Genetic Diversity of 52 Species of Kiwifruit (Actinidia chinensis Planch.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. DNA Extraction, Detection, and Quantitative Dilution
2.2.2. Primer Screening
2.2.3. PCR and Detection
2.2.4. Data Analysis
3. Results
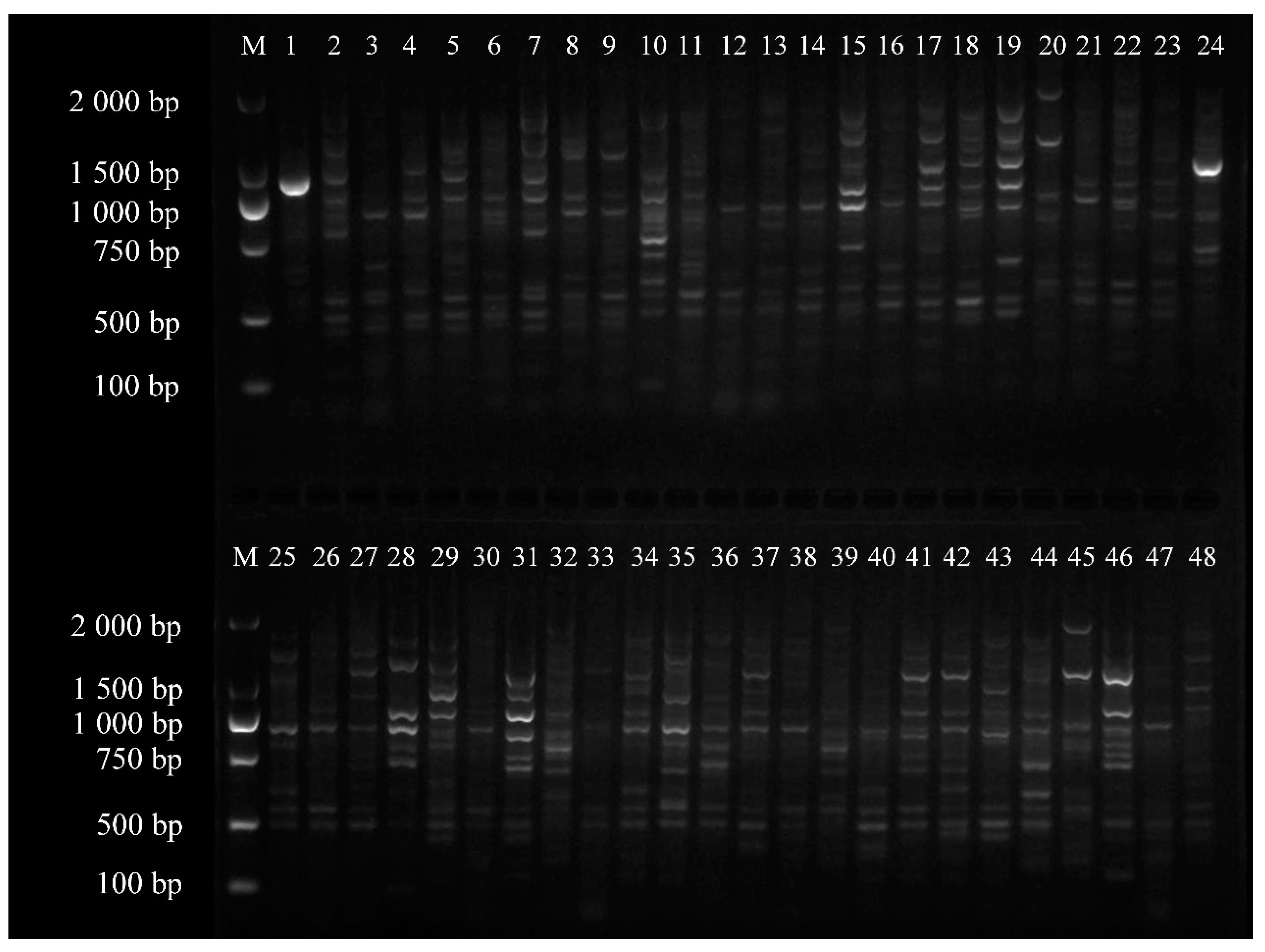
3.1. Genomic DNA Extraction
3.2. Analysis of the Polymorphism of Bands
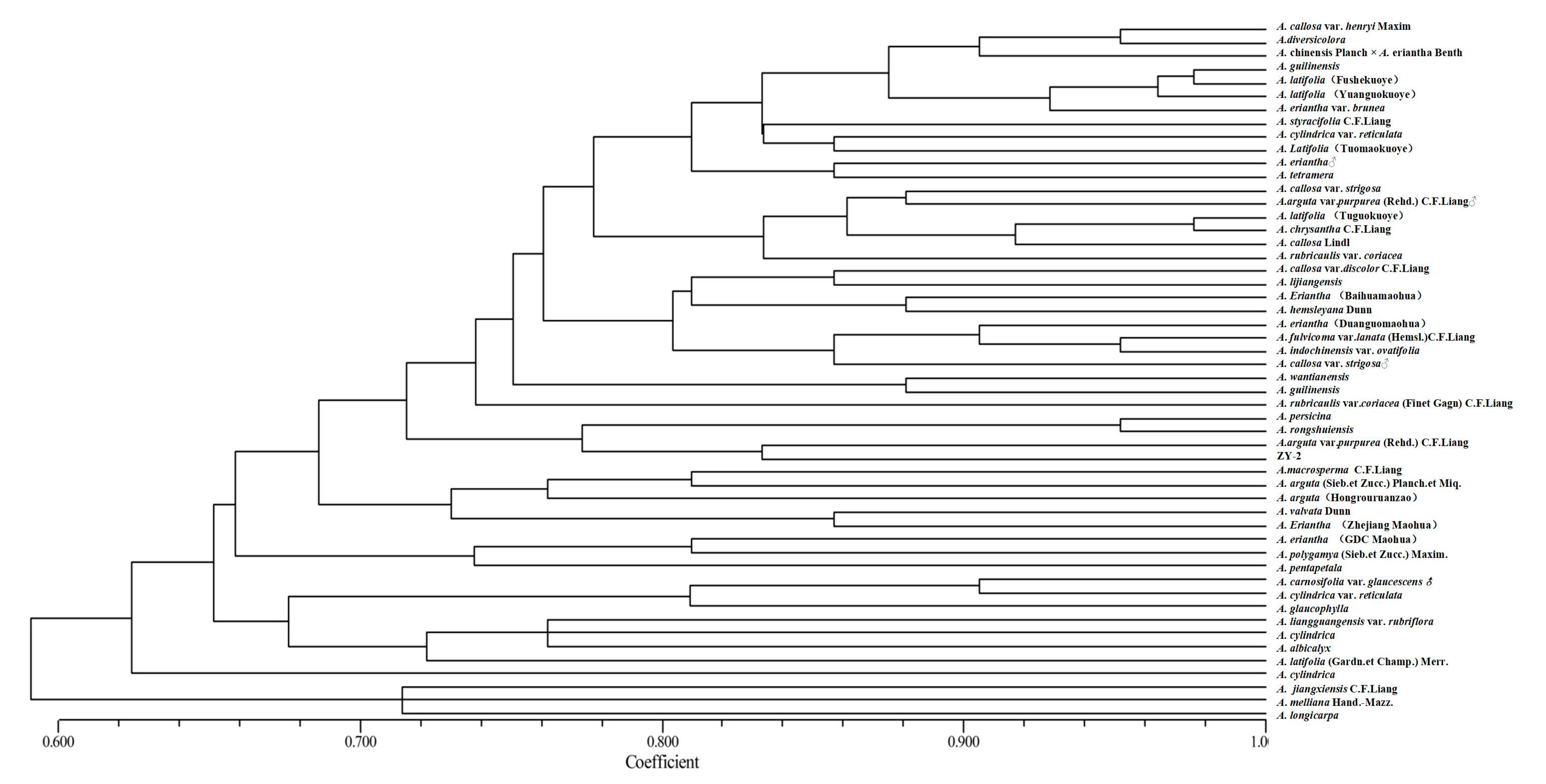
3.3. Genetic Identity, Genetic Distance, and Cluster Analysis
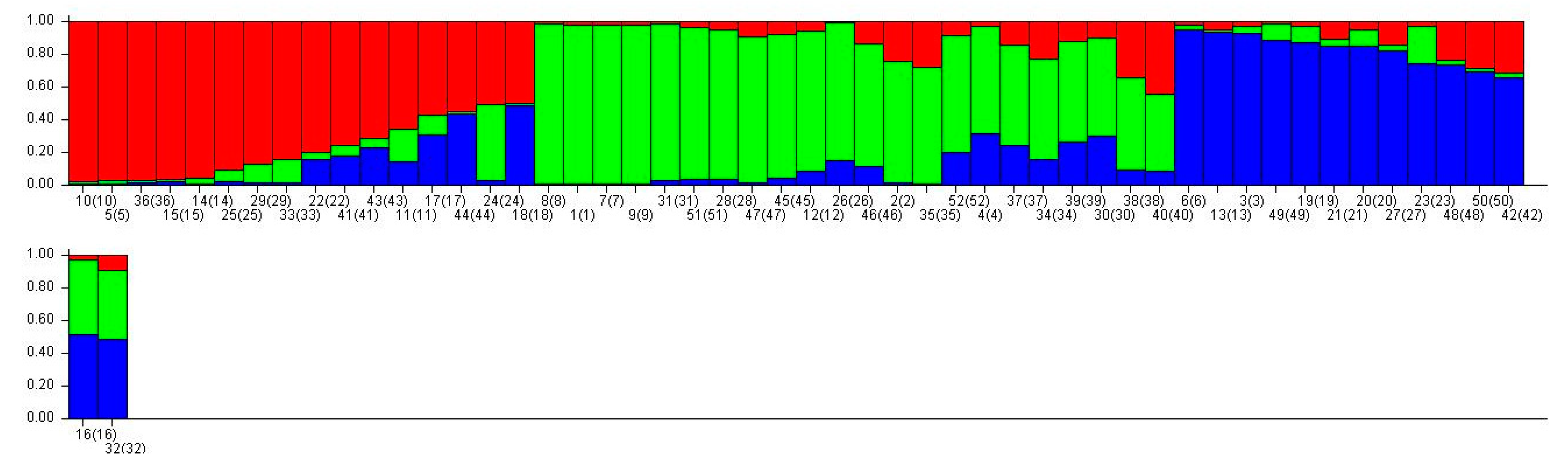
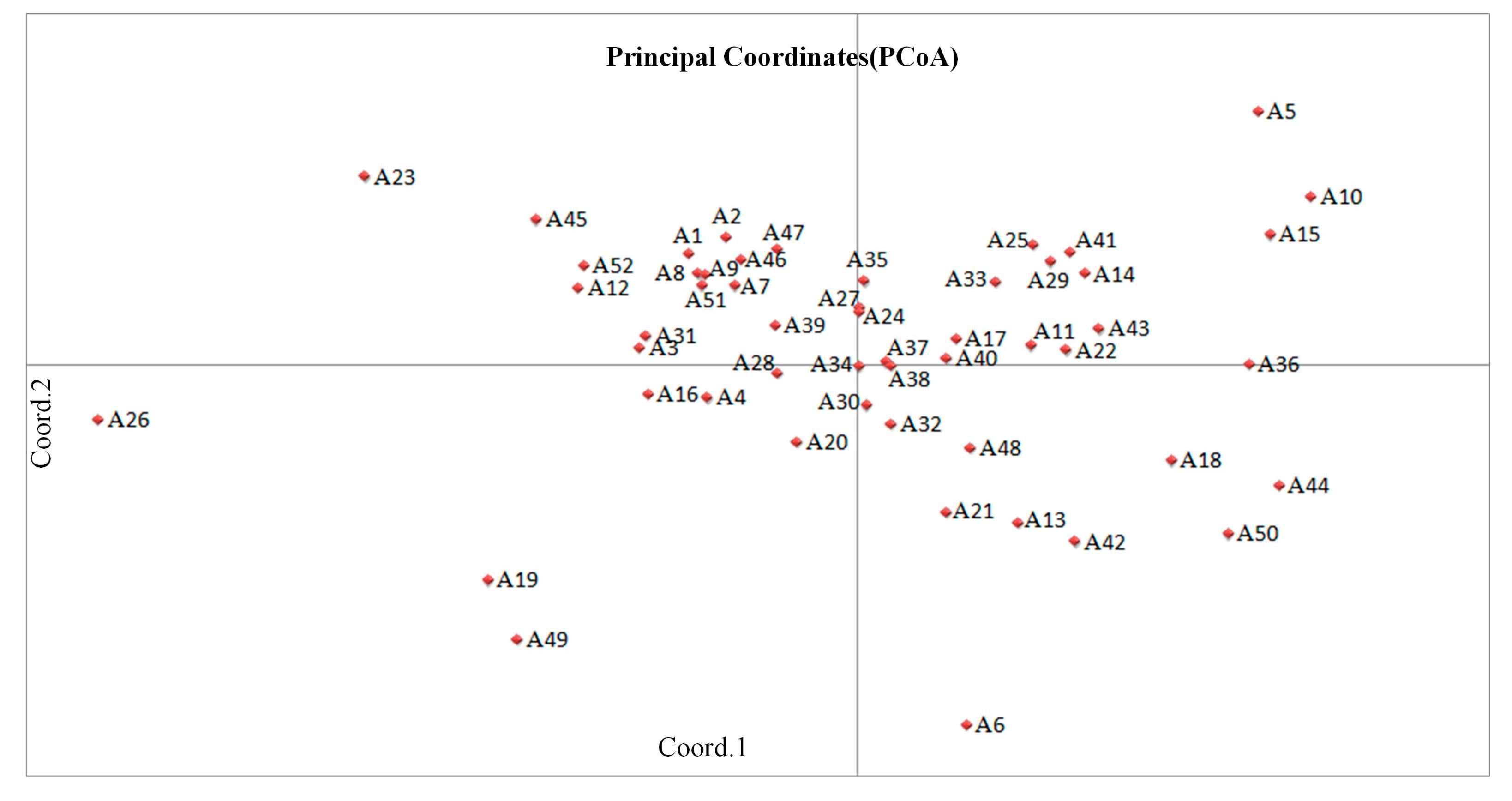
3.4. Population Structure Analysis and Principal Coordinate Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, Z.Z.; Kang, M.; Huang, H.; Testolin, R. Phylogenetic relationships in actinidia as revealed by nuclear DNA genetic markers and cytoplasmic DNA sequence analysis. Acta Hortic. 2007, 753, 45–58. [Google Scholar] [CrossRef]
- Vanneste, J.L. The scientific, economic, and social impacts of the New Zealand outbreak of bacterial canker of kiwifruit (Pseudomonas syringae pv. actinidiae). Annu. Rev. Phytopathol. 2017, 55, 377–399. [Google Scholar] [CrossRef]
- Wu, H.; Ma, T.; Kang, M.; Ai, F.; Zhang, J.; Dong, G.; Liu, J. A high-quality Actinidia chinensis (kiwifruit) genome. Hortic. Res. 2019, 6, 117. [Google Scholar] [CrossRef]
- Li, D.; Liu, Y.; Zhong, C.; Huang, H. Morphological and cytotype variation of wild kiwifruit (Actinidia chinensis complex) along an altitudinal and longitudinal gradient in central-west China. Bot. J. Linn. Soc. 2010, 164, 72–83. [Google Scholar] [CrossRef]
- Huang, H.; Wang, S.; Jiang, Z.; Zhang, Z.; Gong, J. Exploration of actinidia genetic resources and development of kiwifruit industry in China. Acta Hortic. 2003, 610, 29–43. [Google Scholar] [CrossRef]
- Salinger, M.J.; Kenny, G.J. Climate and kiwifruit cv. ‘Hayward’ 2. Regions in New Zealand suited for production. N. Z. J. Crop Hortic. Sci. 1995, 23, 173–184. [Google Scholar] [CrossRef]
- Huang, H.; Ferguson, A.R. Actinidia in China: Natural diversity, phylogeographical evolution, interspecific gene flow and kiwifruit cultivar improvement. Acta Hortic. 2007, 753, 31–40. [Google Scholar] [CrossRef]
- Sun, L.; Fang, J. Conservation, research and utilization of kiwifruit germplasm resources in China. J. Plant Genet. Resour. 2020, 21, 1483–1493. [Google Scholar]
- Kisaki, G.; Shimagami, T.; Matsudaira, K.; Tsugi, Y.; Moriguchi, K.; Nakashima, K.; Morimoto, T.; Sugita-Konishi, S.; Tabuchi, M.; Gomi, K.; et al. A kiwifruit cultivar crossbred with Actinidia chinensis and Actinidia rufa has practical tolerance to Pseudomonas syringae pv. actinidiae biovar 3. J. Plant Pathol. 2019, 101, 1211–1214. [Google Scholar] [CrossRef]
- Huang, H.; Liu, Y. Natural hybridization, introgression breeding, and cultivar improvement in the genus Actinidia. Tree Genet. Genomes 2014, 10, 1113–1122. [Google Scholar] [CrossRef]
- Zhao, C. The genetic diversity of wild actinidia arguta germplasm resources from China and South Korea. J. Fruit Sci. 2018, 35, 1043–1051. [Google Scholar]
- Rai, M.K. Start codon targeted (SCoT) polymorphism marker in plant genome analysis: Current status and prospects. Planta 2023, 257, 34. [Google Scholar] [CrossRef]
- Collard, B.C.Y.; Mackill, D.J. Start codon targeted (SCoT) polymorphism: A simple, novel DNA marker technique for generating gene-targeted markers in plants. Plant Mol. Biol. Rep. 2009, 27, 86–93. [Google Scholar] [CrossRef]
- Dora, S.; Mansour, M.; Aboulila, A.; Abdelwahab, E. Genetic diversity and relationships among some barley genotypes for net blotch disease resistance using RAPD, SCOT and SSR markers. Egypt. J. Genet. Cytol. 2017, 46, 139–165. [Google Scholar] [CrossRef]
- Chen, B.L.; Zhang, J.; Huang, J.K.; Huang, C.M.; Zhou, Q.J.; Wei, T.; Tang, J. Application of SCoT markers on genetic diversity analysis and variation identification of actinidia. J. Agric. Biotechnol. 2018, 26, 77–86. [Google Scholar]
- Long, J.; Fan, L.; Xu, G.; Hu, S.; Han, G. Application advance of SCoT molecular markers in plants. J. Plant Genet. Resour. 2015, 16, 336–343. [Google Scholar]
- Xiong, F.; Tang, R.; Chen, Z.; Pan, L.; Zhuang, W. SCoT: A novel gene targeted marker technique based on the translation start codon. Mol. Plant Breed. 2009, 7, 635–638. [Google Scholar]
- Wang, F.; Li, J.; Ye, K.; Gong, H.; Mo, Q.; Jiang, Q.; Liu, P. Comparative analysis on the genetic diversity of 41 vitis germplasm resources by ISSR and SCoT molecular markers. Guihaia 2017, 37, 1–8. [Google Scholar]
- Guo, D.-L.; Zhang, J.-Y.; Liu, C.-H. Genetic diversity in some grape varieties revealed by SCoT analyses. Mol. Biol. Rep. 2012, 39, 5307–5313. [Google Scholar] [CrossRef]
- Xia, L.; Yang, T.; Yang, Y.; Xia, H.; Zhang, Y.; Wang, R. System optimization of SCot-PCR and analysis on genetic diversity of Persimmion (Diospyros kaki Thunb). Acta Bot. Boreali-Occident. Sin. 2014, 34, 473–480. [Google Scholar]
- Chen, H.; Xin-Hua, H.E.; Luo, C.; Zhu, J.H.; Feng, A.L. Analysis on the genetic diversity of 24 longan (Dimocarpus longan) accessions by SCoT markers. Acta Hortic. Sin. 2010, 37, 1651–1654. [Google Scholar]
- Wang, S.; Jiang, Z.; Huang, H.; Zhang, Z.; Ke, J. Conservation and utilization of germplasm resources of the genus actinidia. Acta Hortic. 2003, 610, 365–371. [Google Scholar] [CrossRef]
- Collard, B.; Mackill, D.J. Conserved DNA-derived polymorphism (CDDP): A simple and novel method for generating DNA markers in plants. Plant Mol. Biol. Report. 2009, 27, 558–562. [Google Scholar] [CrossRef]
- Huang, H.; Li, J.; Lang, P.; Wang, S. Systematic relationships in actinidia as revealed by cluster analysis of digitized morphological descriptors. Acta Hortic. 1999, 498, 71–78. [Google Scholar] [CrossRef]
- He, Z.; Zhong, Y.; Liu, H.; Tang, X.; Ye, L.; Huang, D.; Xu, L. Quantitative taxonomic analyses of actinidia (Actinidiaceae) in China based on micromorphological characters of foliar trichomes. Acta Phytotaxon. Sin. 2000, 38, 121–136. [Google Scholar]
- Li, J.; Huang, H.; Sang, T. Molecular phylogeny and infrageneric classification of actinidia (Actinidiaceae). Syst. Bot. 2009, 27, 408–415. [Google Scholar]
- Huang, H.; Li, Z.; Li, J.; Kubisiak, T.L.; Testolin, R. Phylogenetic relationships in actinidia as revealed by RAPD analysis. J. Am. Soc. Hortic. 2002, 127, 759–766. [Google Scholar] [CrossRef]
- Li, Z.; Huang, H.; Jiang, Z.; Li, J.Q.; Kubisiak, T.L. Phylogenetic relationships in actinidia as revealed by RAPDs and PCR-RFLPs of mtDNA. Acta Hortic. 2003, 610, 387–396. [Google Scholar] [CrossRef]
- Yao, X. The first complete chloroplast genome sequences in actinidiaceae: Genome structure and comparative analysis. PLoS ONE 2015, 10, e0129347. [Google Scholar] [CrossRef]
- Chat, J.L.; Jáuregui, B.; Petit, R.J.; Nadot, S. Reticulate evolution in kiwifruit (Actinidia, Actinidiaceae) identified by comparing their maternal and paternal phylogenies. Am. J. Bot. 2004, 91, 736–747. [Google Scholar] [CrossRef]
- Liang, C.F.; Ferguson, A.R. Emendation of the latin name of actinidia chinensis Pl. var. hispida C.F. Liang. Guihaia 1984, 4, 181–182. [Google Scholar]
- He, T.; Chai, J.; Zhao, N.; Li, X.; Chen, Y. Genetic diversity on its site of Actinidia. Mol. Plant Breed. 2019, 17, 329–336. [Google Scholar]
- Cipriani, G.; Testolin, R.; Gardner, R. Restriction-site variation of PCR-amplified chloroplast DNA regions and its implication for the evolution and taxonomy of Actinidia. Theor. Appl. Genet. 1998, 96, 389–396. [Google Scholar] [CrossRef]
- Kim, S.; Jung, Y.; Misun, C.; Koh, S. Genetic relationships of genus Actinidia based on random amplified polymorphic DNA analysis. Hortic. Environ. Biotechnol. 2003, 44, 340–344. [Google Scholar]
- Lai, J.J.; Li, Z.Z.; Man, Y.P.; Lei, R.; Wang, Y.C. Genetic diversity of five wild Actinidia arguta populations native to China as revealed by SSR markers. Sci. Hortic. 2015, 191, 101–107. [Google Scholar] [CrossRef]
- Wang, F.; Li, J.; Ye, K.; Liu, P.; Gong, H.; Jiang, Q.; Qi, B.; Mo, Q. An in vitro Actinidia bioassay to evaluate the resistance to Pseudomonas syringae pv. actinidiae. Plant Pathol. J. 2019, 35, 372–380. [Google Scholar] [CrossRef]
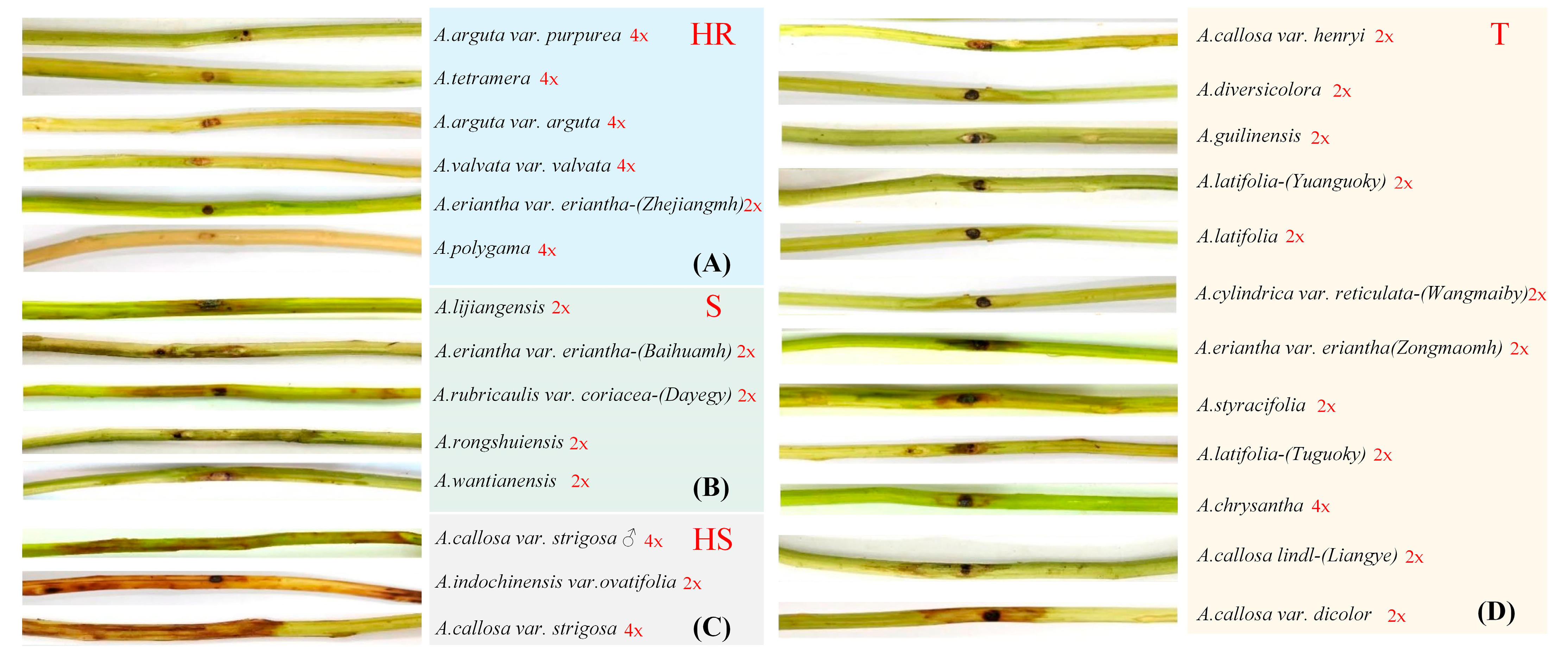
| No. | Species (Genotype) | Distribution/ Origin | Ploidy | Response to pseudomonas syringae pv. actinidia |
|---|---|---|---|---|
| 1 | A. callosa var. henryi Maxim. | Guangxi, China | 2x 4x | T |
| 2 | A.diversicolora | Sichuan, China | 2x | T |
| 3 | A. jiangxiensis C.F.Liang | Jiangxi, China | 2x | T |
| 4 | A.arguta var. purpurea (Rehd.) C.F.Liang | Guangxi, China | 4x 8x | HR |
| 5 | A. pentapetala | Guangxi, China | 2x | S |
| 6 | A.macrosperma C.F.Liang | Zhejiang, China | 4x | HR |
| 7 | A. guilinensis | Guangxi, China | 2x | T |
| 8 | A. latifolia (Fushekuoye) | Shanxi, China | 2x | — |
| 9 | A. latifolia (Yuanguokuoye) | Hubei, China | T | |
| 10 | A.carnosifolia var. glaucescens ♂ | Guangxi, China | — | — |
| 11 | A. Eriantha (Baihuamaohua) | Wuhan, China | 2x | S |
| 12 | A. Latifolia (Tuomaokuoye) | Sichuan, China | 2x | S |
| 13 | A.arguta (Sieb.et Zucc.) Planch.et Miq. | Heilongjiang, China | 4x | HR |
| 14 | A. hemsleyana Dunn | Guangxi, China | 2x | — |
| 15 | A. cylindrica var. reticulata | Guangxi, China | 2x | T |
| 16 | A. tetramera | Guangdong, China | 4x | HR |
| 17 | A. Eriantha (Zhejiang Maohua) | Jiangxi, China | 2x | HR |
| 18 | A.valvata Dunn | Hunan, China | 4x | HR |
| 19 | A. cylindrica | Guangxi, China | 2x | S |
| 20 | A. eriantha (GDC Maohua) | Wuhan, China | 2x | — |
| 21 | A.polygamya (Sieb.et Zucc.) Maxim. | Yunnan, China | 4x | HR |
| 22 | A. indochinensis var. ovatifolia | Guangxi, China | 2x | HS |
| 23 | A. melliana Hand.-Mazz. | Guangxi, China | 2x | HS |
| 24 | A. persicina | Guangxi, China | 2x | T |
| 25 | A. callosa var. strigosa ♂ | Guangxi, China | 4x | HS |
| 26 | A. latifolia (Gardn.et Champ.) Merr. | Guangxi, China | 2x | T |
| 27 | A. longicarpa | Sichuan, China | 2x | T |
| 28 | A. latifolia (Tuguokuoye) | Guangxi, China | 2x | T |
| 29 | A. rongshuiensis | Guangxi, China | 2x | S |
| 30 | A. callosa var. strigosa | Wuhan, China | 4x | HS |
| 31 | A. eriantha ♂ | Guangxi, China | 2x | HR |
| 32 | A.arguta var. purpurea (Rehd.) C.F.Liang ♂ | Jiangxi, China | 4x 8x | HR |
| 33 | A. wantianensis | Guangxi, China | 2x | S |
| 34 | A. eriantha (Duanguomaohua) | Guangxi, China | 2x | — |
| 35 | A. fulvicoma var. lanata (Hemsl.) C.F.Liang | Hunan, China | 2x | — |
| 36 | A. guilinensis | Guangxi, China | 2x | T |
| 37 | A. callosa var. discolor C.F.Liang | Guangxi, China | 2x 4x | T |
| 38 | A. chrysantha C.F.Liang | Guangxi, China | 4x | T |
| 39 | A. rubricaulis var. coriacea | Sichuan, China | 2x | S |
| 40 | A. callosa Lindl | — | 2x | S |
| 41 | A. rubricaulis var. coriacea (Finet Gagn) C.F.Liang | Guangxi, China | 2x | S |
| 42 | A. glaucophylla | Guangxi, China | 2x | HS |
| 43 | A.liangguangensis var. rubriflora | Guangxi, China | 2x | T |
| 44 | A. cylindrica | Guangxi, China | 2x | S |
| 45 | A. eriantha var. brunea | Guangxi, China | 2x | T |
| 46 | A. cylindrica var. reticulata | Guangxi, China | 2x 4x | T |
| 47 | A.chinensis Planch × A.eriantha Benth | Guangxi, China | — | — |
| 48 | A. arguta (Hongrouruanzao) | Guangxi, China | — | — |
| 49 | ZY-2 | Guangxi, China | 4x | T |
| 50 | A. albicalyx | Guangxi, China | 2x | T |
| 51 | A. styracifolia C.F.Liang | Fujian, China | 2x | T |
| 52 | A. lijiangensis | Guangxi, China | 2x | S |
| SCoT Primers | Sequence (5′-3′) | Total Bands | Polymorphic Bands | Percentage of Polymorphic Bands (%) |
|---|---|---|---|---|
| P1 | CAACAATGGCTACCACCA | 9 | 9 | 100.0 |
| P3 | CAACAATGGCTACCACCG | 11 | 10 | 90.9 |
| P4 | CAACAATGGCTACCACCT | 10 | 8 | 80.0 |
| P8 | CAACAATGGCTACCACGT | 9 | 8 | 88.8 |
| P11 | AAGCAATGGCTACCACCA | 11 | 11 | 100.0 |
| P12 | ACGACATGGCGACCAACG | 15 | 14 | 93.3 |
| P13 | ACGACATGGCGACCATCG | 11 | 10 | 90.9 |
| P36 | GCAACAATGGCTACCACC | 9 | 9 | 100 |
| P37 | ACGACATGGCGACCAGCG | 17 | 15 | 88.2 |
| P38 | ACGACATGGCGACCACCG | 11 | 9 | 81.8 |
| Total | 113 | 103 | — | |
| Average | 11.3 | 10.3 | 91.15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, B.; Wang, F.; Ye, K.; Mo, Q.; Gong, H.; Liu, P.; Jiang, Q.; Li, J. Genetic Diversity of 52 Species of Kiwifruit (Actinidia chinensis Planch.). Horticulturae 2023, 9, 753. https://doi.org/10.3390/horticulturae9070753
Qi B, Wang F, Ye K, Mo Q, Gong H, Liu P, Jiang Q, Li J. Genetic Diversity of 52 Species of Kiwifruit (Actinidia chinensis Planch.). Horticulturae. 2023; 9(7):753. https://doi.org/10.3390/horticulturae9070753
Chicago/Turabian StyleQi, Beibei, Faming Wang, Kaiyu Ye, Quanhui Mo, Hongjuan Gong, Pingping Liu, Qiaosheng Jiang, and Jiewei Li. 2023. "Genetic Diversity of 52 Species of Kiwifruit (Actinidia chinensis Planch.)" Horticulturae 9, no. 7: 753. https://doi.org/10.3390/horticulturae9070753
APA StyleQi, B., Wang, F., Ye, K., Mo, Q., Gong, H., Liu, P., Jiang, Q., & Li, J. (2023). Genetic Diversity of 52 Species of Kiwifruit (Actinidia chinensis Planch.). Horticulturae, 9(7), 753. https://doi.org/10.3390/horticulturae9070753






