Combined Analysis of Volatile Compounds and Extraction of Floral Fragrance Genes in Two Dendrobium Species
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Apparatus
2.3. GC-MS Analysis
2.4. Transcriptome Analysis
2.5. RT-PCR Analysis
3. Results
3.1. Analysis the Characteristics of Volatile Constituents in Different Florescences of Two Dendrobium Species
3.2. Analysis of the Characteristics of Volatile Constituents in Different Organs of Two Dendrobium Species
3.3. Identification and Analysis of the DEGs
3.4. KEGG Functional Classifications and Enrichment Analysis of the DEGs
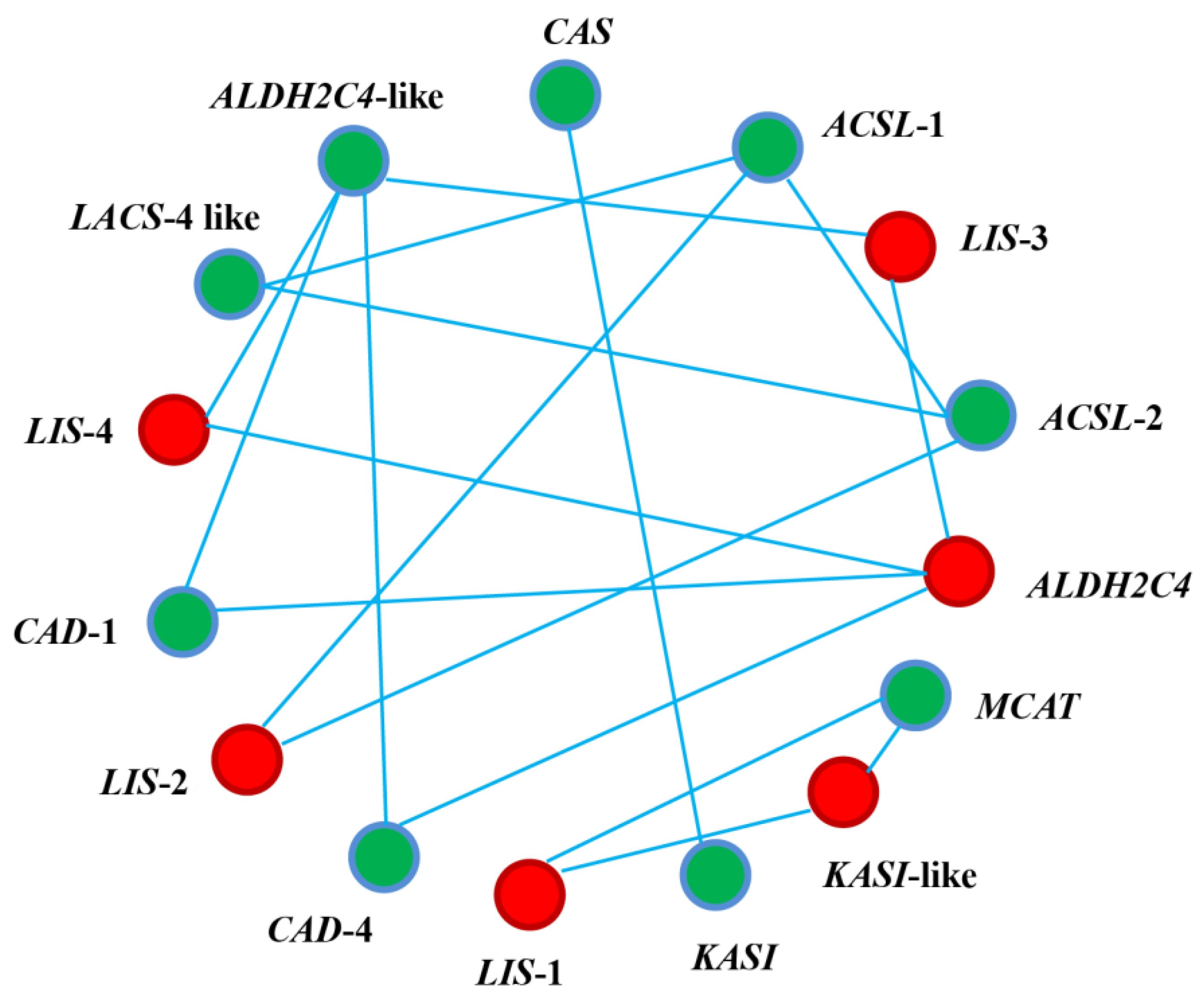
3.5. Protein-Protein Interaction (PPI) Network Analysis of the DEGs
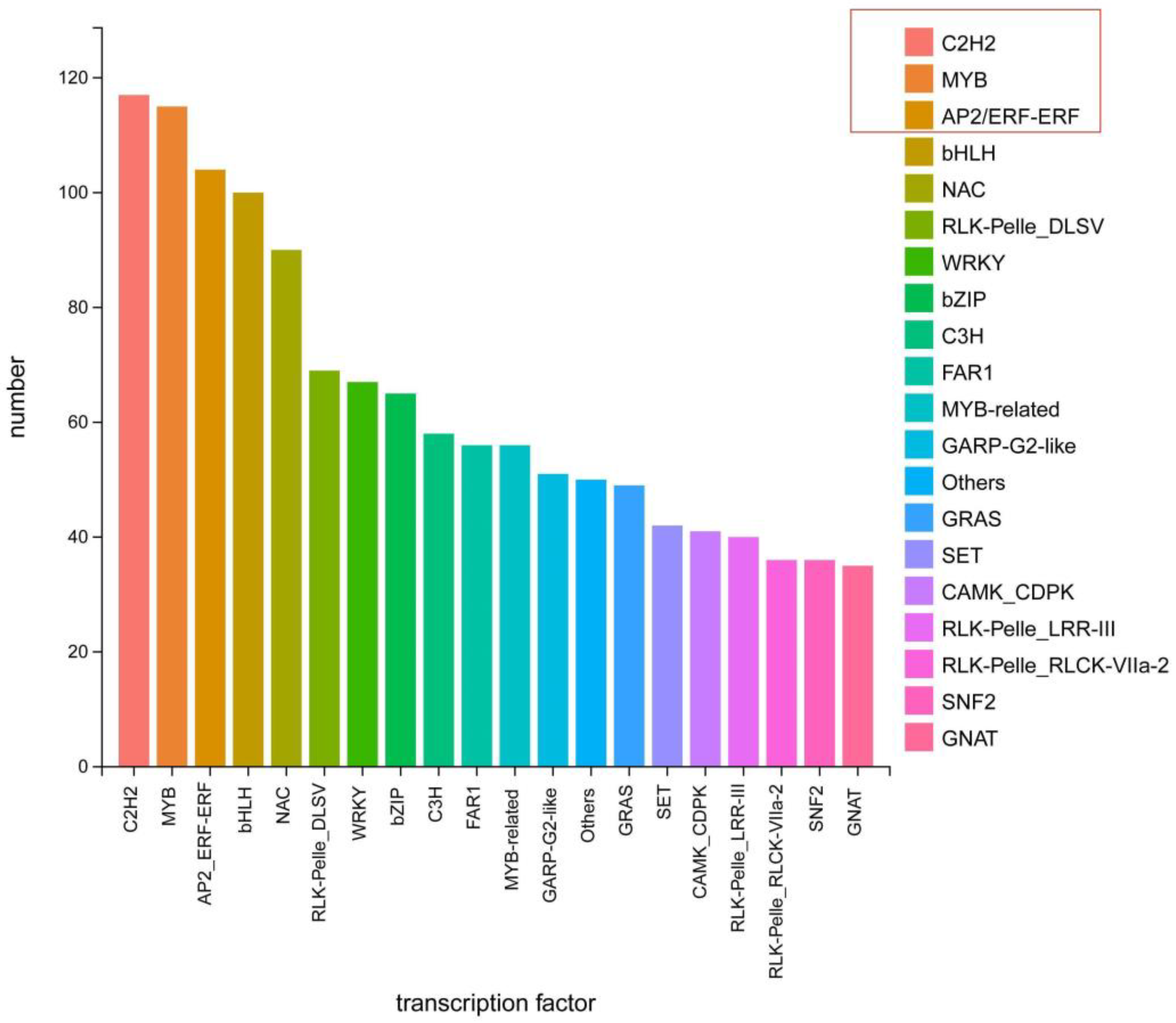
3.6. Transcription Factor Prediction in Two Dendrobium Species
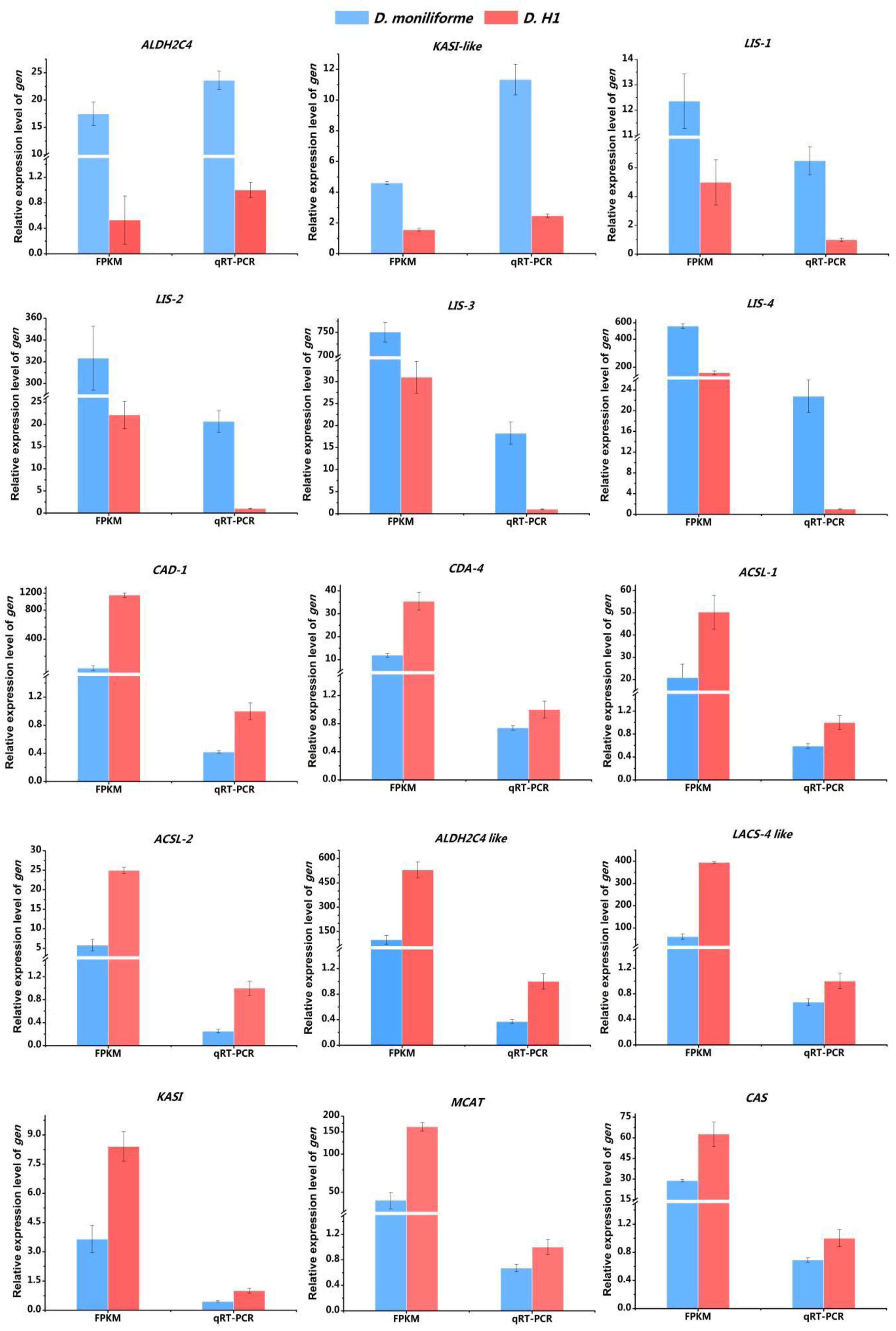
3.7. Validation of the DEGs Expression
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ge, D.; Zhang, D.; Ding, X.; Zhou, Q.; Zhang, W.; Li, X. Genetic variation and conservation of the endangered Chinese endemic herb Dendrobium officinale based on SRAP analysis. Plant Syst. Evol. 2008, 276, 149–156. [Google Scholar]
- Zhang, K.H.; Wang, M.Q.; Wei, L.L. Investigation of the Effects and Mechanisms of Dendrobium loddigesii Rolfe Extract on the Treatment of Gout. Evid.-Based Complement. Altern. Med. 2020, 2020, 4367347. [Google Scholar] [CrossRef]
- Liu, W.M.; Chen, R.; Wei, R.F. Advances in development and utilization of Dendrobium. Subtrop. Agric. Res. 2011, 7, 87–91. [Google Scholar]
- Ai, Y.M. Annals of Medicinal Plants in China; Peking University Medical Press: Beijing, China, 2013. [Google Scholar]
- Sun, Z.; Song, Y.; Zeng, R. Advances in studies on intraspecific and interspecific relationships mediated by plant volatiles. J. South China Agric. Univ. 2019, 40, 166–174. [Google Scholar]
- Wu, J.N.; Liu, Z.Y.; Wu, M.Q. Analysis of Volatile Components in the Flowers of Seven Species of Dendrobium. Mol. Plant Breed. 2023, 3, 1–16. [Google Scholar]
- Kai, L. The Scent of Orchids. Olfactory and chemical investigations. Nord. J. Bot. 2010, 14, 302. [Google Scholar]
- Robustelli della Cuna, F.S.; Calevo, J.; Bari, E.; Giovannini, A.; Boselli, C.; Tava, A. Characterization and Antioxidant Activity of Essential Oil of Four Sympatric Orchid Species. Molecules 2019, 24, 3878. [Google Scholar] [CrossRef]
- Abbas, F.; Ke, Y.G.; Yu, R.C.; Yue, Y.; Amanullah, S.; Jahangir, M.M.; Fan, Y. Volatile terpenoids: Multiple functions, biosynthesis, modulation and manipulation by genetic engineering. Planta 2017, 246, 803–816. [Google Scholar] [CrossRef]
- Yu, F.N.; Utsumi, R. Diversity, regulation, and genetic manipulation of plant mono- and sesquiterpenoid biosynthesis. Cell. Mol. Life Sci. 2009, 66, 3043–3052. [Google Scholar] [CrossRef]
- Qiao, Z.L.; Hu, H.Z.; Shi, S.B.; Yuan, X.; Yan, B.; Chen, L. An Update on the Function, Biosynthesis and Regulation of Floral Volatile Terpenoids. Horticulturae 2021, 7, 451. [Google Scholar] [CrossRef]
- Ramya, M.; Jang, S.; An, H.R.; Lee, S.Y.; Park, P.M.; Park, P.H. Volatile Organic Compounds from Orchids: From Synthesis and Function to Gene Regulation. Int. J. Mol. Sci. 2020, 21, 1160. [Google Scholar] [CrossRef]
- Dinesh, A.; Priyanka, G. Advances in biosynthesis, regulation, and metabolic engineering of plant specialized terpenoids. Plant Sci. 2020, 294, 457–475. [Google Scholar]
- Dudareva, N.; Pichersky, E.; Gershenzon, J. Biochemistry of plant volatiles. Plant Physiol. 2004, 135, 1893–1902. [Google Scholar] [CrossRef]
- DeerInWater, K.M. Plant Volatiles as Information Sources Influencing Herbivore Preference and Performance; University of California: Davis, CA, USA, 2015. [Google Scholar]
- Paschold, A.; Halitschke, R.; Baldwin, I.T. Using ‘mute’ plants to translate volatile signals. Plant J. 2010, 45, 275–291. [Google Scholar] [CrossRef]
- Ali, J.G.; Alborn, H.T.; Campos-Herrera, R.; Kaplan, F.; Duncan, L.W.; Rodriguez-Saona, C.; Koppenhöfer, A.M.; Stelinski, L.L. Subterranean, Herbivore-Induced Plant Volatile Increases Biological Control Activity of Multiple Beneficial Nematode Species in Distinct Habitats. PLoS ONE 2012, 7, 38146. [Google Scholar] [CrossRef]
- Cheng, A.X.; Lou, Y.G.; Mao, Y.B.; Lu, S.; Wang, L.-J.; Chen, X.-Y. Plant Terpenoids: Biosynthesis and Ecological Functions. Acta Bot. 2007, 49, 8. [Google Scholar] [CrossRef]
- Dudareva, N.; Andersson, S.; Orlova, I.; Gatto, N.; Reichelt, M.; Rhodes, D.; Boland, W.; Gershenzon, J. The nonmevalonate pathway supports both monoterpene and sesquiterpene formation in snapdragon flowers. Proc. Natl. Acad. Sci. USA 2005, 102, 933–938. [Google Scholar] [CrossRef]
- Dong, X.X.; Gao, L.; Liu, L.; Chen, L.; Liu, Q.; Zhang, D. Function-specific volatiles and volatilization characteristics of Dendrobium officinale. J. King Saud Univ.-Sci. 2020, 32, 2020–2028. [Google Scholar] [CrossRef]
- Irmisch, S.; Krause, S.T.; Kunert, G.; Gershenzon, J.; Degenhardt, J.; Köllner, T.G. The organ-specific expression of terpene synthase genes contributes to the terpene hydrocarbon composition of chamomile essential oils. BMC Plant Biol. 2012, 12, 84–97. [Google Scholar] [CrossRef]
- Zeng, X.L.; Liu, C.; Zheng, R.R.; Cai, X.; Luo, J.; Zou, J.; Wang, C. Emission and Accumulation of Monoterpene and the Key Terpene Synthase (TPS) Associated with Monoterpene Biosynthesis in Osmanthus fragrans Lour. Front. Plant Sci. 2015, 6, 1232. [Google Scholar] [CrossRef]
- Zhou, F.; Pichersky, E. The complete functional characterisation of the terpene synthase family in tomato. New Phytol. 2020, 226, 1341–1360. [Google Scholar] [CrossRef]
- Yu, Z.M.; Zhao, C.H.; GZhang, G.H. Genome-Wide Identification and Expression Profile of TPS Gene Family in Dendrobium officinale and the Role of DoTPS10 in Linalool Biosynthesis. Int. J. Mol. Sci. 2020, 21, 5419. [Google Scholar] [CrossRef]
- Zhang, C.P.; Zhang, J.L.; Sun, Z.R.; Liu, X.Y.; Shu, L.Z.; Wu, H.; Song, Y.; He, D.H. Genome-wide identification and characterization of terpene synthase genes in Gossypium hirsutum. Gene 2022, 828, 146462. [Google Scholar] [CrossRef]
- Song, W.; Kai, O.Y.; Kai, W. Genome-Wide Identification, Evolution, and Expression Analysis of TPS and TPP Gene Families in Brachypodium distachyon. Plants 2019, 8, 362. [Google Scholar]
- Cao, Z.Q.; Ma, Y.; Weng, J.; Shi, J.; Chen, J.; Hao, Z. Genome-Wide Identification and Expression Analysis of TPS Gene Family in Liriodendron chinense. Genes 2023, 14, 770. [Google Scholar] [CrossRef]
- Chen, F.; Tholl, D.; Bohlmann, J.; Pichersky, E. The family of terpene synthases in plants: A mid-size family of genes for specialized metabolism that is highly diversified throughout the kingdom. Plant J. 2011, 66, 212–229. [Google Scholar] [CrossRef]
- Li, N.Y.; Dong, M.; Lv, L.; Qian, L.; Sun, X.; Liu, L.; Cai, Y.; Fan, H. Combined Analysis of Volatile Terpenoid Metabolism and Transcriptome Reveals Transcription Factors Related to Terpene Synthase in Two Cultivars of Dendrobium officinale Flowers. Front. Genet. 2021, 12, 661296. [Google Scholar] [CrossRef]
- Robert, A.; Flath, K.O. Volatile Components of the Orchid Dendrobium superbum Rchb. f. J. Agric. Food Chem. 1982, 30, 842–845. [Google Scholar]
- Baek, Y.S.; Ramya, M.; An, H.R.; Park, P.M.; Lee, S.Y.; Baek, N.I.; Park, P.H. Volatiles Profile of the Floral Organs of a New Hybrid Cymbidium, ‘Sunny Bell’ Using Headspace Solid-Phase Microextraction Gas Chromatography-Mass Spectrometry Analysis. Plants 2019, 8, 251. [Google Scholar] [CrossRef]
- Bohlmann, J.; Meyer-Gauen, G.; Croteau, R. Plant terpenoid synthases: Molecular biology and phylogenetic analysis. Proc. Natl. Acad. Sci. USA 1998, 95, 4126–4133. [Google Scholar] [CrossRef]
- Chuang, Y.C.; Hung, Y.C.; Tsai, W.C.; Chen, W.H.; Chen, H.H. PbbHLH4 regulates floral monoterpene biosynthesis in Phalaenopsis orchids. J. Exp. Bot. 2018, 69, 4363–4377. [Google Scholar] [CrossRef]
- Gao, F.; Liu, B.; Li, M.; Gao, X.; Fang, Q.; Liu, C.; Ding, H.; Wang, L.; Gao, X. Identification and Characterization of Terpene Synthase Genes Accounting for the Volatile Terpene Emissions in Flowers of Freesia hybrida. J. Exp. Bot. 2018, 69, 4249–4265. [Google Scholar] [CrossRef]
- Sheng, C.Z.; Wei, M.Z. A New Bibenzyl Glycoside from Dendrobium moniliforme. Chin. Chem. Express 2003, 14, 276–277. [Google Scholar]
- Li, N.; Mao, Y.; Zhang, X. Separation and identification of volatile constituents in Artemisia argyi flowers by GC-MS with SPME and steam distillation. J. Chromatogr. Sci. 2008, 46, 401–405. [Google Scholar] [CrossRef]
- Medina, A.; Lucero, M.; Estell, R.; O’CONNELL, M. A comparison of steam distillation and solid- phase microextraction (SPME) for isolation of the anemopsis californica volatiles. Soc. Range Manag. New Mex. Sect. Meet. 2002, 8. Available online: https://www.ars.usda.gov/research/publications/publication/?seqNo115=130925 (accessed on 5 April 2023).
- Wei, S.L.; Liang, X.; Meng, Y.; Li, Y.; Gao, F.; Liu, X.; Wang, S.; Gao, X.; Wang, L. Biochemical and Molecular Characterization of a Flavonoid 3-O-glycosyltransferase Responsible for Anthocyanins and Flavonols Biosynthesis in Freesia hybrida. Front. Plant Sci. 2016, 7, 410. [Google Scholar]
- Feng, L.G.; Chen, C.; Sheng, L.X.; Liu, P.; Tao, J.; Su, J.L.; Zhao, L.Y. Comparative analysis of headspace volatiles of Chinese Rosa rugosa. Molecules 2010, 15, 8390–8399. [Google Scholar] [CrossRef]
- Heller, S.R.; Milne, G.W. EPA/NIH Mass Spectral Data Base. Supplement 1. 1980. U. S. Governwent Printing Office. 1980. Available online: https://www.researchgate.net/publication/235096355_EPANIH_Mass_Spectral_Data_Base_Supplement_1_1980 (accessed on 3 February 2023).
- Zhang, H.; Chen, M.; Wang, X.; Dai, J.; Zhang, X.; Zhang, Z.; Zhang, X.; Tang, M.; Tang, J.; Gong, J.; et al. Transcriptome Analysis of Rhododendron liliiflorum H. Lév. Flower Colour Differences. Horticulturae 2023, 9, 82. [Google Scholar] [CrossRef]
- Bray NL, H.; Pimentel, P.; Melsted, L. Near-optimal RNA-Seq quantification. Comput. Sci. 2015, 11, 48550. [Google Scholar]
- Li, T.; Cai, Z.L.; Wu, H.J. Transcriptome analysis of the Japanese pine sawyer beetle, Monochamus alternatus (Coleoptera: Cerambycidae) by high-throughput Illumina sequencing. J. Asia-Pac. Entomol. 2015, 18, 439–445. [Google Scholar]
- Gaur, R.; Bhatia, S.; Gupta, M. Generation of expressed sequence tags under cadmium stress for gene discovery and development of molecular markers in chickpea. Protoplasma 2014, 251, 955–972. [Google Scholar] [CrossRef]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, 353–361. [Google Scholar] [CrossRef]
- Martin IR, E.; Vigne, V.; Komar, A.; Amandine, V.; Schmitt-Keichinger, C. Transcriptomic analysis of grapevine cv. Gewurztraminer infected by two strains of grapevine fanleaf virus with distinct phenotypes. In Proceedings of the 15th Meeting of the International Council for the Study of Virus and Virus-Like Diseases of the Grapevine (ICVG), Santiago de Chile, Chile, 3–7 April 2018. [Google Scholar]
- Han, X.F.; Ling, Q.F.; Li, C.J.; Wang, G.; Xu, Z.; Lu, G. Characterization of pikeperch (Sander lucioperca) transcriptome and development of SSR markers. Biochem. Syst. Ecol. 2016, 66, 188–195. [Google Scholar] [CrossRef]
- Zhang, G.N.; Zhang, C.F.; Wang, Z.T.; Xu, L.S. Studies on chemical constituents of Dendrobium thyrsiflorum rchb. (I). Chin. J. Nat. Med. 2004, 2, 78–81. [Google Scholar]
- Li, C.H.; Huang, M.Z.; Huang, S.H.; Yin, J. Volatile Components in Flowers of Four Dendrobium Species. J. Trop. Subtrop. Bot. 2015, 23, 454–462. [Google Scholar]
- Wen, X.; Wang, Y.; Chen, L.; Zhang, K.; Zhang, J.; Li, Y. Chemical Constituents from the Flowers of Dendrobium chrysanthum. Agric. Sci. Technol. 2014, 15, 1629–1633. [Google Scholar]
- Hua, L.S.; Meng, X.U.; Zhang, X.F.; Liu, J.J.; Si, J.P. Studies on Volatile Constituents of 11 Families of Dendrobium officinale Flowers. Chin. J. Exp. Tradit. Med. Formulae 2016, 22, 52–57. [Google Scholar]
- Huang, X.Z.; Xiao, Y.T.; Köllner, T.G.; Jing, W.X.; Kou, J.F.; Chen, J.Y.; Liu, D.F.; Gu, S.H.; Wu, J.X.; Zhang, Y.J.; et al. The terpene synthase gene family in Gossypium hirsutum harbors a linalool synthase GhTPS12 implicated in direct defence responses against herbivores. Plant Cell Environ. 2017, 41, 261–274. [Google Scholar] [CrossRef]
- Dudareva, N. Biochemical and Molecular Genetic Aspects of Floral Scents. Plant Physiol. 2000, 122, 627–633. [Google Scholar] [CrossRef]
- Zhou, P.; Du, Y.; Xu, N.; Yue, C.; Ye, L. Improved linalool production in Saccharomyces cerevisiae by combining directed evolution of linalool synthase and overexpression of the complete mevalonate pathway. Biochem. Eng. J. 2020, 161, 107655. [Google Scholar] [CrossRef]
- Zhou, P.; Du, Y.; Fang, X.; Xu, N.; Yue, C.; Ye, L. Combinatorial Modulation of Linalool Synthase and Farnesyl Diphosphate Synthase for Linalool Overproduction in Saccharomyces cerevisiae. J. Agric. Food Chem. 2021, 69, 1003–1010. [Google Scholar] [CrossRef]
- Aharoni, A. Terpenoid metabolism in wild-type and transgenic Arabidopsis plants. Plant Cell 2003, 12, 107655. [Google Scholar] [CrossRef]
- Xiao, Y.; Wang, Q.; Erb, M.; Turlings, T.C.J.; Ge, L.; Hu, L.; Li, J.; Han, X.; Zhang, T.; Lu, J.; et al. Specific herbivore-induced volatiles defend plants and determine insect community composition in the field. Ecol. Lett. 2012, 15, 1130–1139. [Google Scholar] [CrossRef]
- Yang, Z.Z.; Li, Y.Q.; Gao, F.Z.; Jin, W.; Li, S.; Kimani, S.; Yang, S.; Bao, T.; Gao, X.; Wang, L. MYB21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J. Exp. Bot. 2020, 71, 4140–4158. [Google Scholar] [CrossRef]
- Nieuwenhuizen, N.J.; Green, S.A.; Chen, X.; Bailleul, E.J.; Matich, A.J.; Wang, M.Y.; Atkinson, R. Functional genomics reveals that a compact terpene synthase gene family can account for terpene volatile production in apple. Plant Physiol. 2013, 161, 787–804. [Google Scholar] [CrossRef]
- Martin, D.M.; Aubourg, S.; Schouwey, M.B.; Daviet, L.; Schalk, M.; Toub, O.; Lund, S.T.; Bohlmann, J. Functional Annotation, Genome Organization and Phylogeny of the Grapevine (Vitis vinifera) Terpene Synthase Gene Family Based on Genome Assembly, FLcDNA Cloning, and Enzyme Assays. BMC Plant Biol. 2010, 10, 226. [Google Scholar] [CrossRef]
- Hsiao, Y.Y.; Pan, Z.J.; Hsu, C.C.; Yang, Y.P.; Hsu, Y.C.; Chuang, Y.C.; Shih, H.H.; Chen, W.H.; Tsai, W.C.; Chen, H.H. Research on Orchid Biology and Biotechnology. Plant Cell Physiol. 2011, 52, 1467–1486. [Google Scholar] [CrossRef]
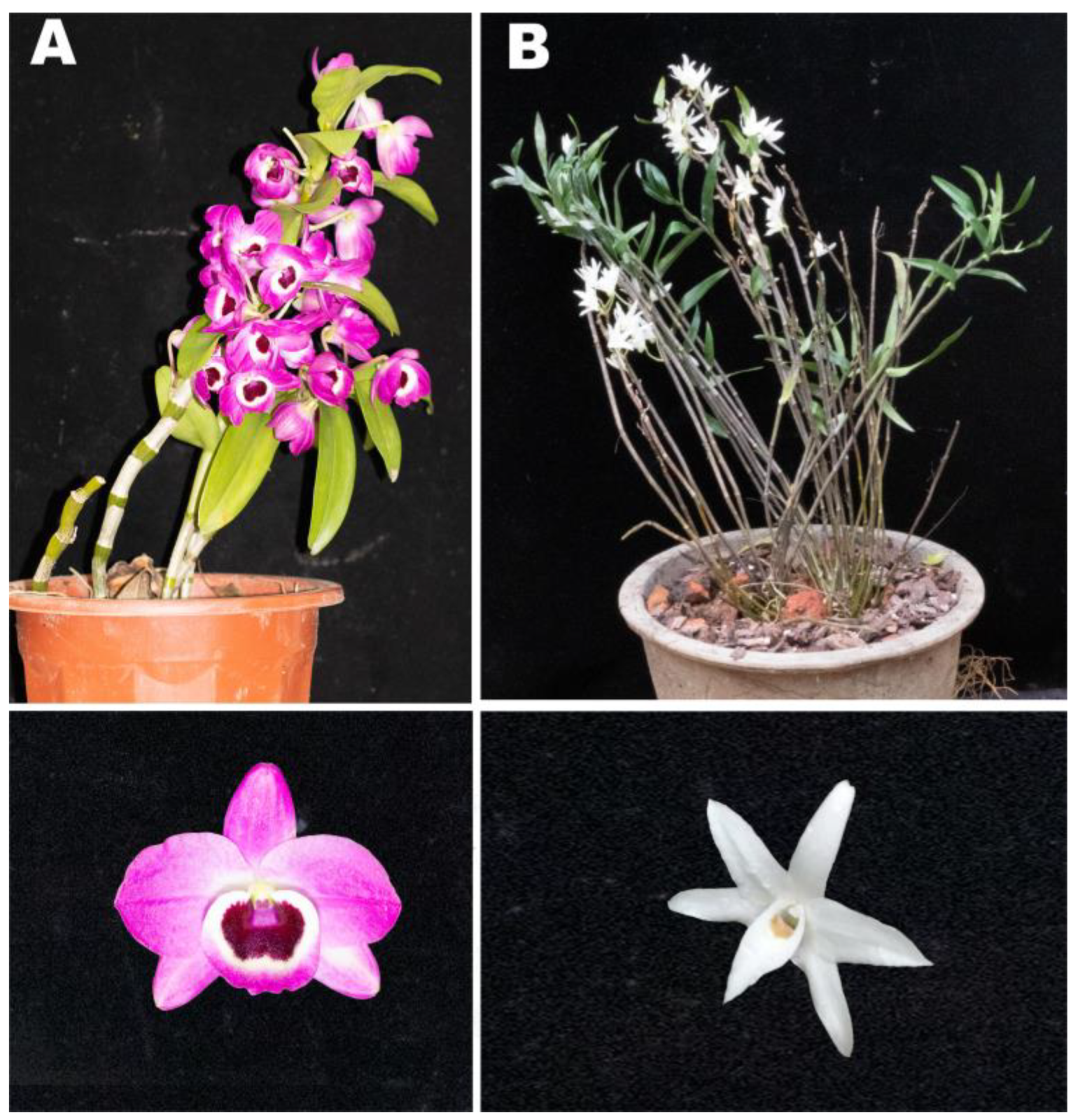
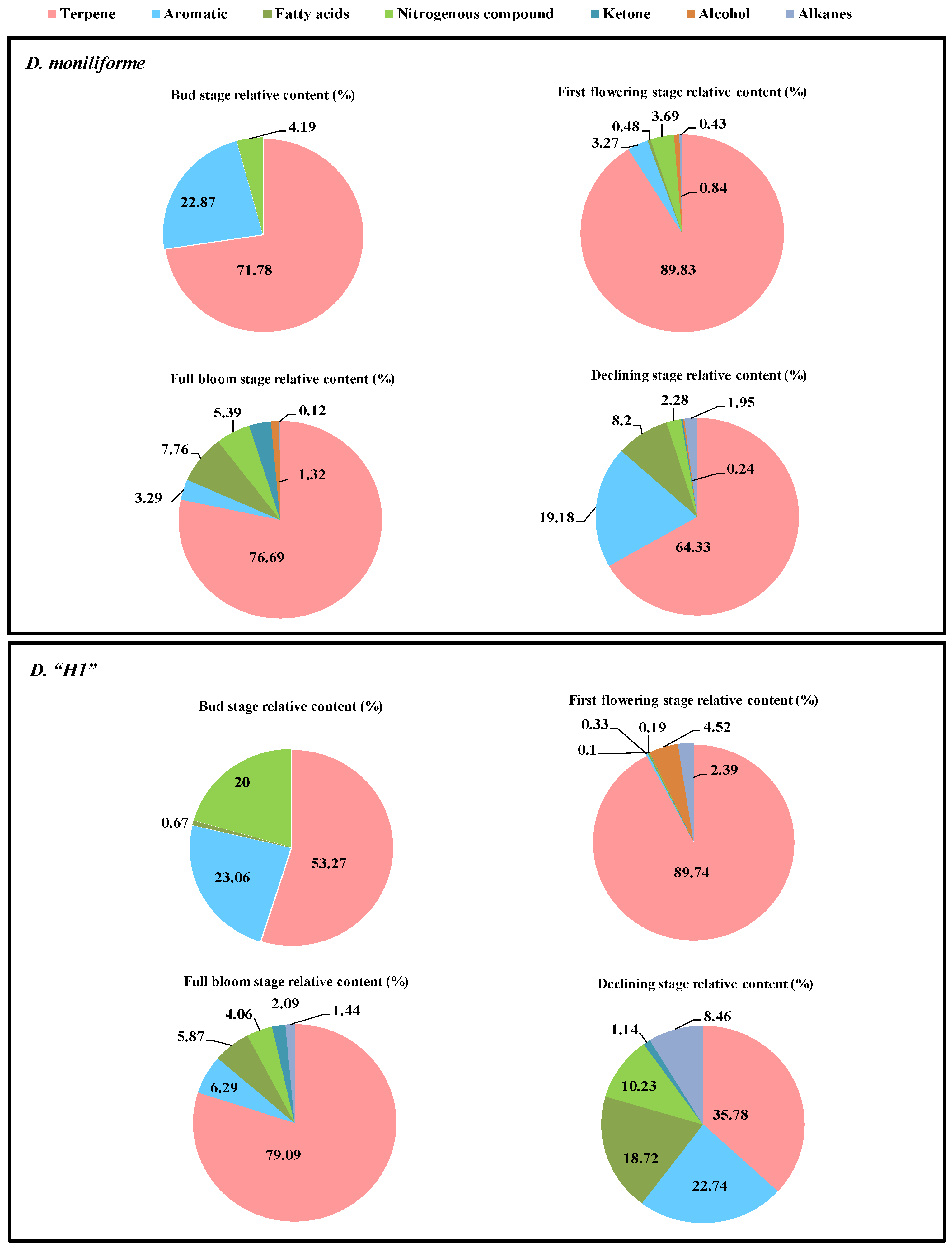
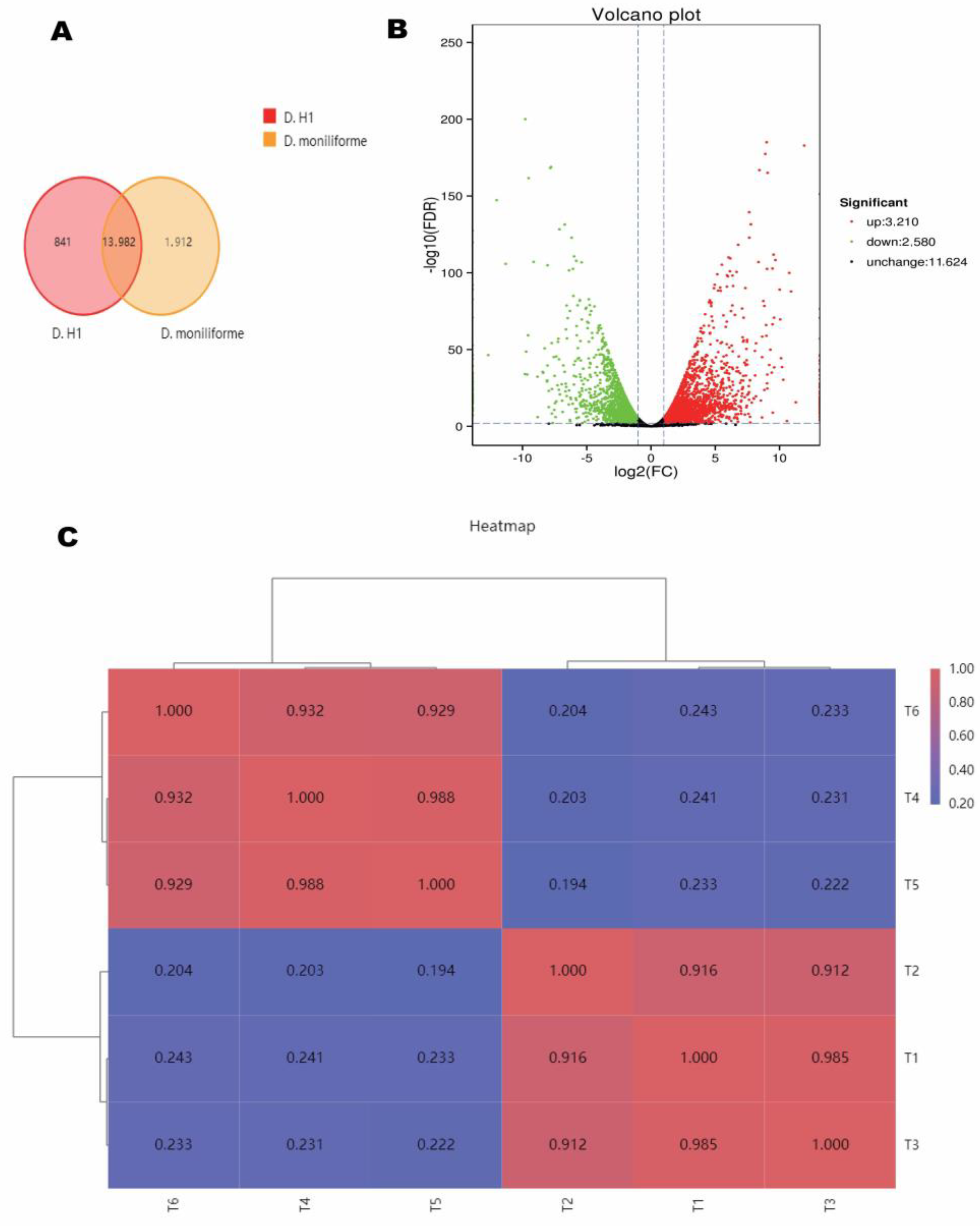
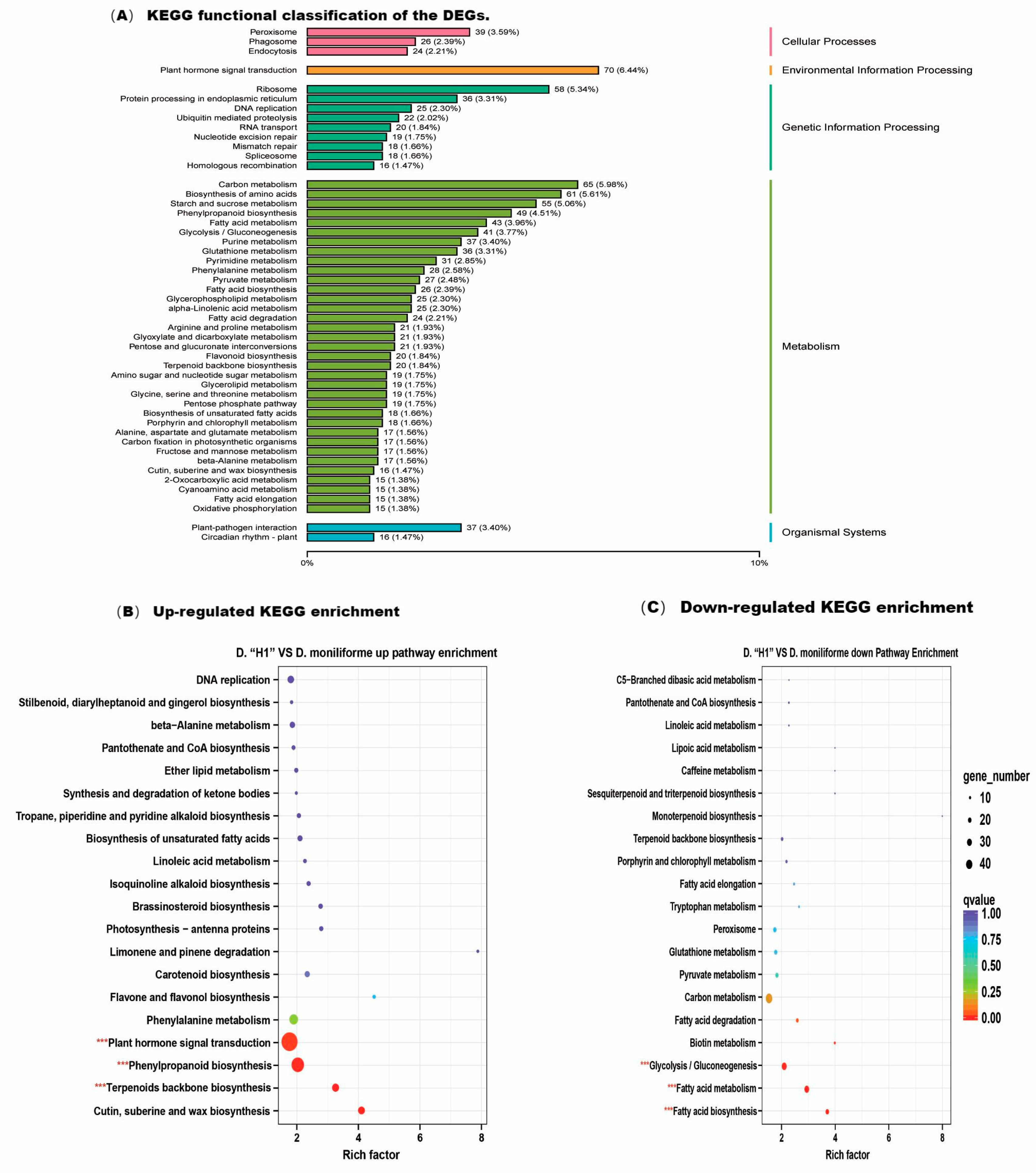
| Cultivar and Flower Part | Compound Name | Variety | Relative Content (%) |
|---|---|---|---|
| D. moniliforme Petals | Terpene | 15 | 77.25 |
| Aromatic | 2 | 2.12 | |
| Fatty acids | 2 | 9.99 | |
| Nitrogenous compound | 6 | 4.94 | |
| Ketone | 0 | 0 | |
| Alcohol | 1 | 1.94 | |
| Alkanes | 0 | 0 | |
| Total | 26 | 96.24 | |
| D. moniliforme Gynandrium | Terpene | 7 | 81.75 |
| Aromatic | 3 | 5.8 | |
| Fatty acids | 0 | 0 | |
| Nitrogenous compound | 4 | 3.03 | |
| Ketone | 3 | 6.19 | |
| Alcohol | 0 | 0 | |
| Alkanes | 0 | 0 | |
| Total | 17 | 96.77 | |
| D. H1 Petals | Terpene | 13 | 50.38 |
| Aromatic | 7 | 18.16 | |
| Fatty acids | 4 | 11.24 | |
| Nitrogenous compound | 7 | 13.1 | |
| Ketone | 1 | 1.5 | |
| Alcohol | 1 | 0.58 | |
| Alkanes | 3 | 2.88 | |
| Total | 36 | 97.84 | |
| D. ‘H1’ Gynandrium | Terpene | 7 | 63.31 |
| Aromatic | 4 | 18.12 | |
| Fatty acids | 1 | 3.36 | |
| Nitrogenous compound | 4 | 7.58 | |
| Ketone | 1 | 1.29 | |
| Alcohol | 1 | 1.67 | |
| Alkanes | 1 | 2.21 | |
| Total | 19 | 97.54 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, Y.; Xia, K.; Wu, Q.; Lu, X.; Lu, S.; Zhao, Z.; Qiu, S. Combined Analysis of Volatile Compounds and Extraction of Floral Fragrance Genes in Two Dendrobium Species. Horticulturae 2023, 9, 745. https://doi.org/10.3390/horticulturae9070745
Yang Y, Xia K, Wu Q, Lu X, Lu S, Zhao Z, Qiu S. Combined Analysis of Volatile Compounds and Extraction of Floral Fragrance Genes in Two Dendrobium Species. Horticulturae. 2023; 9(7):745. https://doi.org/10.3390/horticulturae9070745
Chicago/Turabian StyleYang, Yanni, Ke Xia, Qiaofen Wu, Xi Lu, Shunjiao Lu, Zhiguo Zhao, and Shuo Qiu. 2023. "Combined Analysis of Volatile Compounds and Extraction of Floral Fragrance Genes in Two Dendrobium Species" Horticulturae 9, no. 7: 745. https://doi.org/10.3390/horticulturae9070745
APA StyleYang, Y., Xia, K., Wu, Q., Lu, X., Lu, S., Zhao, Z., & Qiu, S. (2023). Combined Analysis of Volatile Compounds and Extraction of Floral Fragrance Genes in Two Dendrobium Species. Horticulturae, 9(7), 745. https://doi.org/10.3390/horticulturae9070745





