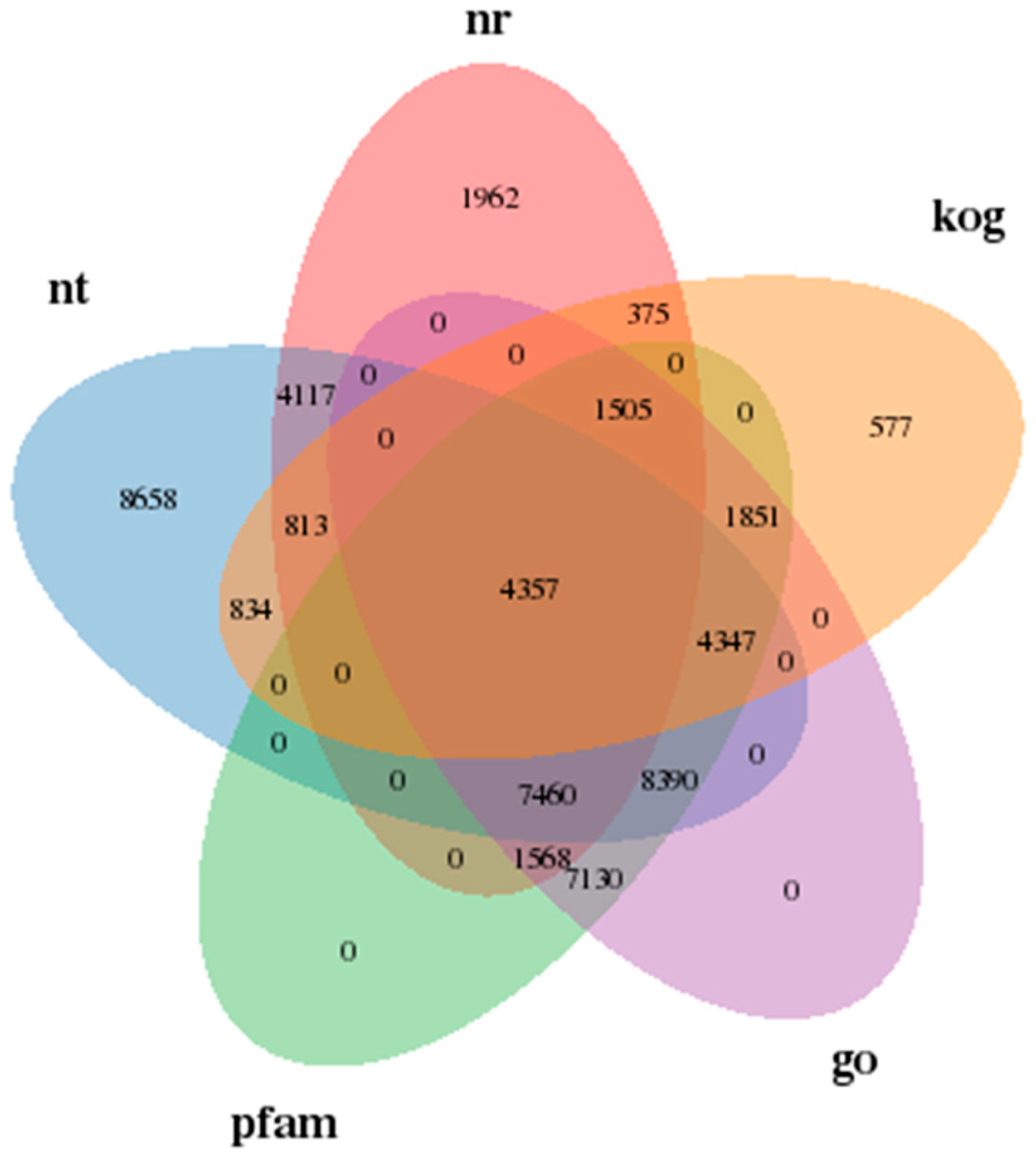
2.4. DEGs’ Annotation and Bioinformatic Analysis
2.5. Quantitative
3. Results
3.1. Appearance Characteristic of S. japonica Flowers
3.2. Quality Assessment of Transcriptome Sequencing Data
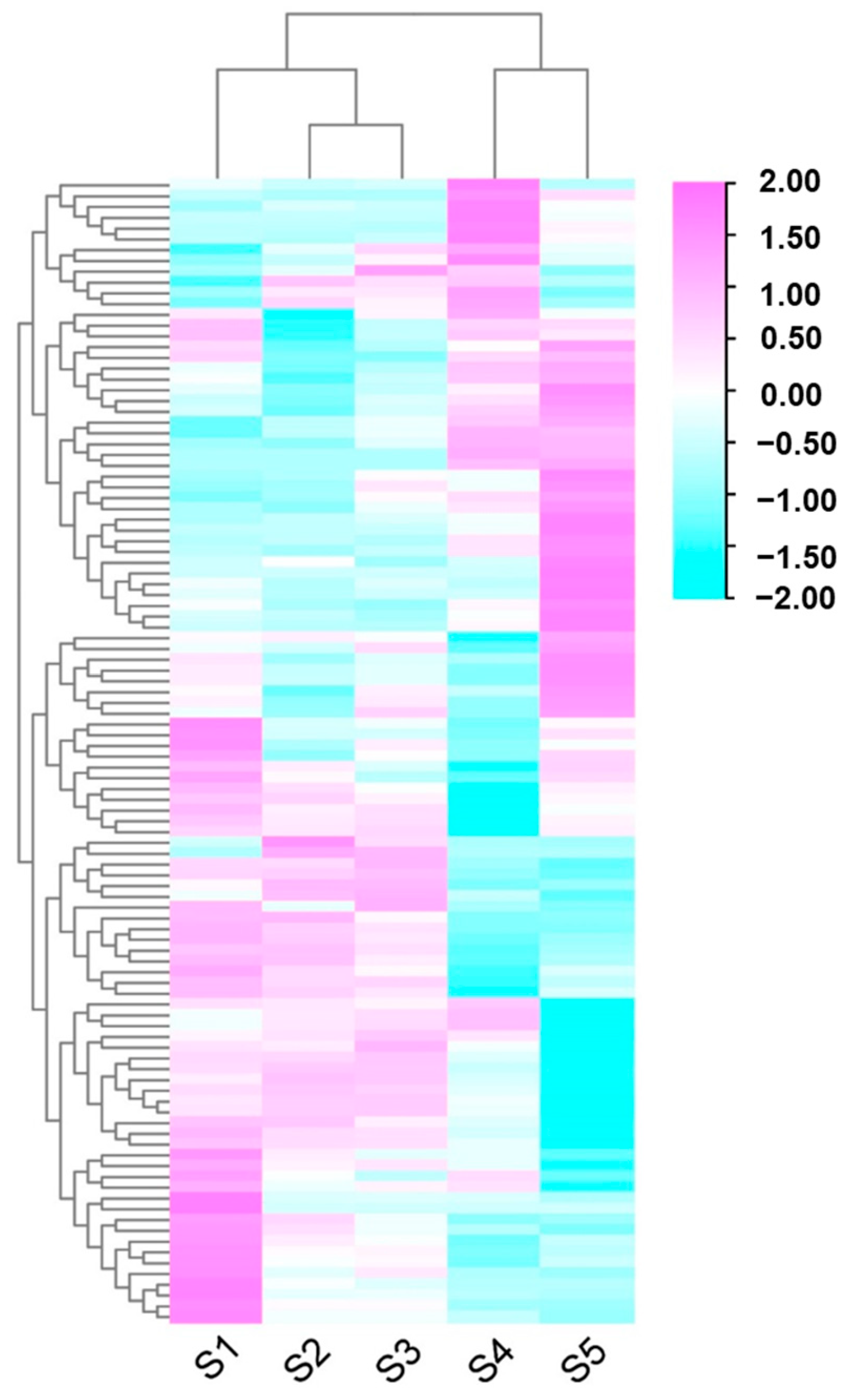
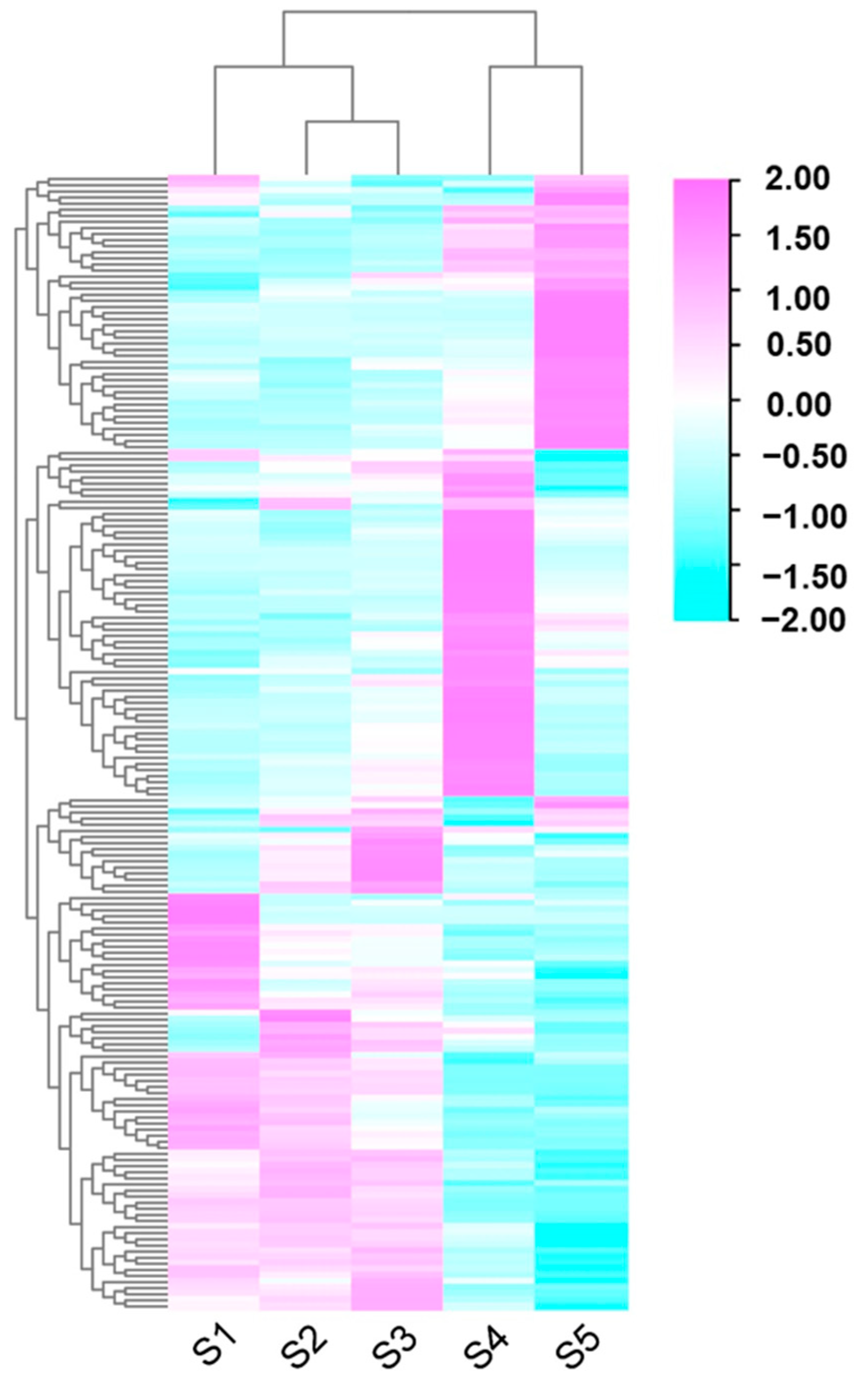
3.3. Differential Expression Gene Analysis of Assembled Unigenes
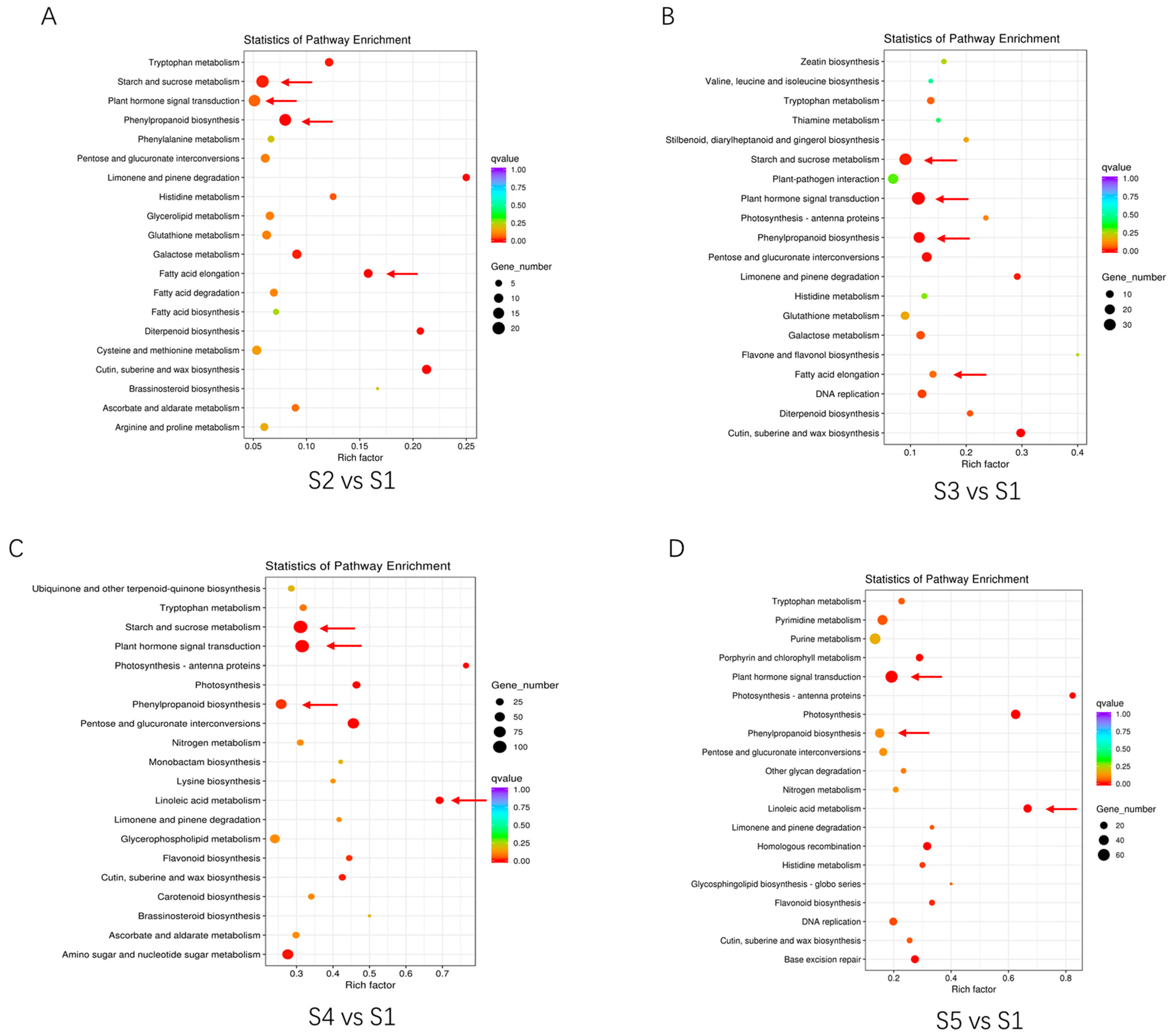
3.4. KEGG Enrichment Analysis of Different Flowering Stages
3.5. Comparative Analysis of Phenylpropanoid Biosynthesis in Huaihua
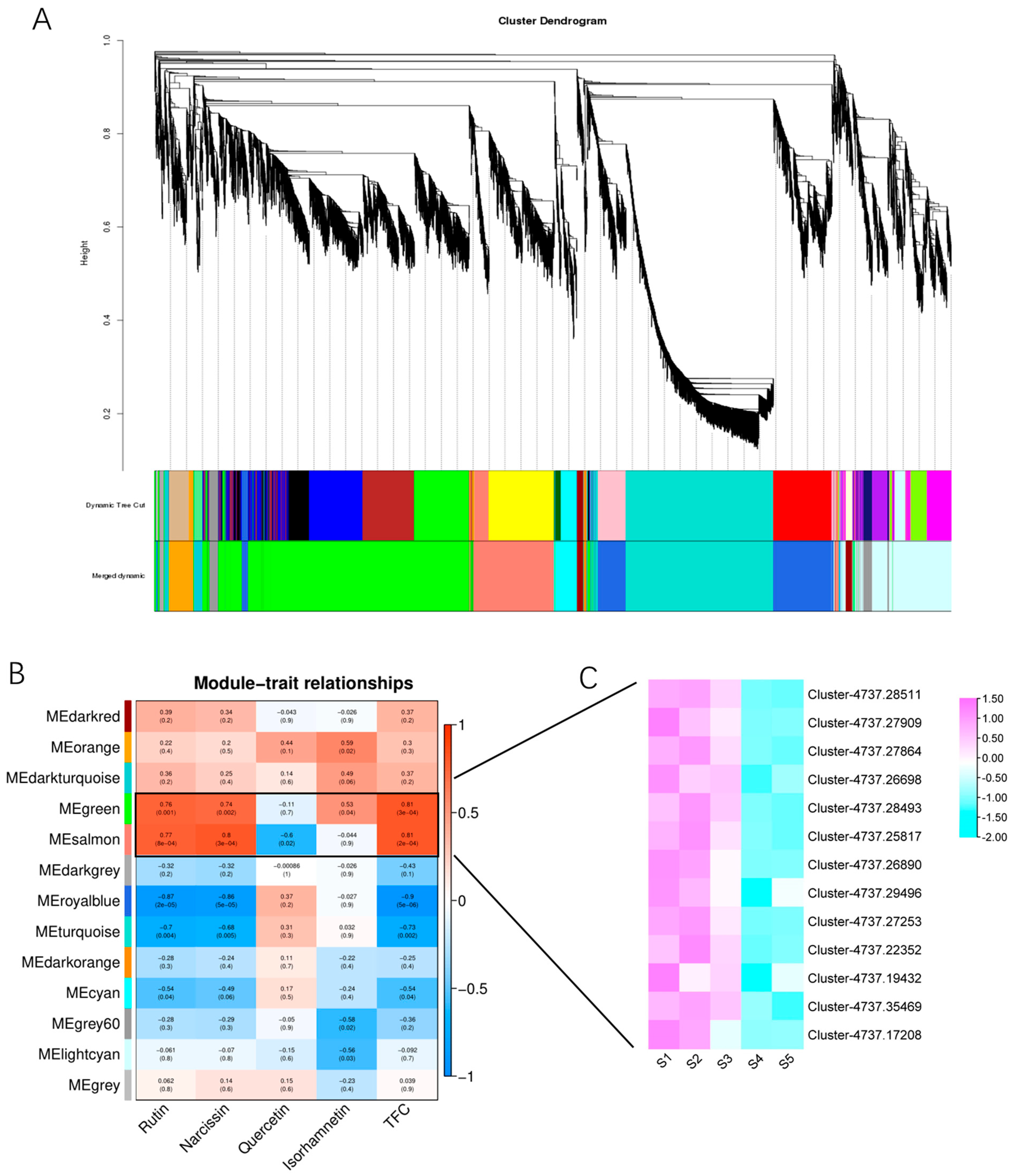
3.6. Weighted Correlation Network Analysis (WGCNA) of Flavonoids in Huaihua
3.7. Comparative Analysis of DEGs Related to Plant Hormones in Huaihua
3.8. The DEGs Related to Starch and Sucrose Metabolism in Huaihua
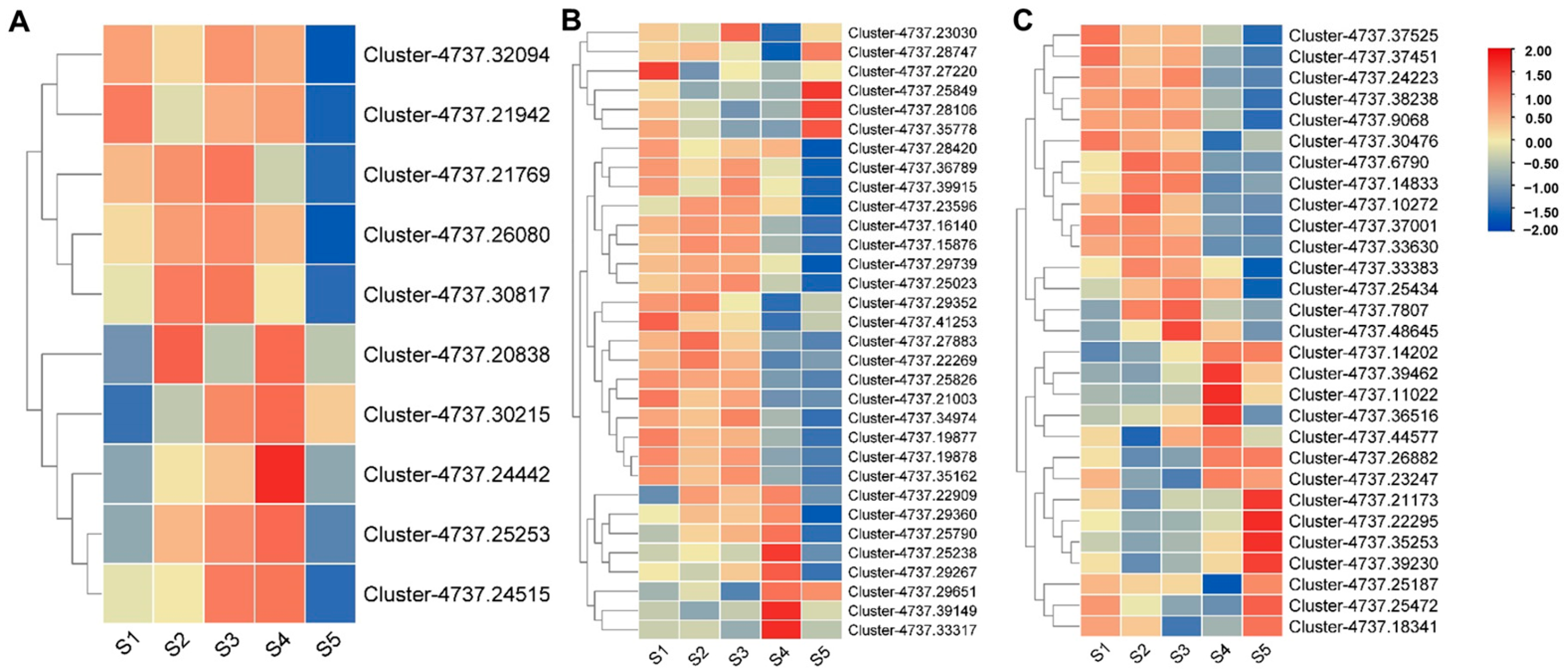
3.9. Quantitative Reverse Transcription PCR (qRT-PCR) Verification
4. Discussion
4.1. Phenylpropanoid Biosynthesis during Development of Huaihua
4.2. Plant Hormone Signal Transduction Related to Huaihua Development
4.3. Starch and Sucrose Metabolism Related to Huaihua Development
4.4. MADS-box, MYB, and bHLH Transcription Factors Related to Huaihua Development
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Park, K.W.; Lee, J.E.; Park, K.M. Diets containing Sophora japonica L. prevent weight gain in high-fat diet-induced obese mice. Nutr. Res. 2009, 29, 819–824. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.R.; Kim, Y.J.; Gwon, A.R.; Lee, J.; Jo, D.G.; Jeon, T.J.; Hong, J.W.; Park, K.M.; Park, K.W. Genistein Mediates the Anti-Adipogenic Actions of Sophora japonica L. Extracts. J. Med. Food 2011, 14, 360–368. [Google Scholar] [CrossRef] [PubMed]
- Abdallah, H.M.; Al-Abd, A.M.; Asaad, G.F.; Abdel-Naim, A.B.; El-Halawany, A.M. Isolation of Antiosteoporotic Compounds from Seeds of Sophora japonica. PLoS ONE 2014, 9, e98559. [Google Scholar] [CrossRef] [PubMed]
- Kim, B.-H.; Lee, S. Sophoricoside from Sophora japonica ameliorates allergic asthma by preventing mast cell activation and CD4+ T cell differentiation in ovalbumin-induced mice. Biomed. Pharmacother. 2021, 133, 111029. [Google Scholar] [CrossRef]
- Rajendran, P.; Rengarajan, T.; Nandakumar, N.; Palaniswami, R.; Nishigaki, Y.; Nishigaki, I. Kaempferol, a potential cytostatic and cure for inflammatory disorders. Eur. J. Med. Chem. 2014, 86, 103–112. [Google Scholar] [CrossRef]
- Zhang, Y.; Gu, D.; He, S.; Meng, J.; Wang, J.; Wang, Y.; Wang, Y.; Tian, J.; Yang, Y. Enzyme reaction-guided identification of active components from the flowers of Sophora japonica var. violacea. Food Funct. 2020, 11, 4356–4362. [Google Scholar] [CrossRef]
- Guan, Y.; Chen, K.; Quan, D.; Kang, L.; Yang, D.; Wu, H.; Yan, M.; Wu, S.; Lv, L.; Zhang, G. The Combination of Scutellaria baicalensis Georgi and Sophora japonica L. ameliorate Renal Function by Regulating Gut Microbiota in Spontaneously Hypertensive Rats. Front. Pharmacol. 2021, 11, 575294. [Google Scholar] [CrossRef]
- Li, L.; Huang, T.; Lan, C.; Ding, H.; Yan, C.; Dou, Y. Protective effect of polysaccharide from Sophora japonica L. flower buds against UVB radiation in a human keratinocyte cell line (HaCaT cells). J. Photochem. Photobiol. B 2019, 191, 135–142. [Google Scholar] [CrossRef]
- Solek, P.; Shemedyuk, N.; Gorka, A.; Bilska-Kos, A.; Shemedyuk, A.; Koziorowski, M. Male reprotoxicity associated with Sophora japonica treatment: Evaluation of cellular and molecular events in vitro. J. Physiol. Pharmacol. 2018, 69, 969–977. [Google Scholar] [CrossRef]
- Chen, H.-N.; Hsieh, C.-L. Effects of Sophora japonica flowers (Huaihua) on cerebral infarction. Chin. Med. 2010, 5, 34. [Google Scholar] [CrossRef]
- Li, W.J.; Gao, Z.H. Comparison of Locust Flos sophorae and Flos sophorae Nutrients and Rutin Content. Food Industry. 2020, 41, 337–339. [Google Scholar]
- Stark, R.; Grzelak, M.; Hadfield, J. RNA sequencing: The teenage years. Nat. Rev. Genet. 2019, 20, 631–656. [Google Scholar] [CrossRef]
- Huang, H.R.; Yan, P.C.; Lascoux, M.; Ge, X.J. Flowering time and transcriptome variation in Capsella bursa-pastoris (Brassicaceae). New Phytol. 2012, 194, 676–689. [Google Scholar] [CrossRef]
- Li, X.; Tang, D.; Du, H.; Shi, Y. Transcriptome Sequencing and Biochemical Analysis of Perianths and Coronas Reveal Flower Color Formation in Narcissus pseudonarcissus. Int. J. Mol. Sci. 2018, 19, 4006. [Google Scholar] [CrossRef]
- Hui, W.; Yang, Y.; Wu, G.; Peng, C.; Chen, X.; Zayed, M.Z. Transcriptome profile analysis reveals the regulation mechanism of floral sex differentiation in Jatropha curcas L. Sci. Rep. 2017, 7, 16421. [Google Scholar] [CrossRef]
- Wang, J.R.; Li, L.Y.; Tan, J.; Song, X.; Chen, D.; Xu, J.; Ding, G. Variations in the Components and Antioxidant and Tyrosinase Inhibitory Activities of Styphnolobium japonicum (L.) Schott Extract during Flower Maturity Stages. Chem. Biodivers. 2019, 16, e1800504. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.D.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef]
- Sims, D.; Sudbery, I.; Ilott, N.E.; Heger, A.; Ponting, C.P. Sequencing depth and coverage: Key considerations in genomic analyses. Nat. Rev. Genet. 2014, 15, 121–132. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Madden, E.; McLachlan, C.; Oketch-Rabah, H.; Calderón, A.I. United States Pharmacopeia comprehensive safety review of Styphnolobium japonicum flower and flower bud. Phytother Res. 2022, 36, 2061–2071. [Google Scholar] [CrossRef]
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.-R.; Song, X.-H.; Li, L.-Y.; Gao, S.-J.; Shang, F.-H.; Zhang, X.-M.; Yang, Y. Metabolomic analysis reveals dynamic changes in secondary metabolites of Sophora japonica L. during flower maturation. Front. Plant Sci. 2022, 13, 916410. [Google Scholar] [CrossRef] [PubMed]
- Lam, P.Y.; Tobimatsu, Y.; Takeda, Y.; Suzuki, S.; Yamamura, M.; Umezawa, T.; Lo, C. Disrupting Flavone Synthase II Alters Lignin and Improves Biomass Digestibility. Plant Physiol. 2017, 174, 972–985. [Google Scholar] [CrossRef] [PubMed]
- Mu, H.; Ci, Z.; Aisajan, M.; Liang, Y.; Liu, X.; DU, X.; Yu, Q.; Li, Q.; Li, Y. Analysis of metabolite differences in skin between Clapp’s Favorite and its mutant Red Clapp’s Favorite through non-targeted metabolomics. Chin. J. Chromatogr. 2021, 39, 1203–1212. [Google Scholar] [CrossRef]
- He, X.; Bai, Y.; Zhao, Z.; Wang, X.; Fang, J.; Huang, L.; Zeng, M.; Zhang, Q.; Zhang, Y.; Zheng, X. Local and traditional uses, phytochemistry, and pharmacology of Sophora japonica L.: A review. J. Ethnopharmacol. 2016, 187, 160–182. [Google Scholar] [CrossRef]
- Potocká, E.K.; Mastihubová, M.; Mastihuba, V. Transrutinosylation of tyrosol by flower buds of Sophora japonica. Food Chem. 2021, 336, 127674. [Google Scholar] [CrossRef]
- Zhang, S.; Chen, C.; Lu, W.; Wei, L. Phytochemistry, pharmacology, and clinical use of Panax notoginseng flowers buds. Phytother. Res. 2018, 32, 2155–2163. [Google Scholar] [CrossRef]
- Shen, T.; Hu, F.; Liu, Q.; Wang, H.; Li, H. Analysis of Flavonoid Metabolites in Chaenomeles Petals Using UPLC-ESI-MS/MS. Molecules 2020, 25, 3994. [Google Scholar] [CrossRef]
- Ke, Y.; Abbas, F.; Zhou, Y.; Yu, R.; Yue, Y.; Li, X.; Yu, Y.; Fan, Y. Genome-Wide Analysis and Characterization of the Aux/IAA Family Genes Related to Floral Scent Formation in Hedychium coronarium. Int. J. Mol. Sci. 2019, 20, 3235. [Google Scholar] [CrossRef]
- Balzan, S.; Johal, G.S.; Carraro, N. The role of auxin transporters in monocots development. Front. Plant Sci. 2014, 5, 393. [Google Scholar] [CrossRef]
- Ghelli, R.; Brunetti, P.; Napoli, N.; De Paolis, A.; Cecchetti, V.; Tsuge, T.; Serino, G.; Matsui, M.; Mele, G.; Rinaldi, G.; et al. A Newly Identified Flower-Specific Splice Variant of AUXIN RESPONSE FACTOR8 Regulates Stamen Elongation and Endothecium Lignification in Arabidopsis. Plant Cell 2018, 30, 620–637. [Google Scholar] [CrossRef]
- Amini, S.; Rosli, K.; Abu-Bakar, M.-F.; Alias, H.; Mat-Isa, M.-N.; Juhari, M.-A.; Haji-Adam, J.; Goh, H.-H.; Wan, K.-L. Transcriptome landscape of Rafflesia cantleyi floral buds reveals insights into the roles of transcription factors and phytohormones in flower development. PLoS ONE 2019, 14, e0226338. [Google Scholar] [CrossRef]
- Davis, S.J. Integrating hormones into the floral-transition pathway of Arabidopsis thaliana. Plant Cell Environ. 2009, 32, 1201–1210. [Google Scholar] [CrossRef]
- Gao, X.; Wang, L.; Zhang, H.; Zhu, B.; Lv, G.; Xiao, J. Transcriptome analysis and identification of genes associated with floral transition and fruit development in rabbiteye blueberry (Vaccinium ashei). PLoS ONE 2021, 16, e0259119. [Google Scholar] [CrossRef]
- Yoon, J.; Cho, L.-H.; Tun, W.; Jeon, J.-S.; An, G. Sucrose signaling in higher plants. Plant Sci. 2020, 302, 110703. [Google Scholar] [CrossRef]
- Porri, A.; Torti, S.; Romera-Branchat, M.; Coupland, G. Spatially distinct regulatory roles for gibberellins in the promotion of flowering of Arabidopsis under long photoperiods. Development 2012, 139, 2198–2209. [Google Scholar] [CrossRef]
- Bartrina, I.; Jensen, H.; Novak, O.; Strnad, M.; Werner, T.; Schmülling, T. Gain-of-Function Mutants of the Cytokinin Receptors AHK2 and AHK3 Regulate Plant Organ Size, Flowering Time and Plant Longevity. Plant Physiol. 2017, 173, 1783–1797. [Google Scholar] [CrossRef]
- Wingler, A.; Fritzius, T.; Wiemken, A.; Boller, T.; Aeschbacher, R.A. Trehalose Induces the ADP-Glucose Pyrophosphorylase Gene, ApL3, and Starch Synthesis in Arabidopsis. Plant Physiol. 2000, 124, 105–114. [Google Scholar] [CrossRef]
- Sánchez-López, Á.M.; Baslam, M.; De Diego, N.; Muñoz, F.J.; Bahaji, A.; Almagro, G.; Ricarte-Bermejo, A.; García-Gómez, P.; Li, J.; Humplík, J.F.; et al. Volatile compounds emitted by diverse phytopathogenic microorganisms promote plant growth and flowering through hcytokinin action. Plant Cell Environ. 2016, 39, 2592–2608. [Google Scholar] [CrossRef]
- Matías-Hernández, L.; Aguilar-Jaramillo, A.E.; Cigliano, R.A.; Sanseverino, W.; Pelaz, S. Flowering and trichome development share hormonal and transcription factor regulation. J. Exp. Bot. 2016, 67, 1209–1219. [Google Scholar] [CrossRef]
- Rounsley, S.D.; Ditta, G.S.; Yanofsky, M.F. Diverse roles for MADS box genes in Arabidopsis development. Plant Cell 1995, 7, 1259–1269. [Google Scholar] [CrossRef] [PubMed]
- Das, A.; Nigam, D.; Junaid, A.; Tribhuvan, K.U.; Kumar, K.; Durgesh, K.; Singh, N.K.; Gaikwad, K. Expressivity of the key genes associated with seed and pod development is highly regulated via lncRNAs and miRNAs in Pigeonpea. Sci. Rep. 2019, 9, 18191. [Google Scholar] [CrossRef]
- Nam, J.; Depamphilis, C.W.; Ma, H.; Nei, M. Antiquity and Evolution of the MADS-Box Gene Family Controlling Flower Development in Plants. Mol. Biol. Evol. 2003, 20, 1435–1447. [Google Scholar] [CrossRef] [PubMed]
- Alvarez-Buylla, E.R.; Pelaz, S.; Liljegren, S.J.; Gold, S.E.; Burgeff, C.; Ditta, G.S.; de Pouplana, L.R.; Martínez-Castilla, L.; Yanofsky, M.F. An ancestral MADS-box gene duplication occurred before the divergence of plants and animals. Proc. Natl. Acad. Sci. USA 2000, 97, 5328–5333. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Yang, L.; Tu, Z.; Zhu, S.; Zhang, C.; Li, H. Genome-wide identification of MIKC-type genes related to stamen and gynoecium development in Liriodendron. Sci. Rep. 2021, 11, 6585. [Google Scholar] [CrossRef]
- Di Marzo, M.; Roig-Villanova, I.; Zanchetti, E.; Caselli, F.; Gregis, V.; Bardetti, P.; Chiara, M.; Guazzotti, A.; Caporali, E.; Mendes, M.A.; et al. MADS-Box and bHLH Transcription Factors Coordinate Transmitting Tract Development in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 526. [Google Scholar] [CrossRef]
- Theissen, G.; Becker, A.; Di Rosa, A.; Kanno, A.; Kim, J.T.; Münster, T.; Winter, K.-U.; Saedler, H. A short history of MADS-box genes in plants. Plant Mol. Biol. 2000, 42, 115–149. [Google Scholar] [CrossRef]
- Sharma, A.; Badola, P.K.; Bhatia, C.; Sharma, D.; Trivedi, P.K. Primary transcript of miR858 encodes regulatory peptide and controls flavonoid biosynthesis and development in Arabidopsis. Nat. Plants 2020, 6, 1262–1274. [Google Scholar] [CrossRef]
- He, L.; Tang, R.; Shi, X.; Wang, W.; Cao, Q.; Liu, X.; Wang, T.; Sun, Y.; Zhang, H.; Li, R.; et al. Uncovering anthocyanin biosynthesis related microRNAs and their target genes by small RNA and degradome sequencing in tuberous roots of sweetpotato. BMC Plant Biol. 2019, 19, 232. [Google Scholar] [CrossRef]
- Wang, L.; Lu, W.; Ran, L.; Dou, L.; Yao, S.; Hu, J.; Fan, D.; Li, C.; Luo, K. R2R3-MYBtranscription factorMYB6 promotes anthocyanin and proanthocyanidin biosynthesis but inhibits secondary cell wall formation in Populus tomentosa. Plant J. 2019, 99, 733–751. [Google Scholar] [CrossRef]
- Ahrazem, O.; Rubio-Moraga, A.; Nebauer, S.G.; Molina, R.V.; Gómez-Gómez, L. Saffron: Its Phytochemistry, Developmental Processes, and Biotechnological Prospects. J. Agric. Food Chem. 2015, 63, 8751–8764. [Google Scholar] [CrossRef]
- Arlotta, C.; Puglia, G.D.; Genovese, C.; Toscano, V.; Karlova, R.; Beekwilder, J.; De Vos, R.C.; Raccuia, S.A. MYB5-like and bHLH influence flavonoid composition in pomegranate. Plant Sci. 2020, 298, 110563. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, X.; Wang, J.; Shang, F.; Ding, G.; Li, L. An Analysis of the Potential Regulatory Mechanisms of Sophora Flower Development and Nutritional Component Formation Using RNA Sequencing. Horticulturae 2023, 9, 756. https://doi.org/10.3390/horticulturae9070756
Song X, Wang J, Shang F, Ding G, Li L. An Analysis of the Potential Regulatory Mechanisms of Sophora Flower Development and Nutritional Component Formation Using RNA Sequencing. Horticulturae. 2023; 9(7):756. https://doi.org/10.3390/horticulturae9070756
Chicago/Turabian StyleSong, Xuhong, Jirui Wang, Fanghong Shang, Gang Ding, and Longyun Li. 2023. "An Analysis of the Potential Regulatory Mechanisms of Sophora Flower Development and Nutritional Component Formation Using RNA Sequencing" Horticulturae 9, no. 7: 756. https://doi.org/10.3390/horticulturae9070756
APA StyleSong, X., Wang, J., Shang, F., Ding, G., & Li, L. (2023). An Analysis of the Potential Regulatory Mechanisms of Sophora Flower Development and Nutritional Component Formation Using RNA Sequencing. Horticulturae, 9(7), 756. https://doi.org/10.3390/horticulturae9070756




