Abstract
Nineteen samples of Gardenia jasminoides Ellis from China were used for genome resequencing. The clean Q30 base rates ranged from 91.19% to 92.94%. When mapping with the reference genome, a total of 7,568,199 inDel sites and 61,535,595 SNP sites were detected, with variations of heterozygosity ratios ranging from 44.86% to 92.69% and from 41.53% to 90.38%, respectively. Genome location and function annotation of SNP and inDel in different samples were carried out. SNP and inDel were mainly located in the intergenic region, with the ratios of 67.2360% and 62.6415%, respectively. Based on the phylogenetic tree constructed by the maximum likelihood method, the 19 samples could be divided into four categories. Among them, Y_10 and Y_38 were put into one category, while Y_2 was put into a separate category. These two were first differentiated from other cultivars and species. Similar conclusions can be drawn from the results of genetic structure and principal component analysis. This study provided sequence foundation for the identifying of the molecular marker and genotype of G. jasminoides. It also provided theoretical foundation for subsequent resource conservation and utilization, the genetic localization of important agronomical traits, and high-quality variety formulation and breeding.
1. Introduction
There are about 250 species of Gardenia Ellis (Rubiaceae), which are mainly distributed in the tropical and subtropical regions of the eastern hemisphere [1]. However, there are only six species of Gardenia in China. Among them, Gardenia jasminoides Ellis is widely distributed in the provinces south of the Yangtze River and is extensively cultivated [2,3]. G. jasminoides is an evergreen shrub with opposite or rarely ternate leaves. The leaf shape changes greatly: it can be oblong-lanceolate, obovate-oblong, obovate, oblanceolate, or elliptic, with dimensions of 3–25 cm × 1.5–8 cm. Flowers can be solitary or terminal, colored corolla white to pale yellow, and be simple or in cultivation sometimes doubled, with their outside glabrous. Fruiting peduncles apparently do not show much elongating. Berries are yellow or orange-yellow, being ovoid, subglobose, or ellipsoid in shape, with dimensions of 1.5–7 cm × 1.2–2 cm; they have 5–9 longitudinal ridges and persistent calyx lobes sized 40 mm × 6 mm. Seeds are suborbicular (ca. 3.5 mm × 3 mm). Flowering is in Mar–Jul, and fruiting in May–Feb [1]. As one of the eight famous fragrant flowers in China, it looks snow-white and smells fragrant [4]. The dried ripe fruit of G. jasminoides is a common raw material in traditional Chinese medicine. It is bitter and cold in nature. It has the functions of clearing fire and overcoming restlessness, clearing heat and promoting diuresis, cooling blood and detoxifying, detumescence, and relieving pain. Therefore, it can improve upset, dryness, restlessness, dampness-heat jaundice, multiple bleeding, eye swelling pain, and other diseases [5]. Modern pharmacological studies have found that G. jasminoides has cholagogic, anti-inflammatory, hepatoprotective, stomach-protecting, analgesic, antipyretic, antibacterial, and cardiovascular and cerebrovascular disease-improving effects [5,6,7]. Apart from being a traditional Chinese medicine, the ripe fruit of G. jasminoides is extensively used in the extraction of pigments and dyes. According to China’s ancient book Han Guan Yi, G. jasminoides was introduced and domesticated as a dye crop by the Han Dynasty, indicating that its cultivation dates back to at least 2100 years ago [8]. During the long-term cultivation, utilization, and breeding of G. jasminoides, rich phenotypic and genetic variations have taken place, and variation types have been developed for ornamental, medicinal, and pigmented purposes [8,9,10,11,12,13].
Featuring double petals, G. jasminoides ‘Fortuneana’ has been cultivated for thousands of years, and has already spread to Japan, South Korea, the Netherlands, and other European countries. Japan, South Korea, the Netherlands, and other countries have also cultivated G. jasminoides ‘Radicans’, G. jasminoides ‘Albomarginata’, G. jasminoides ‘Variegata’, G. jasminoides ‘Ranye’, G. jasminoides ‘Xiao Baichan’, and other cultivars, which has enriched the ornamental resources of G. jasminoides [8]. As traditional Chinese herbal medicine, after long-term artificial breeding, G. jasminoides ‘Yinzhan’, G. jasminoides ‘Zongheng’, G. jasminoides ‘Yuanye’, and other medicinal cultivars of G. jasminoides were cultivated. In addition, G. jasminoides fruit is also a high-grade food-coloring raw material, and so, some cultivars featuring big fruit have also been cultivated, such as G. jasminoides ‘Longicarpa’, G. jasminoides ‘Jinfu Shuizhi’, and G. jasminoides ‘Yuanbao’ [8]. In order to solve the problems of the non-uniform and irregular names of G. jasminoides cultivars, according to the international regulations on the naming of cultivated plants, a search table of cultivar classification was compiled to provide the basis for the phenotypic identification of G. jasminoides cultivars [8].
In order to explore the phylogenetic relationship, 32 morphological characters were used to study the phylogenetic relationship between the cultivars. By using Q-type clustering, the cultivars were divided into three categories: big-fruit cultivars (including G. jasminoides ‘Jinfu Shuizhi’ and other pigmented cultivars), small-fruit cultivars (including G. jasminoides ‘Zongheng’ and other medicinal cultivars), and double-petal cultivars (including G. jasminoides ‘Fortuneana’ and other ornamental cultivars) [9]. However, quantitative classification is highly subjective in the selection and coding of traits. Therefore, based on the ITS2 sequence, the genetic distance and genetic relationship between cultivars were investigated. The results showed that the big-fruit cultivars clustered together, the small-fruit cultivars and G. jasminoides exhibited a relatively close genetic relationship, and the double-petal cultivars were distributed in several branches [10]. Similar results were also obtained when EST-SSR markers were used to study the genetic relationship between the cultivars [14,15].
With the development of molecular technology, multiple studies have reported on G. jasminoides. Utilizing Oxford Nanopore sequencing and Hi-C technology, the whole genome sequencing and chromosome-level genome assembly of G. jasminoides were completed [16]. The complete chloroplast genome of a wild-type gardenia adapted to the island climate was assembled [17]. The variations with the largest content of crocin were sequenced using SMRT sequencing [18]. These studies have partly demonstrated the characteristics of G. jasminoides.
Genome-wide resequencing aims to sequence the genomes of different individuals of a species with known genome sequences and to analyze the differences between individuals or populations [19]. By using this technology, single nucleotide polymorphism (SNP) and insertion deletion polymorphism (inDel) in an individual or a population can be quickly and accurately obtained. Additionally, the genetic relationships and population structure of individuals or populations can be analyzed more accurately through big data analysis [20,21,22,23,24,25].
So far, there has been no report on the analysis of the genome-wide resequencing of the main cultivars of G. jasminoides. In this study, the genome-wide resequencing of 19 samples of G. jasminoides from China was conducted. After the whole genome was compared with the reference genome, SNP and inDel sites were analyzed and detected, the genome location was identified, functional annotation was carried out, and the phylogenetic tree was constructed. Genome-wide resequencing can not only be used to develop polymorphic molecular markers for cultivar identification, the germplasm evaluation and the revelation of the domestication process of G. jasminoides, and the genetic evolution analysis of Gardenia, but also provide research and theoretical bases for genome-wide association and structural variation analysis, functional gene mapping, and the molecular marker-assisted breeding of G. jasminoides.
2. Materials and Methods
2.1. Methods
The experimental materials were collected from the Sichuan, Guangxi, Hunan, Jiangxi, Zhejiang, and Fujian provinces and planted in the traditional Chinese medicine nursery at the Jiangxi Academy of Forestry. There were 19 samples of the Gardenia that were sequenced, including G. jasminoides, 17 cultivars of G. jasminoides, and one outgroup of G. stenophylla Merr. [8,9] (Table 1).

Table 1.
Nineteen samples of Gardenia in this study.
2.2. DNA Extraction and Genome Sequencing
2.2.1. DNA Extraction and Database Establishment
DNA was extracted in line with the Plant Genomic DNA Kit (Tiangen, Beijing, China). Then, sequencing libraries were generated using the Truseq Nano DNA HT Sample Preparation Kit (Illumina, San Diego, California, USA). The DNA sample was fragmented by using sonication to a size of 350 bp, and then, DNA fragments were end-polished, A-tailed, and ligated with the full-length adapter for Illumina sequencing with further PCR amplification. At last, PCR products were purified (AMPure XP system, Beckman, Indianapolis, IN, USA) and the libraries were analyzed for size distribution by Agilent2100 Bioanalyzer and quantified using real-time PCR.
2.2.2. Sequencing
After being approved of in the quality inspection, library products were sequenced and analyzed by HiSeq X Ten high-throughput sequencer. The Fastq data was raw data. Quality control was performed on the raw data to determine whether the sequencing data was suitable for subsequent analysis.
2.3. Resequencing Data Analysis
Fastp 0.20.0 [26] was employed to filter the raw data to obtain clean data. Bwa mem 0.7.17 [27] was employed to align clean data to the reference sequence (https://www.ncbi.nlm.nih.gov/genome/91736?genome_assembly_id=900326 (accessed on 27 September 2020). Duplication reads was removed, and then, GATK HaplotypeCaller 4.1.2.0 [28] was used to detect SNP/inDel. After calling SNP/inDel and Genotyping using GATK software, the genotype of each mutation site in each sample can be obtained. A value of 0 represents consistency with the reference genome sequence, and 1 represents mutation. A genotype of 0/0 or 1/1 indicates homozygosity at this locus, while a genotype of 0/1 indicates heterozygosity. Based on this, the number of homozygous (Ref-Homo, Alt-Homo) and heterozygous (Het) sites in each sample can be counted. Then, SNP/inDel was filtered. The filtered SNP/inDel function was annotated by the ANNOVAR [29] software. Based on SNP markers, principal component analysis (PCA) was carried out by the R software. PLINK was employed to conduct colony formation analysis. The IQ-TREE software (maximum likelihood method) (IQ-TREE; [30]) was employed to model a phylogenetic tree.
3. Results and Analysis
3.1. Quality of Genome Resequencing Data from 19 Samples of G. jasminoides and Comparison with the Reference Genome
In this study, 19 samples of G. jasminoides were resequenced by the Illumina platform (Table 2). Initial data reads ranged from 99,694,304 to 11,5852,754. The amount of filtered Clean reads ranged from 98,716,252 to 114,833,892. Clean read rates ranged from 98.92% to 99.26%. Raw Bases ranged from 14,954,145,600 to 17,377,913,100, and Clean Bases ranged from 14,803,722,258 to 17,220,086,903. The Clean Q30 Bases Rate ranged from 91.19% to 92.94%, indicating that the quality of sample sequencing had reached the resequencing standard.

Table 2.
Quality of resequencing data of 19 samples of G. jasminoides.
See Table 3 for the results of comparisons with the genome of G. jasminoides. Mapped Reads ranged from 94,325,355 to 114,367,172, the Map Rate ranged from 90.84% to 98.29%, and Mean Depth ranged from 24.86 to 30.23. Therefore, the quality value of sequencing data in this project was high enough for subsequent sequence information analysis.

Table 3.
Results of comparisons with the genome of G. jasminoides.
3.2. InDel Site and SNP Site of G. jasminoides
SNP refers to the DNA sequence polymorphism caused by the variation of a single nucleotide at the genome level. InDel is a collective term for miniature Insertions and Deletions. As Table 4 shows, in comparison with the reference genome, a total of 7,568,199 inDel sites (Figure 1) and 61,535,595 SNP sites (Figure 2) were detected. Of the inDel sites, the number of heterozygous sites ranged from 167,512 (Y_40) to 471,006 (Y_4). The heterozygous ratio varied from 41.53% (Y_40) to 90.38% (Y_4), and the number of Deletion sites ranged from 34,175 (Y_4) to 147,405 (Y_37). The total number of sites ranged from 290,157 (Y_40) to 505,181 (Y_4). For the SNP sites, the overall number ranged from 2,392,053 (Y_40) to 4,264,451 (Y_4), the number of heterozygous sites ranged from 1,547,967 (Y_40) to 4,138,168 (Y_4), and the heterozygous ratio varied from 44.86% (Y_50) to 92.69% (Y_5). The number of Deletion sites ranged from 126,283 (Y_4) to 1,073,259 (Y_37). Of the total SNP sites, the number of Y_4 was the largest (4,264,451), while that of Y_40 was the least (2,392,053).

Table 4.
InDel and SNP variation of 19 samples of G. jasminoides in comparison with reference genome.
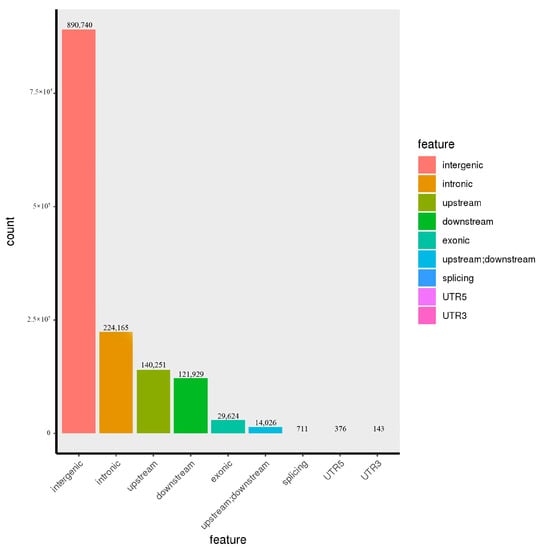
Figure 1.
InDel distribution in 19 samples of G. jasminoides.
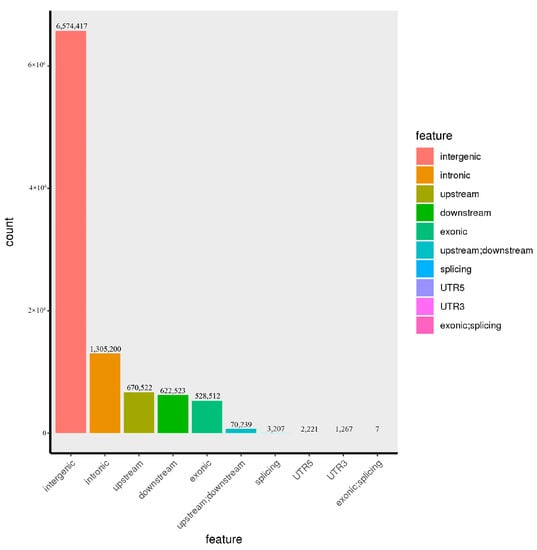
Figure 2.
SNP distribution in 19 samples of G. jasminoides.
3.3. InDel Annotation and SNP Annotation of G. jasminoides Samples
The genome location and function annotation of SNP and inDel from different samples were carried out. As shown in Table 5, SNPs were mainly located in the intergenic region (67.2360%), followed by the intronic region and the upstream, downstream, and exonic regions, which accounted for 13.3482%, 6.8574%, 6.3665%, and 5.4050%, respectively. The numbers of SNPS located in the UTR5 and UTR3 regions were less, accounting for 0.0227% and 0.0130%, respectively. The number of SNPs located within 2bp of the exonic region near the splicing site was the least, being seven (Figure 1). Like SNP, inDels were mainly located in the intergenic region (62.6415%), followed by the intronic region and the upstream, downstream, and exonic regions, which accounted for 15.7645%, 9.8632%, 8.5747%, and 2.0833%, respectively. The numbers of inDels located in the region within 2 bp of the splicing site and in the UTR5 region were less, accounting for 0.0500% and 0.0130%, respectively. The number of inDels located in the UTR3 region was the least, being 143 (Figure 2).

Table 5.
Statistics of the annotation results of inDel and SNP.
3.4. Population Genetic Structure of G. jasminoides Samples
Population structure analysis was performed based on detected genome SNPs (Figure 3). When K = 2 (the number of predefined genetic clades), Y_7, Y_37, and Y_13 were separated from the other cultivars. Y_7 and Y_37 feature double petals and relatively small flower diameters while Y_13 features a simple flower shape. When K = 4, all individuals were subdivided into four clades. Y_10 and Y_38 were separated from the other cultivars; they feature double petals and small flower diameters. When K = 5, the cultivars would be further differentiated: Y_23 and Y_40, Y_29, Y_39, and Y_33 were differentiated from the other cultivars. Y_23 and Y_40 feature big fruit while the others feature small fruit. When K = 7, Y_4, Y_17, Y_20, Y_34, and Y_19, Y_36, Y_22, Y_2 were divided into a subgroup respectively and separated from the other cultivars. Y_4, Y_17, and Y_34 feature large flower diameters while Y_20 features golden-yellow petals and a relatively small flower diameter. Y_19, Y_36, and Y_22 feature medium flower diameters while Y_2 features double petals and a large flower diameter.
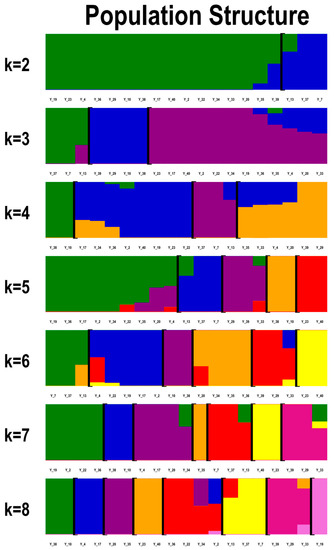
Figure 3.
The population structure of G. jasminoides. Structure results from K = 2 to K = 8 and match to the cluster (black frame) based on the result of STRUCTURE when the value of K corresponds to the Y-axis. The color in the structure plot indicates the component state in each sample.
3.5. The Construction of the Phylogenetic Tree of G. jasminoides
Based on the phylogenetic tree constructed by the maximum likelihood method, similar conclusions can be drawn based on the results of genetic structure and principal component analysis. As Figure 4 shows, the G. jasminoides and the 17 cultivars were put into one category. Y_33 has a closer genetic distance compared to the original species. Y_2 and Y_22 also differentiated early. Featuring double petals, Y_10, Y_38, and Y_2 were put into one category. Y_35, Y_7, Y_37, and Y_13 were differentiated from the other cultivars. Featuring large flower diameters or golden-yellow petals, Y_4, Y_17, Y_20, and Y_34 were put into one category. Featuring medium flower diameters or big fruit, Y_19, Y_36, Y_22, Y_23, and Y_40 were put into one category.
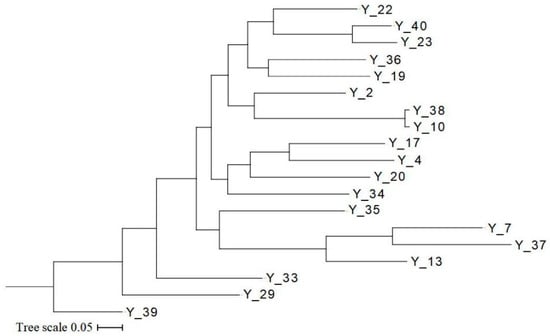
Figure 4.
The phylogenetic tree of the 19 samples of G. jasminoides based on maximum likelihood.
3.6. The Principal Component Analysis (PCA) of the Population of G. jasminoides
PCA was used to obtain the principal component map of the 19 samples of G. jasminoides (Figure 5). According to their positions and distances from each other in the two-dimensional graph, the positions of Y_10 and Y_38, Y_23, and Y_40 almost overlapped with each other, indicating that their genotype difference was small, the genetic relationship was close, and the results of genetic distance were consistent with those of genetic structure and phylogenetic tree clustering. Based on the data from PC1 and PC2, the cultivars were mapped. While the colonies of Y_10 and Y_38, and Y_23 and Y_40, were independent and scattered, and Y_7, Y_37, Y_13, Y_39, and Y_29 were independent and scattered, the other cultivars were clustered. The cultivar was mapped based on PC2 and PC3 data. Y_10 and Y_38, Y_23 and Y_40, Y_4, Y_20 and Y_33 were clustered independently into three groups and dispersed, while Y_29 and Y_39 were still dispersed and distant from other clusters. The cultivar was mapped based on PC1 and PC3 data. Y_4, Y_20, and Y_33 were clustered independently into group and dispersed, while Y_7, Y_37, Y_13, Y_29, and Y_39 were still dispersed individually and far from each other.
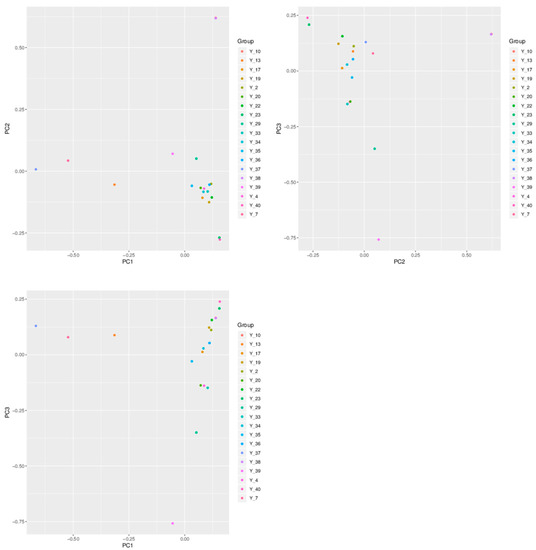
Figure 5.
Principal component analysis of the samples of G. jasminoides using SNP data.
4. Discussion
In this study, 19 samples of Gardenia from China were selected, and various genetic variations including SNP, inDel, etc. were detected among different Gardenia samples by high-depth resequencing. For the SNP sites, the overall number ranged from 2,392,053 to 4,264,451, and the heterozygous ratio varied from 44.86% to 92.69% (Y_5). Of the inDel sites, the total number of sites ranged from 290,157 to 505,181 and the heterozygous ratio varied from 41.53% to 90.38%. The analysis results showed that there were abundant genetic variations among different G. jasminoides at the genome-wide level, which also supported the previous finding that different G. jasminoides species were rich in variation and clustered into different branches based on taxonomy, phenotypic analysis, and various genetic markers [8,9,10,15].
In this study, G. stenophylla, G. jasminoides, and 17 cultivars were resequenced. The clustering results showed that the small-fruit cultivars, such as Y_33, Y_34, and Y_35, had a closer genetic differentiation distance with the original G. jasminoides, indicating that G. jasminoides was earliest used for medicinal purposes. Y_7 and Y_37, which had small flower diameters and double petals, Y_13, which featured big fruits, were differentiated sequentially. According to China’s ancient book Han Guan Yi, dating back to over 2100 years ago, G. jasminoides was introduced and domesticated as a dye crop by the Han Dynasty. During the time of the Song Dynasty, in China’s ancient book Ben Cao Tu Jing, it was discussed that G. jasminoides with small fruit is used as medicine, while G. jasminoides plants with large and long fruit are used for coloring and cannot be used as medicine. In addition, the G. jasminoides garden described by Ban Gu and G. jasminoides flowers are recorded in Jin Shu, written by Fang Xuanling. Moreover, since the Sui Dynasty, G. jasminoides has been presented to visiting envoys in the form of national gifts. The research results of this study are consistent with the long-term use of G. jasminoides in China. The results indicated that in the long-term utilization of G. jasminoides, the artificial selection was mainly based on flower type and fruit type. In this way, humans can obtain cultivars, which are the medicinal types with small fruit, the ornamental types with beautiful flowers and double petals, and the pigment types with big fruit for gardenia cultivation. Featuring big fruits, Y 22, Y 23, and Y 40 have a longer genetic distance from the original G. jasminoides, showing a longer differentiation history. Additionally, the three cultivars clustered together, consistent with the results based on quantitative classification [9] and the ITS2 sequence [10], showing close genetic relationships. Featuring large flower diameters, Y_4, Y_17, and Y_34, and Y_20, featuring golden-yellow petals and a relatively small flower diameter, were put into one category. In addition, Y_10, Y_38, and Y_2 are grouped together. Y 10 and Y 38 feature dwarf plants, smaller leaves, small flower diameters, and double petals, while Y 2 features double petals and a large flower diameter. Y_7, Y_37, and Y_13 were differentiated from the other cultivars. Y_7 and Y_37 feature double petals and relatively small flower diameters while Y_13 features simple flowers. It can be seen that the double petal cultivars are not all separated into a single branch. The clustering results are also controlled by genetic traits such as flower size and leaf size, and the results are not consistent with quantitative classification. However, Y_39, Y_10, and Y_38, with similar leaf shapes, were not clustered. Some of the clustering results were similar to the quantitative classification results based on phenotypic data, but there was no obvious systematic branch formed by the characteristics of having double petals, big fruit, or small fruit [9]. Based on the results of previous studies on quantitative classification [9], the ITS2 sequence [10], EST-SSR [15], etc., the genetic distance and genetic relationships between cultivars were further verified and clarified based on the results of whole genome resequencing. The big-fruit cultivars, like the double-petal cultivars, have a long history of genetic differentiation. Moreover, long-term artificial selection results in smaller genetic differentiation and closer genetic relationships among big-fruit cultivars. In order to meet different aesthetic needs, the genetic differentiation of double-petal cultivars was greater, with more abundant variations in leaf size, corolla size, and plant type. Similarly, ‘Yinzhan‘, ‘Hengsheng’, and ‘Yuanye’ cultivated in recent years are close in terms of genetic distance to G. jasminoides. This indicates that after long-term natural and artificial selection, the cultivars have not only formed stable phenotypic characteristics significantly different from those of the original species, but also formed stable genetic variations at the molecular level. The results of genetic structure also showed that when K = 2–7, except for Y_2, Y_4, Y_13, Y_17, Y_20, Y_22, Y_33, Y_34, Y_35, Y_36, and Y_39, all other cultivars maintained the same color, indicating that the cultivars formed native cultivars with high purities formed through long-term evolution, cultivation, and domestication. Judging from comparisons between the phenotypic information of the cultivars and the phylogenetic tree, there was a significant close relationship between the cultivars in terms of grouping, which may be attributed to the time-honored history of the cultivation and breeding of G. jasminoides in China or the frequent exchange of local cultivars and the close genetic relationship between the species used in the breeding of different cultivars. Some of the areas of several groups that were differentiated by different two-dimensional maps overlapped with each other, indicating that all kinds of cultivars are of the same origin, or that there are more gene exchanges between cultivars and the genetic relationship is more complex. The results of structural analysis, principal component analysis, and the phylogenetic tree were consistent with each other.
Since G. jasminoides was domesticated for ornamental, medicinal, pigment, and oil resource plant purposes, it has spread to various places and adapted itself to different natural environments, and its phenotype and genotype have undergone different degrees of differentiation under the pressure of artificial and natural selection. In this study, the resequencing of G. jasminoides and cultivars with different utilization purposes and ecological adaptations was analyzed, which provided important clues for analyzing the origin and genetic basis of G. jasminoides and theoretical guidance for subsequent genetic breeding and sustainable conservation and utilization.
Author Contributions
Conceptualization—S.D.; methodology—S.D. and K.L.; software—K.L. and L.G.; validation—L.G., X.Y., M.W. and X.F.; data curation, K.L., X.Y., M.W. and X.F.; writing: original draft preparation—K.L.; writing: review and editing—S.D.; funding acquisition—S.D., K.L. and L.G. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 32060356), Key Research and Development Projects of Jiangxi Province, China (Grant No. 20203BBF63024), and Doctoral Program at the Jiangxi Academy of Forestry, China (Grant No. 2023522802).
Data Availability Statement
Data sharing is not applicable to this article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Chen, T.; Taylor, C.M. Gardenia. In Flora of China; Wu, Z.Y., Raven, P.H., Hong, D.Y., Eds.; Science Press: Beijing, China; Botanical Garden Press: St. Louis, MO, USA, 2011; Volume 19, pp. 141–144. [Google Scholar]
- Ye, X.E.; Xis, N.H. Gardenia reflexisepala (Rubiaceae), a New Species from Hainan Province, China with Typification of G. angkorensis and G. cambodiana. Phytotaxa 2016, 257, 193–197. [Google Scholar] [CrossRef]
- Chen, Y.L. Study on Resources Survey of Gardenia and Herbalism of Gardenia jasminoides J. Ellis. Master’s Thesis, Peking Union Medical College, Beijing, China, 2018. [Google Scholar]
- Yu, X.S.; Zhou, Q. Aesthetic Culture of Gardenia Flower in China. J. Beijing For. Univ. (Soc. Sci.) 2010, 9, 6–12. [Google Scholar]
- National Pharmacopoeia Committee. Pharmacopoeia of the People’s Republic of China, 1st ed.; China Medical Science Press: Beijing, China, 2020; pp. 259–260. [Google Scholar]
- Ni, H.Y.; Zhang, Z.H.; Fu, H.Z. Research and development of Fructus Gardeniae. China J. Chin. Mater. Med. 2006, 31, 538–541. [Google Scholar]
- Li, X.Y.; Wu, M.; Wang, S.Z.; Liu, L.T. Pharmacological effects of Gardenia jasminoside Ellis on cardiovascular system. Chin. J. Integr. Tradit. West. Med. 2022, 42, 373–378. [Google Scholar]
- Deng, S.Y.; Zhu, P.L.; Wang, X.R. Cultivar classification of Gardenia plants. South China For. Sci. 2018, 46, 13–18. [Google Scholar]
- Deng, S.R.; Yang, H.; Zhu, P.L.; Chen, Y.J.; Li, T.; Li, K.Q. Numerical Taxonomy of Cultivars and Their Related Wild Species in the Genus Gardenia. Acta Agric. Univ. Jiangxiensis 2020, 42, 92–100. [Google Scholar]
- Deng, S.Y.; Li, K.Q.; Jia, Q.Q.; Chen, Y.J.; Zhu, P.L.; Yang, C.X. ITS2 sequence analysis of cultivars and their related wild species in Gardenia genus. Acta Agric. Univ. Jiangxiensis 2021, 43, 1140–1148. [Google Scholar]
- Xie, Z.W. Research on variety and quality evaluation of Gardenia jasminoides Ellis. J. Chin. Med. Mater. 1991, 14, 45–46. [Google Scholar]
- Zhou, H.C.; Zhang, X.C.; Luo, J.; Yu, H.M.; Hu, K.Z. Comprehensive Evaluation of Gardenia jasminoides Ellis Variety Resources. China J. Chin. Mater. Med. 1998, 23, 141. [Google Scholar]
- Cao, L.; Liu, D.W.; Dai, Z.X. Investigation and Analysis of Gardenia jasminoides Ellis Resources in Jiangxi Province. Li Shi Zhen Med. Mater. Madica Res. 2008, 2, 288–290. [Google Scholar]
- Deng, S.Y.; Wang, X.R.; Zhu, P.L.; Wen, Q.; Yang, C.X. Development of polymorphic microsatellite markers in the medicinal plant Gardenia jasminoides (Rubiaceae). Biochem. Syst. Ecol. 2015, 58, 149–155. [Google Scholar] [CrossRef]
- Deng, S.Y.; Zhu, P.L.; Li, K.Q.; Chen, Y.J.; Zhu, P.L.; Wang, X.R.; Li, T.; Tang, S. Genetic relationship analysis and fingerprint construction of Gardenia jasminoides cultivars based on EST-SSR markers. Chin. Tradit. Herb. Drugs 2022, 53, 2795–2802. [Google Scholar]
- Xu, Z.C.; Pu, X.D.; Gao, R.R.; Demurtas, O.C.; Fleck, S.J.; Richter, M.; He, C.; Ji, A.; Sun, W.; Kong, J.; et al. Tandem gene duplications drive divergent evolution of caffeine and crocin biosynthetic pathways in plants. BMC Biol. 2020, 18, 63. [Google Scholar] [CrossRef]
- Zhang, M.; Chen, W.D.; Li, Y.Y.; Zhang, C.; Chai, Z.-H.; Li, Y.-F.; Xie, S.-W.; Deng, S.-Y.; Duan, Y.-F.; Wang, X.-R. Complete chloroplast genome of a wild-type Gardenia jasminoides ellis (rubiaceae) adapted to island climate. Mitochondrial DNA Part B 2021, 6, 313–314. [Google Scholar] [CrossRef] [PubMed]
- Shen, T.F.; Zheng, Y.J.; Liu, Q.; Chen, C.H.; Huang, L.L.; Deng, S.Y.; Xu, M.; Yang, C.X. Integrated SMRT and Illumina Sequencing Provide New Insights into Crocin Biosynthesis of Gardenia jasminoides. Int. J. Mol. Sci. 2022, 23, 6321. [Google Scholar] [CrossRef] [PubMed]
- Ley, T.J.; Mardis, E.R.; Li, D.; Fulton, B.; Wilson, R.K. DNA sequencing of a cytogenetically normal acute myeloid leukaemia genome. Nature 2018, 456, 66–72. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Yao, S.L.; Cheng, X.H.; Liu, Y.Y.; Ma, L.X.; Xiang, Y.; Huang, J.Y.; Tong, C.B.; Liu, S.Y. Genomic variation of spring, semi-winter and winter Brassica napus by high-depth DNA re-sequencing. Chin. J. Oil Crop Sci. 2018, 40, 469–478. [Google Scholar]
- Chen, X.; Guo, R.; Wang, L.; Liu, Y.H.; Guo, M.B.; Xu, Y.P.; Guo, H.Y.; Yang, M.; Zhang, Q.Y. SNP analysis of wild cultivation cannabis based on whole genome resequencing. Mol. Plant Breed. 2018, 16, 893–897. [Google Scholar]
- Fan, W. Population Genetic Analysis of Castor (Ricinus communis) Based on Whole Genome Resequencing. Ph.D. Thesis, Chinese Academy of Agricultural Sciences, Beijing, China, 2020. [Google Scholar]
- Yin, M.H.; Wang, Q.; Zhang, H.L.; Cai, X.H.; Xu, C.Q.; Chen, F.L.; Liu, S.Y.; Zhang, Q.W.; Cai, H.; Chen, R.H. Whole genome re-sequencing analysis of alpine potato and local farm potato in Huaiyu Mountain under high altitude habitats. Genom. Appl. Biol. 2020, 39, 1198–1207. [Google Scholar]
- Shen, Y.Y.; Jin, Q.L.; Cai, W.M.; Fan, L.J.; Feng, W.L.; Song, T.T. Analyses of population diversity and structure of Flammulina filiformis strains based on whole genome resequencing data. Mycosystema 2020, 39, 1016–1028. [Google Scholar]
- Du, H.D.; You, X.; Li, Y.F.; Wang, S.; Mao, X.J.; Zhang, N. Genomic variation detection analysis of 3 self-pruning Tomato Lines Based on Resequencing. Mol. Plant Breed. 2022, 20, 756–764. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef] [PubMed]
- Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv 2013, arXiv:13033997. [Google Scholar]
- Mckenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M. The genome analysis toolkit: A mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef]
- Yang, H.; Wang, K. Genomic variant annotation and prioritization with ANNOVAR and wANNOVAR. Nat. Protoc. Erecipes Res. 2015, 10, 1556–1566. [Google Scholar] [CrossRef]
- Nguyen, L.T.; Schmidt, H.A.; Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2014, 32, 268–274. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).