1. Introduction
Auxin was the earliest plant hormone discovered, and it has recently played a notable role in the regulation of fruit ripening. Applying exogenous auxin during the early ripening stage of strawberry, grape, and tomato fruit can inhibit the ripening process [
1,
2,
3]. However, applying auxin to treat apples, pears, and plums before fruit ripening can induce ethylene synthesis, thus promoting fruit softening and ripening [
4,
5,
6]. Peach is a typical climacteric fruit. Studies have shown that the softening after peach fruit maturation is related to ethylene synthesis, which is regulated by endogenous auxin [
7]. The abrupt change in ethylene at the mature stage of peach fruit was regulated by IAA (indole-3-acetic acid) [
8]. Other studies have shown that inhibiting IAA synthesis during the ripening of hard peaches leads to a decrease in ethylene synthesis, which causes the fruit to soften [
9,
10]. At the same time, studies have shown an ethylene-independent auxin regulation pathway in peach fruit ripening [
11].
Auxin regulates diverse physiological and developmental processes through the perception and transduction of auxin signals. The canonical auxin signaling pathway is comprised of the SCFTIR1/AFBs complex, which includes SKP1, Cullin, and the auxin signaling F-box protein. Additionally, it involves the participation of the transcriptional suppressor Aux/IAA (Auxin/Indole Acetic Acid) and the transcription factor ARF (Auxin Response Factor) [
12]. In tomatoes, the transgenic line with overexpression of the
SlTIR1 gene manifests changes in leaf morphology and fruit setting compared with the wild type [
13]. Other studies on cucumbers have found that the auxin receptor gene may play an important role in plant height, leaf morphology, and parthenocarpy [
14]. Overexpression of the plum
PslTIR1 gene in tomatoes decreased the height of transgenic plants and altered fruit development and fruit softening by controlling genes related to cell wall decomposition [
15]. TIR1-like auxin receptors are involved in regulating plum fruit development [
6].
Degradation of the Aux/IAA repressors is critical for auxin signaling. At present, 25 Aux/IAA genes have been identified in tomatoes that are involved in regulating auxin-mediated multiple signaling pathways [
16]. Among them, SlIAA3 is an important factor in the cross-regulation of physiological responses by auxin and ethylene, which regulate tomato leaf morphogenesis, floral organ development, fruit set, and fruit development [
17]. SlIAA9 resulted in abnormal leaf shape and parthenocarpy of tomato [
18]. SlIAA17 plays a role in regulating fruit quality, and it is found that the SlIAA17 silencing line has larger fruit and thicker pericarp [
19].
The regulatory mechanisms of Aux/IAAs on peach fruit development and maturation have also attracted much attention. The transient overexpression of the
PpIAA1 (
Ppa010303m) gene in peach fruit can promote the expression of the
PpPG1 and
PpACS1 genes and result in earlier ripening and shorter postharvest storage, which indicates that PpIAA1 acts as a positive regulator to promote fruit ripening and softening. The overexpression of the peach
PpIAA19 (
Ppa011935m) gene in tomatoes resulted in an increase in plant height, the number of lateral roots, and changes in parthenogenesis and fruit morphology [
20]. The latest study showed that high levels of PpIAA13 (
Ppa010871m) resulted in high expression of PpACS1, which increased ethylene production and peach fruit softening [
21].
Although many studies have documented the influence of auxin on fruit ripening, the auxin signaling genes have not been investigated more. The whole genome analysis of the Aux/IAA and ARF gene families in peach fruit was conducted in our previous study [
22,
23,
24]. A total of 4 TIR1/AFBs were identified in peach fruit. The transcript of PpTIR1 (
ppa003344m) responds to exogenous auxin, and the expression level differs in peach fruit with different melting characteristics [
22]. When PpIAAs were identified, the expression levels of 14 genes were higher in the melting “Okubo” than in the stony hard “Jing Yu” during almost all developmental stages. This strongly suggests that these genes may be related to auxin signaling during peach fruit ripening [
23]. In this current study, we treated peach fruits with NAA (1-naphthylacetic acid). We found that low-concentration treatment delayed fruit softening and decreased the expression of cell wall-disassembling genes and ethylene biosynthetic genes. We also found that overexpression of the
PpTIR1 gene caused significant down-regulation of the expression of early auxin-responsive genes and cell wall metabolic genes related to fruit firmness. A combinatorial TIR1-Aux/IAA co-receptor system may be involved in this process. Therefore, the regulation of auxin on peach fruit softening is concentration-dependent. This study can enrich the theoretical research of drupe fruit ripening and lay a theoretical foundation for the hormone regulation measures of peach fruit softening.
2. Materials and Methods
2.1. Plant Materials and Treatments
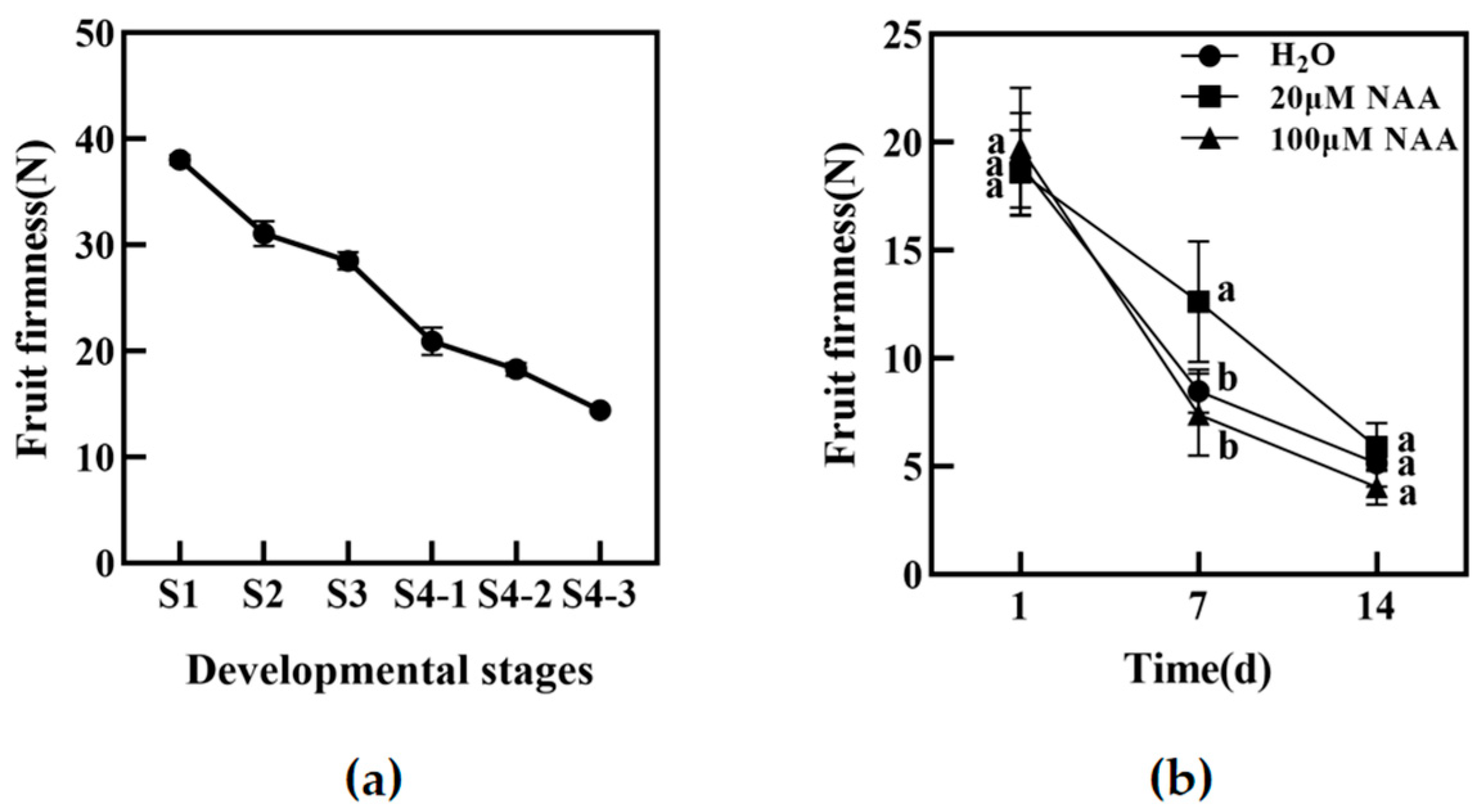
Experimental samples of the melting peach “Okubo” were picked from the experimental orchard of the Beijing University of Agriculture (Changping District, Beijing, China). The fruits are at 37, 46, 55, 63, 70, 78, 84, 92, 98, and 110 days after full bloom (DAFB). We divided the development and maturation of peach fruit into four periods: the first rapid growth period (1 to 37 DAFB, S1), the hardcore stage (37 to 63 DAFB, S2), the second rapid growth period (63 to 84 DAFB, S3), and the mature period (after 84 DAFB, S4). The mature period was further divided into S4-1 (84 to 92 DAFB), S4-2 (92 to 98 DAFB), and S4-3 (after 98 DAFB).
Peach fruits “Okubo” at the developmental stage of S4-1 were selected and treated with deionized water (H2O), 20 μM NAA, and 100 μM NAA, according to the preliminary results of our group. After washing with water and drying naturally, the peach fruit was soaked in the above three solutions, respectively. The solution was placed in a vacuum for 30 min, and the vacuum was slowly released to help the solution enter the peach fruit. After natural drying at room temperature, the treated peach fruits were stored under natural light for 14 days at 20 ± 2 °C. Samples were taken on the 1st, 7th, and 14th days after treatment, and the firmness of the fruits was measured.
2.2. Quantitative RT-PCR
Total RNA was extracted from peach fruit using the EASYspin reagent (Biomen, Beijing, China). Quantitative RT-PCR (qRT-PCR) was performed as described by Guan et al. [
23]. The first-strand complementary DNA (cDNA) was synthesized using the TransScript First-Stand cDNA Synthesis SuperMix kit (TransGen Biotech, Beijing, China). The cDNA template for qRT-PCR analysis was diluted ten times with RNase-free water before use. TB Green Premix Ex Taq II (Tli RNaseH Plus) (Takara, Kusatsu Shiga, Japan) reagent was used to perform qRT-PCR analysis on the Applied Biosystems StepOnePlus system (Thermo Fisher Technologies, Waltham, MA, USA).
PpTEF-2 (Translation Elongation Factor 2) was used as an internal reference gene. Each line of treated peach fruit pieces was used to represent one biological replicate, and at least three technical replicates were analyzed for each biological replicate. The gene-specific primers used to detect the transcriptional level are listed in
Table S1.
2.3. Agrobacterium-Mediated Infiltration
For overexpression of
PpTIR1, the CDS fragment was ligated into the pCAMBIA3301-121 vector by the Seamless Cloning Kit (catalog no. D7010M; Beyotime, Shanghai, China) to generate overexpression constructs. The primers used are listed in
Table S1. The resulting constructs were transferred into competent cells of
Agrobacterium tumefaciens (strain GV3101). A transient expression followed previously published methods [
23]. The fruit of “Okubo” in the developmental stage of S3 was used for infection. Six fruit pieces with a volume of about 1 cm
3 were taken on both sides of the ventral suture of peach fruit and cultured on MS medium for 24 h. Then, the pieces were soaked in the treatment solution and vacuum treated (−70 Kpa). The vacuum is slowly released to help the solution enter the pulp cells. After vacuum infiltration, the fruit pieces were washed three times with sterile water and cultured on MS medium in the growth chamber (20 °C, R.H. 85%) for 2 days, then quickly frozen in liquid nitrogen and stored at −80 °C for later use. Every single infection of peach pieces was used as one biological replicate, and three biological replicates were analyzed. The empty vector solution was used as a negative control.
2.4. GUS Histochemical Staining
The transiently overexpressed peach fruit pieces were cut into thin slices (1–3 mm). GUS staining solution was added to cover the material completely and placed at room temperature overnight. After that, the material was transferred to anhydrous ethanol for decolorization 2–3 times. The positive blue spots stained by the GUS solution were stable and did not fade with alcohol. The negative control was untreated peach fruit pieces, and the positive control was pieced transiently expressing pCAMBIA3301-121.
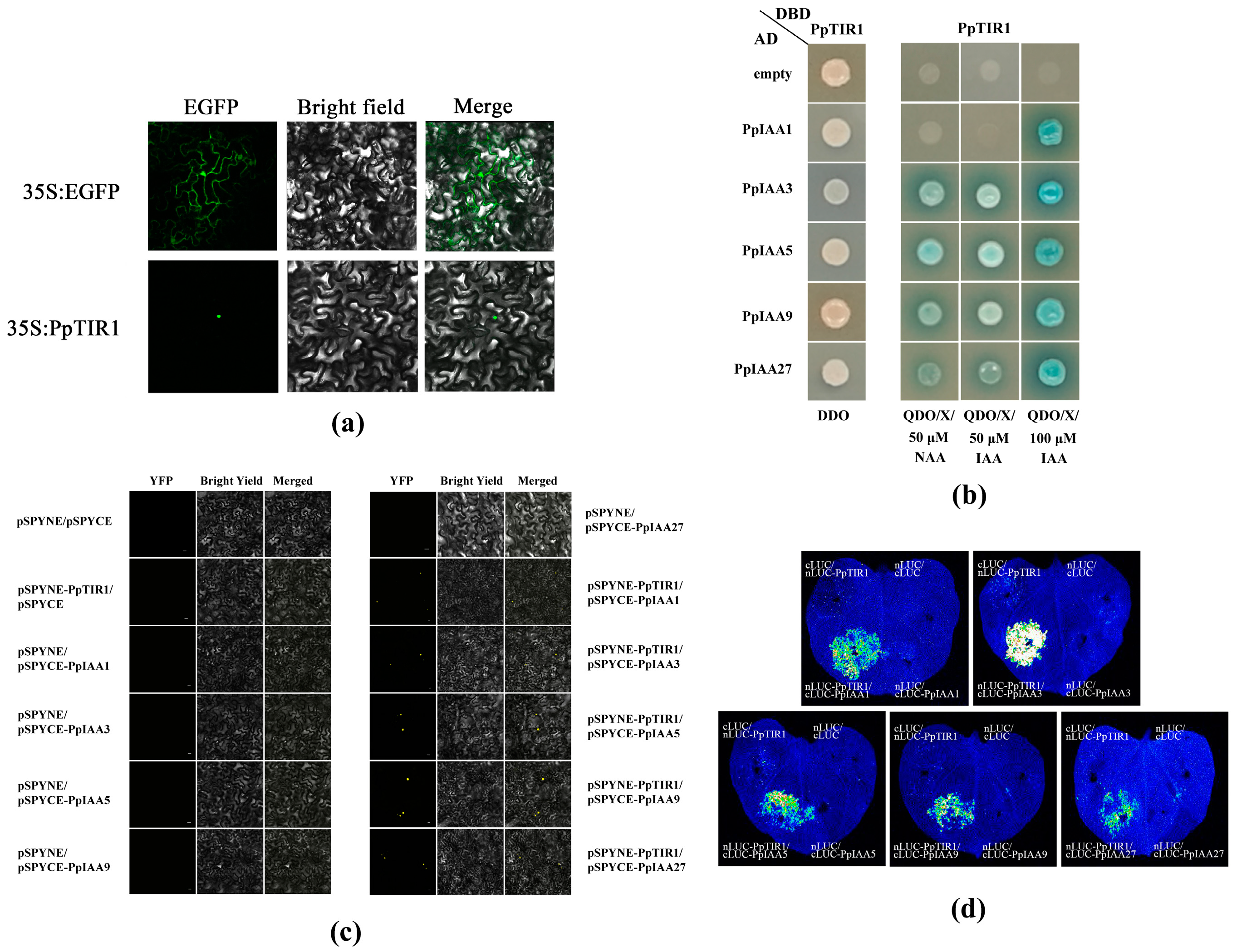
2.5. Subcellular Localization Analysis
The construction of the subcellular localization analysis was based on the cDNA of peach mesocarp. The CDS fragment of PpTIR1 was amplified by primers (
Table S1). PpTIR1 without a stop codon and the full-length coding sequences of the genes were amplified by PCR and constructed into pBI121-GFP vectors by viscous terminal ligation. The successfully sequenced PpTIR1-GFP plasmids were transformed into the competent state of GV3101
Agrobacterium tumefaciens, and the bacteria identified were selected for an expanded culture so that the final value of OD600 was 0.4. Tobacco leaves were injected with solution after being kept in darkness for 2–3 h at room temperature and cultured for 2 days. The marked areas of tobacco leaves were cut, and the GFP fluorescence signals were detected and photos taken by laser scanning confocal microscopy (Leica SP5, Leica, Wetzlar, Germany).
2.6. Yeast Two-Hybrid
The construction of a yeast two-hybrid vector was based on the cDNA of peach mesocarp. The CDS fragments of PpTIR1, PpIAA1, PpIAA3, PpIAA5, PpIAA9, and PpIAA27 were amplified by primers (
Table S1). PpTIR1 without a stop codon and the full-length coding sequences of the genes were amplified by PCR and constructed into pGADT7 and pGBKT7 vectors by viscous terminal ligation. The PpTIR1-DBD and PpIAA1/3/5/9/27-AD plasmids were co-transformed into Saccharomyces cerevisiae strain AH109, and the yeast cells that contained these two vectors were screened on SD/-Trp-Leu media. When the transformed cells were inoculated on the strict four-deficiency plate SD/-Trp-Leu-His-Ade/X-α-Gal/auxin, different concentrations of NAA, 2,4-D, and IAA were added to check the effect according to previous research [
6]. The colonies grew and turned blue, indicating that the plasmid was successfully constructed and that proteins interacted with each other. In addition, pGADT7 and pGBKT7 were used as negative controls.
2.7. Bimolecular Fluorescence Complementarities
The bimolecular fluorescence complementary vector was constructed using primers (
Table S1) to amplify the CDS fragments, PCR to amplify the full-length coding sequence of the non-stop codon gene, and construction in pSPYNE173 and pSPYCE (M) vectors by viscous terminal ligation. The successfully sequenced PpTIR1-YNE and PpIAA1/3/5/9/27-YCE plasmids were transformed into competent
Agrobacterium tumefaciens cells, respectively, to produce fusion proteins. Two types of bacteria containing different plasmids were mixed in an equal volume, and only 100 μM IAA was added simultaneously, according to the above results of the yeast two-hybrid. The bacterial solution was injected into tobacco leaves and cultured at room temperature for 2 days. A confocal microscope (Leica SP5, Leica, Wetzlar, Germany) was used to observe and take images.
2.8. Firefly Luciferase Fragment Complementary Image Technique (LCI)
The firefly luciferase fragment complementary image technology vector was constructed using primers (
Table S1) to amplify the CDS fragments and PCR to amplify the full-length coding sequence of the non-stop codon gene. The vector was constructed in pCAMBIA1300-nLUC and pCAMBIA1300-cLUC vectors using a Seamless Cloning Kit (catalog no. D7010M; Beyotime, Shanghai, China). The PpTIR1-nLUC and PpIAA1/3/5/9/27-cLUC constructs successfully sequenced were transformed into
Agrobacterium tumefaciens (strain GV3101), respectively. The suspension was prepared by mixing two types of construct solution with an equal volume, and only 100 μM IAA was added simultaneously, according to the above results of yeast two-hybrid. The suspension was injected into the back of tobacco leaves with a 1 mL syringe (without the needle). After culturing at room temperature for 2 days, the presence of fluorescence in the area where tobacco leaves were injected was determined by imaging in vivo (Tanon-5200muli, Tanon Science & Technology, Inc., Shanghai, China).
2.9. Statistical Analyses
All experiments were performed at least three times. All qRT-PCR reactions and other quantitative analyses were repeated at least three times. The Student’s t-test was used to evaluate the significant differences.
4. Discussion
Peach is a kind of respiratory climacteric fruit, and its development and ripening processes experience a series of complex physiological and biochemical changes related to size, color, texture, flavor, and fragrance smell. Softening is the most significant textural change during the ripening and postharvest storage of peach fruit, which will affect the taste and shelf life of the fruit and the economic benefits of the peach industry. Therefore, a study on the mechanism of peach fruit softening has theoretical and practical significance.
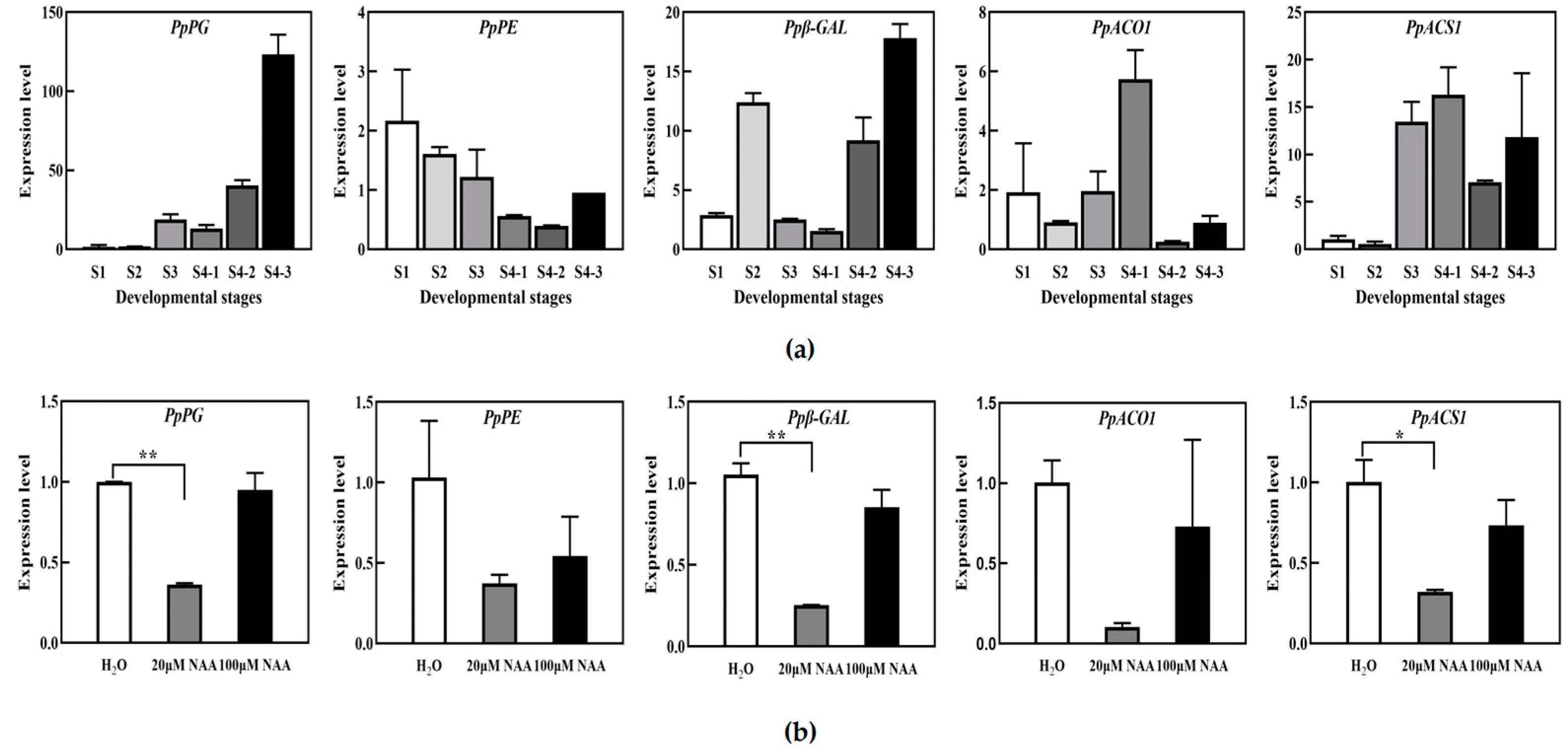
It has been reported that there is a relationship between auxin and peach fruit development and softening during ripening. Previous research results from our group showed that the content of IAA in the hard fruit “Jingyu” was very low and did not increase during the late ripening stage. In contrast, the content of IAA in the rapidly dissolving fruit “Okubo” was significantly higher, which preliminarily revealed that the non-softening of hard fruit was related to low levels of IAA [
23]. Therefore, our research focused on regulating peach fruit softening by auxin. To explore the relationship between auxin and softening, “Okubo” peach fruits were treated in vitro with exogenous NAA at different levels. The results showed that the firmness of the peach decreased after treatment with 20 μM NAA. However, 100 μM NAA had no obvious effect compared with H
2O treatment. These results indicated that low NAA concentrations could delay peach fruit’s softening. Still, a report indicated a higher accumulation of auxin triggered the fast softening of peach fruit [
25]. For other flesh fruits, previous studies have reported that exogenous auxin treatment can promote fruit ripening in pears [
5]. NAA treatment accelerated the onset of ripening at a time when apple fruit could not ripen naturally [
4,
26]. However, exogenous auxin treatment can inhibit fruit ripening and softening in strawberries [
1] and grapes [
27,
28]. Therefore, the regulatory effect of exogenous auxin on fruit softening may be related to the type of fruit and the concentration of exogenous auxin.
In the auxin signal transduction pathway, after the TIR1/AFBs protein binds to auxin, the Aux/IAAs protein can be degraded through the ubiquitin degradation pathway. This degradation process alleviates the inhibition of transcription factor ARFs, allowing them to regulate the expression of a series of downstream auxin response genes. Therefore, it is of substantial significance to study the interaction between TIR1/AFBs and Aux/IAAs proteins to reveal the physiological function of auxin. Different TIR1/AFBs-Aux/IAAs co-receptors have different results in response to auxin. In
Arabidopsis thaliana, rice, tomato, and plum, the interaction between TIR1/AFBs and Aux/IAAs proteins was found to depend on auxin. Still, there was also an auxin-independent interaction between
Arabidopsis thaliana and rice [
29,
30]. In
Arabidopsis thaliana, the interaction between AtTIR1, AtAFB1, AtAFB2, and AtAFB3 and the AtIAA3/5/7/8/12/28/29/31 protein depends on different concentrations of IAA [
31]. In plums, PslTIR1, PslAFB2, and PslAFB5 can interact with the AtIAA7 protein in the presence of 100 μM IAA [
6]. In this study, there was no interaction between PpTIR1 and the PpIAA1/3/5/9/27 protein in the absence of IAA and 50 μM 2,4-D, according to the yeast two-hybrid experiment. There was an interaction between PpTIR1 and the PpIAA3/5/9/27 protein, but the strength of the interaction differed depending on the concentration of NAA and IAA. In physiological experiments, 20 μM NAA had an obvious effect on the softening of peach fruits. In yeast two-hybridization, there was no effect of 20 μM NAA on the interaction between TIR1 and Aux/IAA. Compared with 50 μM IAA, 100 μM IAA can lead to an interaction between PpTIR1 and the Pp/IAA1 protein. Bimolecular fluorescence complementary and firefly luciferase fragment complementary image techniques were also used to prove the interaction between PpTIR1 and the PpIAA1/3/5/9/27 proteins under the condition of 100 μM IAA. These results showed that the response of different Aux/IAA factors to NAA or IAA varies in concentration and auxin type. Perhaps this difference enriches the multifunctional nature of Aux/IAA proteins. The interaction between PpTIR1 and PpIAA proteins may be an important regulatory process involved in activating downstream gene expression, thus realizing the biological function of auxin regulation.
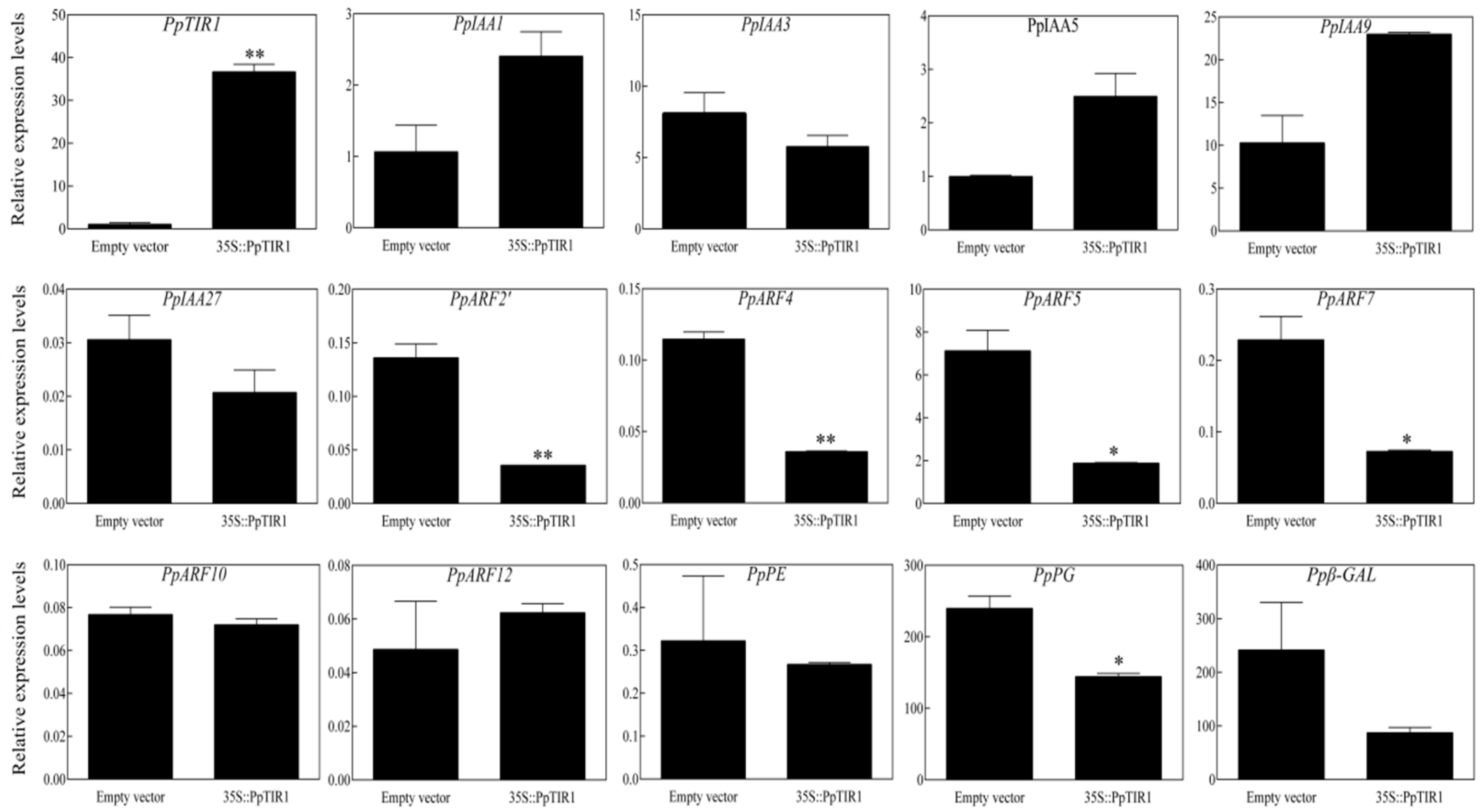
The involvement of TIR1/AFBs in fruit ripening and softening has been reported in some studies. Currently, research on the TIR1/AFBs gene in fruit development, ripening, and softening is primarily focused on tomato fruit. In tomatoes, the overexpression of SlTIR1A affected flower morphology and fruit development, resulting in parthenocarpy formation. This led to the conclusion that SlTIR1A could interact with the SlIAA9 protein and regulate the expression of SlIAA9 and SlARF7 genes at the transcriptional level, thus affecting fruit setting. The overexpression of SlTIR1B would affect apical dominance, leaf morphology, and fruit formation. Other studies on SlTIR1 also showed that the overexpression of the SlTIR1 gene caused dwarfing, leaf morphological changes, and parthenocarpy in tomato plants. Simultaneously, the overexpression of the SlTIR1 gene led to a decrease in the expression of some early auxin response genes, such as SlIAA9, SlARF6, and SlARF7, while the level of expression of SlIAA3 increased [
17]. A study on plums showed that the overexpression of the PslTIR1 gene led to early fruit setting before flowering, resulting in parthenocarpy and a decrease in the transcription of the IAA9 and ARF7 genes. It is hypothesized that PslTIR1 positively regulates auxin response and fruit set by mediating the degradation of Aux/IAA proteins, especially IAA9. In peaches, lower TIR1 protein levels trigger the stabilization of PpIAA13, leading to the accumulation of PpIAA13 protein, thus activating the expression of PpACS1 and promoting peach fruit softening [
21]. In this study, we analyzed the expression of some auxin response genes and cell wall metabolic genes related to fruit firmness by transiently overexpressing the PpTIR1 gene in peach fruit. After overexpressing
PpTIR1 in peach fruit, the expression of the PpIAA1/5/9 gene increased (
Figure 3). However, it did not reach a significant level, preliminarily indicating that these factors respond quickly to auxin and enhance auxin signaling. The results indicated that the overexpression of the PpTIR1 gene in peach fruit resulted in a decrease in the expression of
PpARF2′,
PpARF4,
PpARF5, and
PpARF7 genes, suggesting that the PpTIR1 gene may affect fruit development, ripening, and softening by regulating the downstream
PpARFs genes. These
PpARFs genes may play a negative feedback regulation of auxin signaling. Simultaneously, the overexpression of the PpTIR1 gene caused a significant decrease in the expression of PpPG, a gene related to fruit softening, indicating a close relationship between the PpTIR1 gene and peach fruit softening. In a study on plums, it was found that the firmness of plum fruit obtained by the overexpression of the
PslTIR1 gene was lower than that of wild-type fruit, and the expression level of soften-related genes in transgenic fruit was higher than that in wild-type fruit. These results show that PslTIR1 regulates fruit softening by controlling the level of enzymes related to cell wall decomposition [
6]. However, whether PpTIR1 can promote or inhibit peach fruit softening needs further investigation.