Abstract
The Cucurbitaceae family comprises economically valuable vegetables such as cucumber, melon, and pumpkin. GRAS proteins, which are crucial transcription factors, play diverse roles in plant growth and development. However, comparative investigations of GRAS proteins across Cucurbitaceae species are limited. Here, we identified 241 GRAS family genes in six cucurbit crops. The number of GRAS genes in cucumber, melon, wax gourd, watermelon, and bottle gourd ranged from 36 to 37, while the pumpkin genome contained 57 GRAS genes, possibly due to a recent whole-genome duplication. We classified cucurbit GRAS genes into 16 subfamilies and identified species-specific motifs and specific-expression patterns in the SCLB and RAD1 subfamilies. Notably, we identified 38 tissue-specific expressed genes, particularly fruit-specific genes potentially involved in fruit development. Additionally, we predicted the role of GRAS genes in regulating hypocotyl elongation under weak or dark light conditions in cucurbit plants. These findings enhance our understanding of the characteristics, evolution, and potential functions of GRAS genes in six cucurbit crops, providing valuable resources for genetic research in the Cucurbitaceae family as well as important agronomic traits.
1. Introduction
Transcription factors play important roles in plant development and resistance to abiotic/biotic stresses. The GRAS gene family, named after its first three founding members: GAI (Gibberellic acid insensitive) [1], RGA (Repressor of GAI) [2], and SCR (Scarecrow) [3], is a plant-specific transcription factor family. GRAS consists of five conserved and ordered domains in C-termini, including two LHRs (leucine heptad repeats), LHR I and LHR II, possibly involved in protein–protein interactions; VHIID; PFYRE; and SAW, and an N-terminus region highly variable in length and sequence [4,5,6].
GRAS genes have been reported in many species, including 32–34 in Arabidopsis (Arabidopsis thaliana L.) [7,8], 60 in rice (Oryza sativa L.) [8,9], and 53–54 in tomato (Solanum lycopersicum L.) [10,11]. The GRAS gene family can be divided into 8–17 subfamilies [7,8,9,11,12], including DELLA, HAM (Hairy meristem), PAT1 (Phytochrome-A signal transduction 1), DLT (Dwarf and low-tillering), SCR (Scarecrow), and SHR (Short root), etc. GRASs have been reported to regulate multiple biological processes and molecular functions, including hormone-related [1,2,13,14], abiotic stress responses [15,16,17,18], cell cycle [19,20], grain size [21,22], fruit development and ripening [23,24,25], and hypocotyl elongation [26]. For instance, previous studies have reported the involvement of SlGRAS2 (Solyc07g063940) [23], SlGRAS4 (Solyc01g100200) [25], and SlFSR (Solyc07g052960) [24] in fruit development or ripening in tomato. ZmGRAS11 has been shown to promote the enlargement of endosperm cells, and ultimately increase grain size and weight in maize [22]. SCARECROW-LIKE28 (SCL28) has been identified as a role to inhibit the G2-to-M phase cell cycle transition in Arabidopsis [19,20].
Light is a crucial factor in the growth and development of plants. Seedlings grown under dark conditions exhibit longer hypocotyls than those grown under light conditions [26]. Recent studies have reported that cucumber buffers against the high-light stress-induced accumulation of CsGA2ox8 transcripts through alternative splicing, which finely tunes gibberellin levels and maintains hypocotyl elongation [27]. DELLAs are nuclear transcriptional regulators that belong to the GRAS family of plant-specific nuclear proteins. They possess a distinctive N-terminal GA perception region that binds the GA receptor GID 1 (Gibberellin-insensitive dwarf 1) and a C-terminal GRAS domain that interacts with several regulatory proteins, leading to GA repression activity [6,28]. Members of the DELLA protein family play essential roles in regulating various aspects of plant growth and development, including hypocotyl elongation [6,26,28].
The Cucurbitaceae (cucurbit) family encompasses several significant crops, including cucumber (Cucumis sativus L.), melon (Cucumis melo L.), watermelon (Citrullus lanatus L.), wax gourd (Benincasa hispida L.), bottle gourd (Lagenaria siceraria L.), pumpkin (Cucurbita moschata L.), etc. These crops display distinct characteristics, such as varied floral sex types and melon fruit [29]. The GRAS gene family has been previously identified in several cucurbit crops, such as cucumber (37 GRASs) [16], melon (37 GRASs) [15], watermelon (37 GRASs) [30], and bottle gourd (37 GRASs) [31]. However, these studies mainly focused on a single species, and comparative studies among multiple cucurbit crops remain unexplored. In this study, we conducted comprehensive analyses of GRAS genes in six cucurbit crops: cucumber, melon, watermelon, wax gourd, bottle gourd, and pumpkin. We investigated their characteristics, cis-acting elements, evolutionary history, and expression patterns within these crops. Moreover, we predicted the potential roles of GRAS genes in early fruit development and hypocotyl growth response to light conditions. Our findings contribute to understanding the evolutionary and functional characteristics of GRAS genes in cucurbit crops, emphasizing the significance of comparative studies for comprehending the roles of gene families in plant evolution and development.
2. Materials and Methods
2.1. Identification and Phylogenetic Analysis of GRAS Genes
The genome sequences, protein sequences, and annotation information for cucumber [32], melon [33], wax gourd [29], watermelon [34], bottle gourd [35], and pumpkin [36] were acquired from the CuGenDB database (http://cucurbitgenomics.org accessed on 31 August 2018). The GRAS hidden Markov model (HMM) (PF03514) was retrieved from the Pfam database (http://pfam.xfam.org/ accessed on 31 August 2018) [37] and used to conduct HMM searches against annotated protein databases from different genomes with an E-value cutoff of 0.01, using HMMER (version: 3.1) (http://hmmer.org/ accessed on 2 September 2018). To ensure the accuracy of GRAS gene identification, a high-quality protein set (E-value < 1 × 10−20) was aligned and utilized to construct a species-specific GRAS HMM using hmmbuild from the HMMER v3 suite. The resulting species-specific HMM was employed to select all the proteins with an E-value lower than 0.01 [38]. Moreover, the presence of GRAS domains in amino-acid sequences was verified using the NCBI Conserved Domain Search (https://www.ncbi.nlm.nih.gov/Structure/cdd/ accessed on 5 September 2018). In total, 241 GRAS genes were identified across the six cucurbit crops. To further verify the accuracy of the method, GRAS genes were also identified from the Arabidopsis and tomato genomes, and the results were compared with the number of GRAS genes previously reported [7,8,10,11]. Additionally, the ExPASy proteomics server (http://web.expasy.org/protparam/ accessed on 7 September 2018) [39] was utilized to extract the chemical characteristics of each candidate GRAS protein, such as isoelectric point (pI) and molecular weight (MW).
Furthermore, after subjecting the initial 241 GRAS candidate genes to further filtration, a refined selection of 237 genes was generated based on the ratio of GRAS-conserved domain length to GRAS HMM length being greater than 60%. The four genes (MELO3C014029, Bhi09P000670, CmoCh06G007340, and CmoCh16G010540) that did not meet this criterion were filtered out. These 237 genes were subsequently analyzed in detail.
Phylogenetic analysis was conducted using full protein sequences of 237 GRASs from six cucurbit crops, 33 GRASs from Arabidopsis, 49 GRASs from tomato, 24 GRASs from rice, 29 GRASs from grape, and 30 GRASs from Amborella [12]. Firstly, all protein sequences were aligned using the MUSCLE software (version 3.8.1551) [40], with default parameters. Secondly, a neighbor-joining (NJ) consensus tree was constructed using the MEGA software (version 6.06) [41], with the Jones–Taylor–Thornton (JTT) amino acid substitution model, partial deletion, and 1000 bootstrap replicates’ parameters. Finally, the members were grouped into different subfamilies based on previous reports [12], and the phylogenetic trees were modified using iTOL (https://itol.embl.de accessed on 28 September 2018) [42].
2.2. Duplication and Synteny Analysis of GRAS Genes in Six Cucurbit Crops
We employed the Multiple Collinearity Scan (MCScanX) toolkit [43] to identify potential paralogous genes (E-value < 1 × 10−5, top five matches) within each of the six cucurbit crops. The identified paralogous genes were then subjected to the duplicate_gene_classifier program to classify them into whole-genome duplication (WGD) and tandem duplication (TD) [44]. We mapped a total of 237 GRAS genes (excluding BhiUN50P4, BhiUN508P6, and CmoCh00G001590, whose chromosomal locations remain unknown) to the chromosomes of the six cucurbit crops using their genome annotation files. We plotted diagrams of the chromosomal locations using Mapchart software [45]. Additionally, we calculated the non-synonymous (Ka) and synonymous substitution (Ks) between duplicated GRAS gene pairs using the add_ka_and_ks_to_collinearity program of the MCScanX toolkit.
We visualized the schematic representations of syntenic relationships of pumpkin duplicated GRAS genes using Circos software (version: 0.69-8) [46] based on the syntenic blocks. Similarly, we identified orthologous genes among the six cucurbit plants using MCScanX software. We then identified each interspecies GRAS orthologous gene among the six cucurbit crops based on the syntenic blocks. Finally, we merged the orthologous gene pairs’ information using a Python script. We constructed the plots of chromosome-scale synteny blocks for the six cucurbit genomes using the Python version of MCScan (https://github.com/tanghaibao/jcvi/wiki/MCscan-(Python-version) accessed on 25 January 2019).
2.3. Characteristics of the GRAS Genes in Six Cucurbit Crops
To further understand the characteristics of each subfamily, a tree-motif structural map was constructed for all GRASs in the six cucurbit crops. Firstly, an NJ tree was performed for all GRAS protein sequences. Secondly, the motifs for all GRAS protein sequences were annotated using the Multiple Expectation for Motif Elicitation (MEME) tool (version: 4.9.1) (http://meme-suite.org/ accessed on 7 December 2018) [47] with the parameters “minsites 6, maxsites 149, minw 6, maxw 200, and nmotifs 20”. Thirdly, the CDS/UTR organization of the predicted GRASs was investigated based on the GFF/GTF annotation files of the six cucurbit genomes. Finally, the NJ tree, motif, and coding-sequence-untranslated region (CDS-UTR) structural patterns were visualized using TBtools software [48].
2.4. The Prediction of Cis-Acting Elements of GRAS Gene Promoter Regions
The 2000 bp upstream sequences from the promoters of GRAS genes in the six cucurbit crops were extracted from the assembly file using a Python script. Subsequently, PlantCARE (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/ accessed on 7 April 2023) [49,50] was employed to analyze the cis-acting elements present in the putative promoter regions.
2.5. RNA-seq Analysis of GRAS Genes in Various Tissues
The expression patterns of GRAS genes in various tissues, including roots, stems, leaves, flowers, and fruits, were analyzed utilizing publicly available RNA-seq data [36,51,52,53,54,55,56,57,58]. The RNA-seq data were processed using Hisat2 (version: 2.1.0) [59] to align to the six cucurbit plant genomes and assembled using StringTie (version: 1.3.4) [60]. Uniquely mapped reads were utilized as inputs for the StringTie software, and gene expression levels were measured using fragments per kilobase million (FPKM) values. The tissue specificity of GRAS gene expression patterns was evaluated by calculating the tissue specificity index, τ. Genes with 0.85 ≤ τ ≤ 1 were considered tissue-specific genes, while those with 0 ≤ τ ≤ 0.15 were classified as housekeeping genes. The pheatmap package in R software was used to plot heatmaps for GRAS genes, with FPKM values transformed to log2. The expression levels of SCL28 in fruits at different stages were extrated from CuGenDBv2 database (http://cucurbitgenomics.org/v2/ accessed on 10 April 2023).
2.6. RNA-seq and RT-qPCR of GRAS Genes in Hypocotyl Tissues under Dark and Normal Light Conditions
To investigate the differential expression of GRAS genes in cucumber, we utilized RNA-seq data, obtained from cucumber gene expression under weak light (40 μmol·m−2·s−1 PPFD) and strong light (180 μmol·m−2·s−1 PPFD) conditions [27]. FPKM values were calculated, and a heatmap was generated for the GRAS genes in cucumber using the method described in the previous section.
To validate the expression patterns of the candidate genes in the six cucurbit crops under different light conditions, we sterilized and soaked the six cucurbit crops’ seeds in water for 3 h before transferring them to Petri dishes lined with wet filter papers, and then they were germinated at 28 °C. After germination, we selected single seedlings with cotyledons emerging from the soil for normal light (without tin foil covering) and dark light (with tin foil covering) treatments that lasted for 56 h. We harvested the hypocotyls for RNA extraction using the SV Total RNA Isolation System (Promega) and synthesized cDNA using the GoScriptTM Reverse Transcriptase Kit (Applied Biosystems). We performed quantitative real-time RT-PCR (RT-qPCR) using the SYBR® Premix Ex TaqTM Kit (TaKaRa, China) and an Applied Biosystems 7500 real-time PCR system. For the RT-qPCR reaction, we mixed 20 ng of reverse-transcribed RNA, 0.3 µM of each primer, and 10 µL of SYBR Buffer (2×) to develop a 20 µL reaction mixture. We performed the PCR reaction with one cycle at 95 °C for 80 s, followed by 40 cycles at 95 °C for 15 s and 60 °C for 1 min. To check the specificity of PCR amplification, we used a melting curve after the final PCR cycle, heating it from 65 °C to 95 °C at a rate of 0.5 °C for 0.05 s. Each treatment was replicated three times, and the 2−ΔΔCt method [61] was used to calculate the relative transcript levels of genes. Student’s t-test was used to determine significant changes (p < 0.05). The primers used are listed in Table S8.
3. Results
3.1. Identification and Phylogenetic Analysis of GRAS Genes in Six Cucurbit Crops
A total of 241 GRAS genes were identified in the genomes of the six cucurbit crops (Table S1). The number of GRAS genes was relatively consistent among these species, with 36–37 in cucumber, melon, wax gourd, watermelon, and bottle gourd, and a larger number of 57 in pumpkin, possibly due to a lineage-specific recent WGD event [36]. The identified GRAS genes in cucumber, melon, watermelon, and bottle gourd were consistent with previous reports [15,16,30,31], indicating the reliability of our methods.
To investigate the phylogenetic relationships among the GRAS genes of the six cucurbit crops, a phylogenetic tree was constructed using the complete set of 402 GRAS proteins. Specifically, 237 GRAS proteins were obtained from the six cucurbit crops, while the remaining 165 GRAS proteins were selected from five other representative angiosperms, including Arabidopsis (33), tomato (49), rice (24), grape (29), and Amborella (30), which were used as an outgroup. These GRAS members were classified into 17 subfamilies and 30 orthologous groups (OGs) (Figure 1a), according to a previous classification. Among the 17 subfamilies, the PAT1 subfamily contained the largest number of cucurbit GRAS genes, followed by DELLA (27), HAM (26), LISCL (21) subfamilies (Figure 1a,b), etc. Certain orthologous genes, namely those belonging to the SCLA (Scarecrow–Like A) subfamily, RAD1-2 (Required for Arbuscule Development 1–2) subfamily, OG-NSP2-Amb, and OG-NSP2-3 OG (Figure 1b), were not identified in any of the six cucurbit crops. This suggests that these genes may not be essential and have been lost during evolution, or that they were not detected in the current version of the genome due to genome sequencing or genome structural annotation errors.
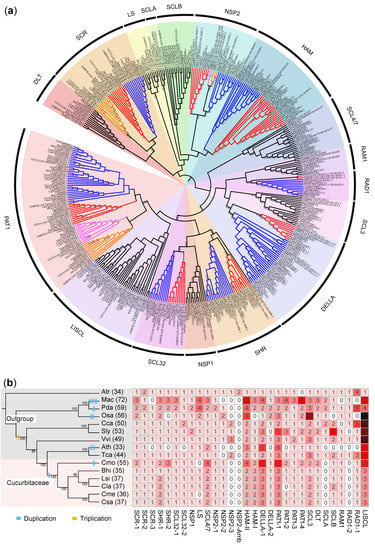
Figure 1.
Phylogenetic relationships and copy number variation of GRAS genes in Cucurbitaceae and outgroup species: (a) An unrooted neighbor-joining phylogenetic tree was constructed using 402 GRAS protein sequences from Amborella, grape, rice, Arabidopsis, tomato, cucumber, melon, wax gourd, watermelon, bottle gourd, and pumpkin. All GRAS genes were divided into 17 subfamilies and labeled outside the outermost circle of the phylogenetic tree. The 17 subfamilies of GRAS genes were further partitioned into 30 OGs, with several OGs existing within each subfamily. The branches of the evolutionary tree are color-coded as blue (representing “OG-1”), red (representing “OG-2”), orange (representing “OG-3”), and magenta (representing “OG-4” or “OG-amb”). However, in cases where only one OG existed within a subfamily, the corresponding branch is labeled in black; (b) The distribution of copy number of GRAS genes in cucurbit and outgroup species. The species tree of Amborella (Atr), grape (Vvi), rice (Os), Arabidopsis (Ath), tomato (Sly) and cucumber (Csa), melon (Cme), wax gourd (Bhi), watermelon (Cla), bottle gourd (Lsi), and pumpkin (Cmo) is displayed on the left. The abundance of genes is illustrated by a gradient color from white to black in the matrix, which shows the number of genes by subfamilies or OGs specified in the header.
3.2. Evolution of GRAS Genes in Six Cucurbit Crops
To explore the evolutionary history of GRAS genes among the six cucurbit crops, we conducted a synteny analysis. A total of 39 syntenic blocks containing GRAS genes were identified (Figure 2a; Table S2). The majority of highly conserved syntenic blocks were shared by all six cucurbit genomes, with 87.34% (207/237) of GRAS genes mapping to 32 syntenic blocks. Notably, some conserved syntenic blocks exhibited double the number of GRAS genes in pumpkin compared with the five other Benincasae species, as indicated by the red lines in Figure 2a. We also analyzed the TD and WGD events of the GRAS gene family in the six cucurbit crops (Table S3; Figure S1a–f). TD events were only found in cucumber, melon, watermelon, and bottle gourd, each with one TD event. A total of 38 WGD events were identified, with the highest number observed in pumpkin, which accounted for 57.89% (22/38) of the WGD events. These results suggest that the GRAS gene family diverged with the evolutionary separation of the Cucurbita and Benincasae groups, and a WGD event was the main reason for the difference in the number of GRAS genes between the two groups [36]. Furthermore, the number of GRAS genes in subgenome A of pumpkin (32 GRAS genes) was greater than that in subgenome B (22 GRAS genes) (Figure 2b and Figure S1f), indicating a higher rate of gene loss within this family in subgenome B than in subgenome A.
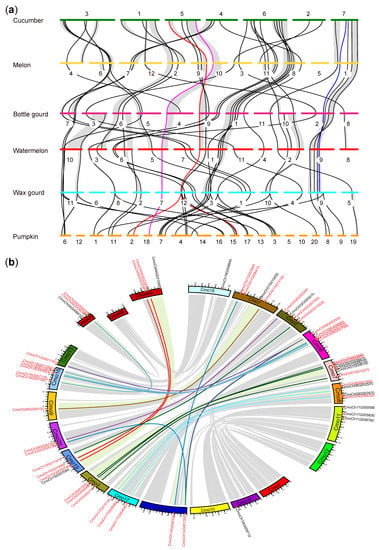
Figure 2.
Evolution of GRAS genes in six cucurbit crops: (a) Synteny analysis of GRAS genes among six cucurbit crops. The collinear GRAS genes are connected by black lines. Grey ribbons represent pairwise collinear blocks that connect the GRAS genes between the genomes of six cucurbit species. The red (2:1 ratio), magenta (1:1 ratio), and blue lines (0:1 ratio) indicate collinear GRAS genes that have maintained different ratios in pumpkin and Benincasae crops; (b) WGD events of GRAS genes in the pumpkin genome. The chromosomes of pumpkin are shown in different colors and subgenome A and subgenome B represent the outer and inner rings. The approximate positions of WGD GRAS genes are marked with the red font on the circle, and colored curves denote the WGD GRAS genes in pumpkin. The grey lines in the background represent the regions of the collinear blocks in pumpkin.
To investigate the selective evolutionary pressure on the divergence of WGD and TD GRAS gene pairs, we calculated the Ka/Ks parameters for the WGD and TD GRAS gene pairs (Table S4). We observed that all WGD and TD gene pairs exhibited Ka/Ks ratios lower than 0.5, indicating that these genes underwent strong purifying selection during genome evolution.
3.3. Characteristics of GRAS Genes in Six Cucurbit Crops
To investigate the characteristics of the GRAS genes, we recorded detailed information for each gene, including its genomic position, intron number, and protein characteristics such as size, molecular weight, and isoelectric point (Table S5; Figure S2). Our results showed that the majority of GRAS genes in the six cucurbit crops were intronless, with percentages ranging from 50% to 78.95% (Figure S2). These findings are consistent with previous studies on tomato [10] and Arabidopsis [8]. We also analyzed the distribution patterns of the conserved motifs and CDS-UTR structures for all GRAS members (Figures S3 and S4). Our results showed consistent distribution patterns within most subfamilies or OGs in terms of their motif and CDS-UTR distribution, indicating the reliability of the classification of GRAS members and the potential functional similarity of genes within the same subfamilies or OGs.
Notably, we conducted a comparative analysis of predicted motifs distribution patterns in the six cucurbit crops and non-cucurbit crops, including Arabidopsis and tomato, in each subfamily (Figure S5). Interestingly, we found that certain motifs were exclusively identified in the SCLB and RAD1 subfamilies of the six cucurbit crops (Figure S5). To confirm whether these specific motifs were unique to cucurbits, we expanded the outgroup range to include rice, grape, Amborella, Theobroma cacao, Coffea canephora, Phoenix dactylifera, and Musa acuminata and obtained similar results (Figure 3). For instance, motifs 12 (SANQSGFYQQDISKIGDQTNYQQPNSDCLIFDELLFGNDF), 18 (GIHEKQETEQRGKEDDCPFRMDEFGDFNF), and 19 (TISELIQEKSKISENLAADSI) were only found in the six cucurbit crops in the SCLB clade when compared with nine other species (Figure 3a). Similarly, motifs 12 (DGDGGSFFSTDFTSVGKEDED), 15 (SRVDHSSDRKK), and 20 (GAAHWLSLL) only appeared in the six cucurbit crops in the RAD1 clade (Figure 3b). These findings suggest that certain motifs may be unique to specific subfamilies within the GRAS gene family in cucurbits.
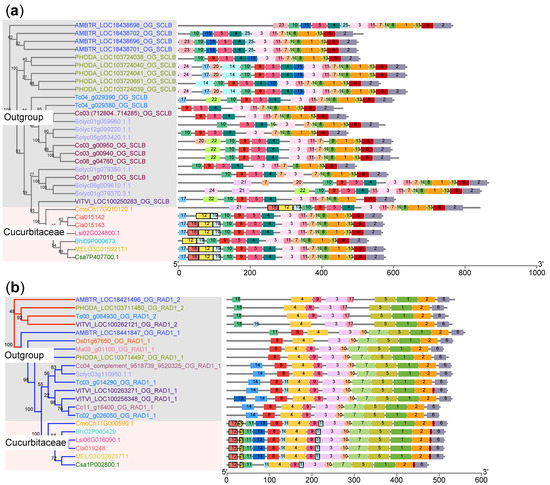
Figure 3.
Cucurbit-specific motifs of GRAS genes in the SCLB and RAD1 subfamilies. The neighbor-joining trees of the SCLB (a) and RAD1 (b) subfamilies of GRAS proteins are shown on the left. The horizontal, colored boxes indicate conserved motifs within each protein.
3.4. Cis-Acting Elements of GRAS Genes in Six Cucurbit Crops
To analyze the characteristics of GARS gene promoters, we predicted a total of 6991 cis-acting elements for all GRAS genes in the six cucurbit crops (Table 1; Figure S6). These elements were further classified into three categories based on the annotation information of their cis-acting elements and an earlier report [62]: abiotic and biotic stress (42.08%, 2942/6991), phytohormone-responsive (25.95%, 1814/6991), and plant growth and development (31.97%, 2235/6991) categories.

Table 1.
Summary of cis-acting elements of GRAS genes in six cucurbit crops.
In the abiotic and biotic stress category, we observed stress-related elements responses such as oxidation (ARE, GC motif), defense (TC rich), drought (MBS), wounding (WUN motif), heat shock (STRE), and low temperature (LTR). In the phytohormone-responsive category, we observed many hormone-related responses reflected in cis-acting elements, such as jasmonic acid (TGACG motif and CGTCA motif), abscisic acid (ABRE), salicylic acid (TCA element), gibberellin responses (GARE motif, P box, and TATC box), and auxin (AuxRR core, TGA box, and TGA element). ABRE, TGACG motif, CGTCA motif, and as-1 elements occupied the largest proportion in this group, especially in DLT, SCL4/7, DELLA, and LISCL subfamilies. In the plant growth and development category, we identified 2235 cis-acting elements, 89.84% (2008/2235) of which were involved in light responses, including Box-4 (ATTAAT), G box (TACGTG), GT1 motif, AE box, and MRE. We also observed interesting elements such as cell cycle, circadian control, and tissue-specific motifs like the MSA-like (AACGG), CAT box (GCCACT, specific for meristem), and the GCN4_motif (TGA[CG]TCA, specific for endosperm) (Table 1; Figure S6), etc. These findings suggest that the GRAS gene family plays a broad role in multiple processes.
3.5. Gene Expression Profiles Unveil Potential Roles of GRAS Genes in Fruit Development
To characterize the expression patterns of GRAS genes in the six cucurbit crops, we calculated their expression levels (FPKM values) and tissue-specific expression index (τ) in various tissues including roots, stems, leaves, flowers, and fruits. Our results indicated that 90.30% (214/237) of the analyzed genes were expressed in at least one of the analyzed tissues (FPKM >1) (Table S6). Among the expressed gene, two GRAS genes (CmoCh16G009830 and CmoCh08G008320) were identified as housekeeping genes (0.15 > τ > 0), while 35 GRAS genes exhibited tissue-specific expression (τ > 0.85), including 16 in roots, 1 in stems, 7 in leaves, 2 in flowers, and 9 in fruits (Figures S7 and S8; Table S6). For instance, we observed that one DLT member (Lsi07G014150), six PAT members (Bhi10P001034, CmoCh07G003440, CmoCh09G009100, CmoCh01G012140, MELO3C018144, and CmoCh15G011370), one SCL4/7 member (Bhi03P000998), and one SCR-1 member (MELO3C025282) were relatively highly expressed in fruits.
Notably, Lsi07G014150, Bhi11G000315, Cla004422, Csa2G043910, MELO3C014056, Cmoch16G010470, and CmoCh06G007250 were found to be conserved in the six cucurbit crops and were clustered into DLT subfamilies in the phylogenetic tree (Figure 4a) and orthologous gene analyses (Table S7). A recent study has demonstrated that SCARECROW-LIKE28 (AtSCL28, AT1G63100), a homologous gene for DLT members in Arabidopsis, is a mitotic gene that is directly regulated by MYB3Rs and plays a critical role in determining cell size by negatively regulating cell cycle progression at the G2-to-M cell cycle transition [20]. Similarly, the so-called mitosis-specific activator (MSA) element that serves as a binding motif for MYB3Rs [63,64] was repeatedly present within the proximal promoter regions of SCL28 in the six cucurbit species (Figure 4a). To investigate whether SCL28 plays a role in fruit development, we analyzed the expression levels of this gene in fruits at different stages using publicly available transcriptome data. All SCL28 genes exhibited high expression levels during the early stages of fruit development, with their expression levels gradually decreasing in the later stages (Figure 4b). These findings suggest that SCL28 genes are conserved in cucurbit crops and may be involved in the early stages of fruit development.
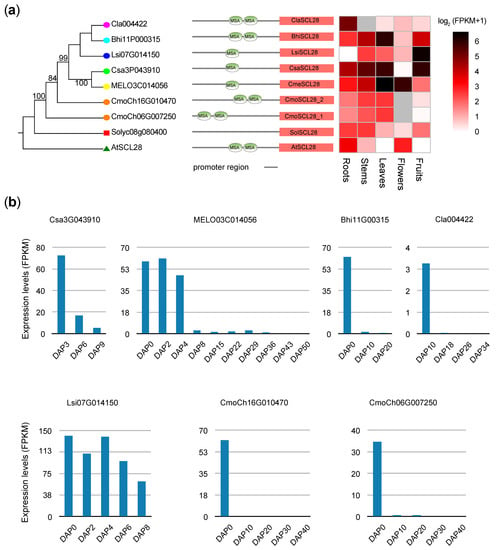
Figure 4.
Expression profiles of DLT members in Arabidopsis, tomato, and six cucurbit crops: (a) Heatmaps of the expression levels of DLT members in various tissues, including roots, stems, leaves, flowers, and fruits. Missing values are indicated by the grey color. The core motifs (MSA elements and AACGG) are represented by green ovals; (b) Expression levels of cucurbit crops’ SCL28 genes in fruits at different stages.
3.6. Differential Expression Patterns of GRAS Genes in Hypocotyl Tissues under Dark Conditions
As previously stated, a significant proportion of light-responsive cis-acting elements, including Box-4, G box, GT1 motif, and TCCC motif, were identified in the promoter regions of GRAS genes in the six cucurbit crops (Table 1). Hypocotyl growth is known to be influenced by light conditions, with dark or weak light conditions promoting hypocotyl elongation. To further investigate the potential roles of GRAS genes in hypocotyl elongation under dark or weak conditions, we analyzed the expression patterns of GRAS genes in cucumber using publicly available transcriptome data. Our analysis revealed that three genes (Csa4P038690, Csa1P616330, and Csa1P408720) exhibited differential expression between weak and normal light conditions (Figure 5a). Subsequently, we utilized RT-qPCR to further investigate the expression levels of these genes in hypocotyl tissues under dark and normal light conditions (Figure 5b). Our results revealed that Csa1P408720 and its homologous genes from five other cucurbit crops were located in the DELLA-2 clades and exhibited an ascending trend under dark treatment. Importantly, all six genes contained light-responsive elements, including G box and Box-4, in their promoters, suggesting that changes in transcript abundance were likely influenced by light conditions (Figure 5c). The findings of this study suggest that these DELLA-2 genes might participate in the reaction to dark conditions during hypocotyl elongation and potentially function as a protective mechanism against hypocotyl overgrowth in cucurbit crops.
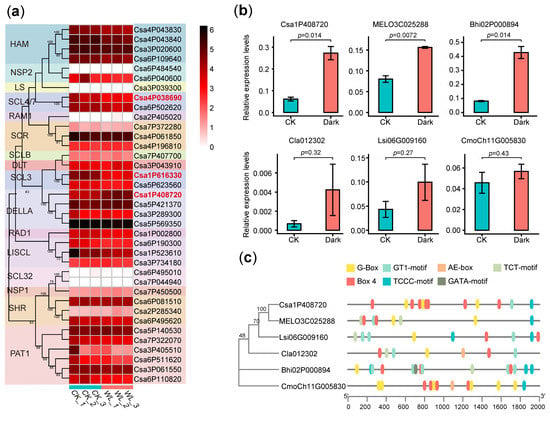
Figure 5.
Expression levels and cis-acting elements of GRAS genes: (a) The differential expression levels of GRAS genes in cucumber under strong light (control check, CK) and weak light (WL) conditions were analyzed using RNA-seq; (b) Relative expression levels of DELLA-2 genes from six cucurbit crops, namely Csa1P408720, MELO3C025288, Bhi02P000894, Cla012302, Lsi06G009160, and CmoCh11G005830 in hypocotyl under normal light (CK) and dark light (Dark) conditions. P values obtained using Student’s tests are indicated; (c) The cis-acting elements related to light responses in the promoters of the DELLA-2 gene from the six cucurbit crops.
4. Discussion
Recent studies in various plants, such as Arabidopsis [1,2,7,19,65,66,67,68,69,70], rice [9,13,14,71,72,73], and tomato [10,23,25,74,75,76,77], have demonstrated that GRAS genes play crucial roles in multiple biological processes. Although the GRAS gene family has been studied in cucumber [16], melon [15], watermelon [30], and bottle gourd [31], limited information is available for the comparative analysis of the GRAS gene family in multiple cucurbit crops. In this study, we identified the GRAS gene family and conducted a comprehensive comparison of their classification, characteristics, evolution, cis-acting elements, and expression patterns among six cucurbit crops. Additionally, we predicted the involvement of the GRAS gene in early fruit development and the response of hypocotyl growth to light.
4.1. Duplication Events Drive the Evolution of GRAS Genes in Six Cucurbit Crops
The GRAS gene family exhibits variable member counts across different species, with Arabidopsis [7,8], tomato [10,11], and rice [8,9] having 32–34, 53–54, and 60 members, respectively. Previous investigations of cucurbit crops, including cucumber [16], melon [15], watermelon [30], and bottle gourd [31], have reported 37 GRAS genes, which aligns with our findings. The number of GRAS genes in cucumber, melon, wax gourd, watermelon, and bottle gourd of the Benincasae group are comparable, while the Cucurbita species pumpkin genome contains 57 GRAS genes (Figure 1b). The presence of 8 TD genes and 66 WGD genes in 4 TD and 38 WGD events across the six cucurbit crops (Table S3) suggests that TD and WGD are the driving forces behind the expansion of the GRAS gene family. The synteny analysis of the six cucurbit crops (Figure 2a) revealed that the GRAS gene family diverged with the evolutionary separation of the Cucurbita and Benincasae groups in the Cucurbitaceae family, and a WGD event was the primary cause for the difference in the number of GRAS genes between the two groups [36]. Furthermore, the number of GRAS genes in subgenome A of pumpkin (32 GRAS genes) were greater than that in subgenome B (22 GRAS genes) (Figure 2b and Figure S1f), indicating a higher rate of gene loss within this family in subgenome B than in subgenome A.
4.2. Diverse Patterns of Characteristics Evolved in the GRAS Genes of Six Cucurbit Crops
The scope of our characterization of GRAS family members expanded through the analysis of gene structures and motifs in their encoded proteins (Figures S3–S5). Homologous GRAS genes that clustered together in a subfamily or clade tended to share similar structure distribution patterns in CDS-UTR and motifs (Figures S3 and S5). However, some genes within the same subfamilies or OGs exhibited different structure patterns, reflecting the evolutionary differences among these GRAS members and suggesting that they might have different functions. Our analysis revealed that genes from the SCLB and RAD1 subfamily exhibited unique motifs in the six cucurbit crops (Figures S3 and S5). Additionally, we also observed that genes from the SCLB and RAD1 subfamily exhibited unique expression patterns in the tested tissues of the six cucurbit crops. Specifically, we observed that most genes belonging to the SCLB and RAD1 subfamilies showed broad expression patterns across various tissues of these six cucurbit crops. Conversely, the homologous genes of these subfamilies were found to be barely expressed in the tested tissues of tomato species, except for Solyc06g009610 and Solyc01g07937, which exhibited low expression levels in roots and stems (Figure S8; Table S6). Previous studies have shown that GRAS genes induced by arbuscular mycorrhizal (AM) fungi are commonly found in SCLB and RAD1 subfamilies. These genes have been reported to play a crucial role in regulating AM development, including SlGRAS33 (Solyc06g009610), SlGRAS23 (Solyc01g079380), SlGRAS22 (Solyc01g079370), SlGRAS21 (Solyc01g059960), and SlGRAS28 (Solyc03g11095) in tomato [78]. Based on this information, we hypothesize a potential association between the specific motifs and the observed expression patterns in the six cucurbit crops within the SCLB and RAD1 subfamilies, and that these changes may have led to the emergence of new functions in these genes within the Cucurbitaceae family. However, we refrain from further exploration of this matter in this study and leave it for future investigations.
4.3. Gene Expression Profiles Unveiled Potential Roles of GRAS Genes in Fruit Development
Based on our findings, the expression patterns of GRAS genes in the six cucurbit crops exhibited significant variation among different members, even between orthologous pairs (Figures S7 and S8; Table S6). This result is consistent with previous studies in tomato [10] and Rosaceae species [17], suggesting that GRAS genes may undergo neo-functionalization or sub-functionalization. In this study, we observed a typical example, where most OG-SCR-1 and OG-SCR-2 members were relatively highly expressed in all the tested tissues, while most OG-SCR-3 members were hardly expressed in any analyzed tissues, except for roots (Figure S8). However, some subfamilies or OGs exhibited conserved expression patterns, indicating their retention due to genetic redundancy and selection for their contributions to the robustness of the genetic network. For example, almost all Cucurbitaceae SCL32 members were expressed in leaves, while hardly any SCL32 members were expressed in flowers and fruits (Figure S8).
Previous research has established the involvement of GRAS genes in various biological processes, including cell cycle regulation, grain size determination, fruit development, ripening [19,21,22,23,24,25], etc. For instance, a recent report showed that AtSCL28 is a crucial component of a transcriptional regulatory network downstream of the central MYB3Rs that controls the G2-to-M phase cell cycle transition and plays a pivotal role in determining cell size in Arabidopsis [19,20]. In this study, we predicted a total of nine GRAS genes that exhibited fruit-specific expression patterns (Table S6) and found that Lsi07G014150, which is homologous to AtSCL28 (Figure 4a; Table S7), was highly expressed in fruits (Figure 4a), indicating its potential involvement in fruit development. We also observed the repeated occurrence of MSA elements in the promoter regions of SCL28 in the six cucurbit species (Figure 4a), and all showed high expression levels in the early stages of fruit development (Figure 4b). Further in-depth studies are required to clarify whether fruit development is affected by expression levels of SLC28 in these six cucurbit crops.
4.4. Differential Expression Patterns of DELLA-2 Genes in Response to Dark Conditions
The GRAS members have been widely reported to be involved in regulating plant growth and development [6,26]. In a recent study, light-related cis-acting elements were identified in the promoter regions of GRAS genes in six Rosaceae species [17]. Our findings also found light-related cis-acting elements in the GRAS genes of the six cucurbit crops (Table 1; Figure S5), indicating that these GRAS genes may regulate light responses in these six cucurbit crops. It is known that seedlings grown under dark conditions exhibit longer hypocotyls than those grown under light conditions [26]. Based on publicly available transcriptome data, we observed significant changes in the expression levels of three GRAS genes in hypocotyl under weak and strong light conditions in cucumber (Figure 5a). Furthermore, qRT-PCR experiments conducted under dark and normal light conditions showed that the expression levels of Csa1P408720 and its orthologous genes in five other cucurbit crops were upregulated in hypocotyl tissue under dark light conditions (Figure 5b). These findings suggest that Cucurbitaceae DELLA–2 genes may be involved in responses to dark conditions and regulate hypocotyl elongation.
5. Conclusions
In this study, we conducted a comprehensive analysis of GRAS genes in six cucurbit crops. Using phylogenetic relationships and previous classifications, we divided the identified GRAS genes into 16 subfamilies. Cucurbit-specific motifs and expression patterns in the SCLB and RAD1 clades suggest their potential roles in cucurbit crops. The analysis of duplication events revealed that WGD and TD contributed to the expansion of the GRAS family, particularly in pumpkin. We examined tissue-specific expression patterns using publicly available transcriptome data and discussed the potential functions of GRAS genes, with a focus on fruit development. Additionally, we found that GRAS genes might potentially regulate hypocotyl elongation under weak or dark light conditions. Overall, our findings enhance our understanding of the characteristics, evolutionary history, expression patterns, and functional roles of GRAS genes in the Cucurbitaceae family.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae9060717/s1, Figure S1. Chromosomal location of GRAS genes on six cucurbit crops; Figure S2. GRAS gene numbers and intron number distribution patterns in six cucurbit crops; Figure S3. Phylogenetic tree, GRAS gene structure, and GRAS protein motifs from six cucurbit crops; Figure S4. Sequence analysis and weblog of 20 identified motifs of 237 GRAS proteins from six cucurbit crops; Figure S5. Phylogenetic tree, GRAS gene structures, and GRAS protein motifs from Arabidopsis, tomato, and six cucurbit crops from different clades; Figure S6. Cis-acting element number analysis in the GRAS gene family in six cucurbit crops; Figure S7. Expression profiles of 237 cucurbit GRAS genes in different tissues; Figure S8. Expression profiles of 318 GRAS genes in Arabidopsis, tomato, and six cucurbit plants from different clades; Table S1. The identified GRAS genes in six cucurbit crops; Table S2. List of GRAS genes within the same syntenic blocks among six cucurbit crops; Table S3. The duplicated GRAS genes from the six cucurbit crops; Table S4. The Ka, Ks, and Ka/Ks values for TD and WGD GRAS genes; Table S5. The detailed information about physiochemical characteristics of GRAS genes in six cucurbit crops; Table S6. Expression profiles of 319 GRAS genes in Arabidopsis, tomato, and six cucurbit crops from different tissues; Table S7. List of orthologous gene groups among six cucurbit crops; Table S8. Primers used for RT-qPCR analysis in this study; Table S9. The accession number of RNA-seq data referred to in this study.
Author Contributions
Z.Z. conceived and designed the research. Q.Z. performed the data analysis and wrote the manuscript. C.W. performed the qRT-PCR experiments. Z.Z., J.H., T.S., Y.X., H.L., S.C., H.X., K.X. and S.M. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (32225044 to Z.Z. and 32130093 to Z.Z.), This work was also supported by the “Taishan Scholar” Foundation of the People’s Government of Shandong Province.
Data Availability Statement
All needed genome sequences and genome annotation files of cucurbit crops were obtained from the Cucurbit Genome Database (http://cucurbitgenomics.org accessed on 31 August 2018). The genome assemblies and annotations of Arabidopsis and tomato are available at the TAIR database (https://www.arabidopsis.org accessed on 31 August 2018) and Sol Genomics Network (https://solgenomics.net accessed on 31 August 2018). The RNA sequencing data required for this study are available in the NCBI (https://www.ncbi.nlm.nih.gov/ accessed on 8 September 2018) Sequence Read Archive (SRA) database and can be accessed using the accession numbers provided in Table S9.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Peng, J.; Carol, P.; Richards, D.E.; King, K.E.; Cowling, R.J.; Murphy, G.P.; Harberd, N.P. The Arabidopsis GAI gene defines a signaling pathway that negatively regulates gibberellin responses. Genes Dev. 1997, 11, 3194–3205. [Google Scholar] [CrossRef]
- Silverstone, A.L.; Ciampaglio, C.N.; Sun, T.-P. The Arabidopsis RGA gene encodes a transcriptional regulator repressing the gibberellin signal transduction pathway. Plant Cell 1998, 10, 155–169. [Google Scholar] [CrossRef]
- Laurenzio, L.D.; Wysocka-Diller, J.; Malamy, J.E.; Pysh, L.; Helariutta, Y.; Freshour, G.; Hahn, M.G.; Feldmann, K.A.; Benfey, P.N. The SCARECROW gene regulates an asymmetric cell division that is essential for generating the radial organization of the Arabidopsis root. Cell 1996, 86, 423–433. [Google Scholar] [CrossRef] [PubMed]
- Hofmann, N.R. A structure for plant-specific transcription factors: The GRAS domain revealed. Plant Cell 2016, 28, 993–994. [Google Scholar] [CrossRef] [PubMed]
- Hakoshima, T. Structural basis of the specific interactions of GRAS family proteins. FEBS Lett. 2018, 592, 489–501. [Google Scholar] [CrossRef] [PubMed]
- Jaiswal, V.; Kakkar, M.; Kumari, P.; Zinta, G.; Gahlaut, V.; Kumar, S. Multifaceted roles of GRAS transcription factors in growth and stress responses in plants. iScience 2022, 25, 105026. [Google Scholar] [CrossRef]
- Lee, M.H.; Kim, B.; Song, S.K.; Heo, J.O.; Yu, N.I.; Lee, S.A.; Kim, M.; Kim, D.G.; Sohn, S.O.; Lim, C.E.; et al. Large-scale analysis of the GRAS gene family in Arabidopsis thaliana. Plant Mol. Biol. 2008, 67, 659–670. [Google Scholar] [CrossRef]
- Liu, X.; Widmer, A. Genome-wide comparative analysis of the GRAS gene family in Populus, Arabidopsis and Rice. Plant Mol. Biol. Report. 2014, 32, 1129–1145. [Google Scholar] [CrossRef]
- Tian, C.; Wan, P.; Sun, S.; Li, J.; Chen, M. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 2004, 54, 519–532. [Google Scholar] [CrossRef] [PubMed]
- Huang, W.; Xian, Z.; Kang, X.; Tang, N.; Li, Z. Genome-wide identification, phylogeny and expression analysis of GRAS gene family in tomato. BMC Plant. Biol. 2015, 15, 209. [Google Scholar] [CrossRef]
- Niu, Y.; Zhao, T.; Xu, X.; Li, J. Genome-wide identification and characterization of GRAS transcription factors in tomato (Solanum lycopersicum). PeerJ 2017, 5, e3955. [Google Scholar] [CrossRef]
- Cenci, A.; Rouard, M. Evolutionary analyses of GRAS transcription factors in Angiosperms. Front. Plant Sci. 2017, 8, 273. [Google Scholar] [CrossRef]
- Tong, H.; Jin, Y.; Liu, W.; Li, F.; Fang, J.; Yin, Y.; Qian, Q.; Zhu, L.; Chu, C. DWARF AND LOW-TILLERING, a new member of the GRAS family, plays positive roles in brassinosteroid signaling in rice. Plant J. 2009, 58, 803–816. [Google Scholar] [CrossRef]
- Li, W.; Wu, J.; Weng, S.; Zhang, Y.; Zhang, D.; Shi, C. Identification and characterization of dwarf 62, a loss-of-function mutation in DLT/OsGRAS-32 affecting gibberellin metabolism in rice. Planta 2010, 232, 1383–1396. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Fu, Q.; Zheng, J.; Zhang, A.; Wang, H. Transcriptomic and metabolomic analyses reveal that melatonin promotes melon root development under copper stress by inhibiting jasmonic acid biosynthesis. Hortic. Res. 2020, 7, 79. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Liu, W.; Xiang, C.; Li, X.; Wang, Q.; Wang, T.; Liu, Z.; Zhang, J.; Gao, L.; Zhang, W. Genome-wide characterization of GRAS family and their potential roles in cold tolerance of cucumber (Cucumis sativus L.). Int. J. Mol. Sci. 2020, 21, 3857. [Google Scholar] [CrossRef] [PubMed]
- Bai, Y.; Liu, H.; Zhu, K.; Cheng, Z.M. Evolution and functional analysis of the GRAS family genes in six Rosaceae species. BMC Plant Biol. 2022, 22, 569. [Google Scholar]
- Luo, W.; Zhao, Z.; Chen, H.; Ao, W.; Lu, L.; Liu, J.; Li, X.; Sun, Y. Genome-wide characterization and expression of DELLA genes in Cucurbita moschata reveal their potential roles under development and abiotic stress. Front. Plant Sci. 2023, 14, 1137126. [Google Scholar] [CrossRef]
- Goldy, C.; Pedroza-Garcia, J.A.; Breakfield, N.; Cools, T.; Vena, R.; Benfey, P.N.; De Veylder, L.; Palatnik, J.; Rodriguez, R.E. The Arabidopsis GRAS-type SCL28 transcription factor controls the mitotic cell cycle and division plane orientation. Proc. Natl. Acad. Sci. USA 2021, 118, e2005256118. [Google Scholar] [CrossRef]
- Nomoto, Y.; Takatsuka, H.; Yamada, K.; Suzuki, T.; Suzuki, T.; Huang, Y.; Latrasse, D.; An, J.; Gombos, M.; Breuer, C.; et al. A hierarchical transcriptional network activates specific CDK inhibitors that regulate G2 to control cell size and number in Arabidopsis. Nat. Commun. 2022, 13, 1660. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Li, X.; Fu, Y.; Zhu, Z.; Tan, L.; Liu, F.; Sun, X.; Sun, X.; Sun, C. GS6, a member of the GRAS gene family, negatively regulates grain size in rice. J. Integr. Plant Biol. 2013, 55, 938–949. [Google Scholar] [CrossRef] [PubMed]
- Ji, C.; Xu, L.; Li, Y.; Fu, Y.; Li, S.; Wang, Q.; Zeng, X.; Zhang, Z.; Zhang, Z.; Wang, W.; et al. The O2-ZmGRAS11 transcriptional regulatory network orchestrates the coordination of endosperm cell expansion and grain filling in maize. Mol. Plant 2022, 15, 468–487. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Wang, X.; Li, C.; Li, H.; Zhang, J.; Ye, Z. Silencing GRAS2 reduces fruit weight in tomato. J. Integr. Plant Biol. 2018, 60, 498–513. [Google Scholar] [CrossRef]
- Zhang, L.; Zhu, M.; Ren, L.; Li, A.; Chen, G.; Hu, Z. The SlFSR gene controls fruit shelf-life in tomato. J. Exp. Bot. 2018, 69, 2897–2909. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, Y.; Su, D.; Lu, W.; Li, Z. SlGRAS4 accelerates fruit ripening by regulating ethylene biosynthesis genes and SlMADS1 in tomato. Hortic. Res. 2021, 8, 3. [Google Scholar] [CrossRef]
- Xiong, H.; Lu, D.; Li, Z.; Wu, J.; Ning, X.; Lin, W.; Bai, Z.; Zheng, C.; Sun, Y.; Chi, W.; et al. The DELLA-ABI4-HY5 module integrates light and gibberellin signals to regulate hypocotyl elongation. Plant Commun. 2023. [Google Scholar] [CrossRef] [PubMed]
- Liu, B.; Zhao, S.; Li, P.; Yin, Y.; Niu, Q.; Yan, J.; Huang, D. Plant buffering against the high-light stress-induced accumulation of CsGA2ox8 transcripts via alternative splicing to finely tune gibberellin levels and maintain hypocotyl elongation. Hortic. Res. 2021, 8, 2. [Google Scholar] [CrossRef]
- Xue, H.; Gao, X.; He, P.; Xiao, G. Origin, evolution, and molecular function of DELLA proteins in plants. Crop J. 2022, 10, 287–299. [Google Scholar] [CrossRef]
- Xie, D.; Xu, Y.; Wang, J.; Liu, W.; Zhou, Q.; Luo, S.; Huang, W.; He, X.; Li, Q.; Peng, Q.; et al. The wax gourd genomes offer insights into the genetic diversity and ancestral cucurbit karyotype. Nat. Commun. 2019, 10, 5158. [Google Scholar] [CrossRef] [PubMed]
- Lv, G.; Zheng, X.; Duan, Y.; Wen, Y.; Zeng, B.; Ai, M.; He, B. The GRAS gene family in watermelons: Identification, characterization and expression analysis of different tissues and root-knot nematode infestations. PeerJ 2021, 9, e11526. [Google Scholar] [CrossRef]
- Sidhu, N.S.; Pruthi, G.; Singh, S.; Bishnoi, R.; Singla, D. Genome-wide identification and analysis of GRAS transcription factors in the bottle gourd genome. Sci. Rep. 2020, 10, 14338. [Google Scholar] [CrossRef]
- Huang, S.; Li, R.; Zhang, Z.; Li, L.; Gu, X.; Fan, W.; Lucas, W.J.; Wang, X.; Xie, B.; Ni, P.; et al. The genome of the cucumber, Cucumis sativus L. Nat. Genet. 2009, 41, 1275–1281. [Google Scholar] [CrossRef]
- Garcia-Mas, J.; Benjak, A.; Sanseverino, W.; Bourgeois, M.; Mir, G.; Gonzalez, V.M.; Henaff, E.; Camara, F.; Cozzuto, L.; Lowy, E.; et al. The genome of melon (Cucumis melo L.). Proc. Natl. Acad. Sci. USA 2012, 109, 11872–11877. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Zhang, J.; Sun, H.; Salse, J.; Lucas, W.J.; Zhang, H.; Zheng, Y.; Mao, L.; Ren, Y.; Wang, Z.; et al. The draft genome of watermelon (Citrullus lanatus) and resequencing of 20 diverse accessions. Nat. Genet. 2013, 45, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Shamimuzzaman, M.; Sun, H.; Salse, J.; Sui, X.; Wilder, A.; Wu, Z.; Levi, A.; Xu, Y.; Ling, K.S.; et al. The bottle gourd genome provides insights into Cucurbitaceae evolution and facilitates mapping of a Papaya ring-spot virus resistance locus. Plant J. 2017, 92, 963–975. [Google Scholar] [CrossRef]
- Sun, H.; Wu, S.; Zhang, G.; Jiao, C.; Guo, S.; Ren, Y.; Zhang, J.; Zhang, H.; Gong, G.; Jia, Z.; et al. Karyotype stability and unbiased fractionation in the paleo-allotetraploid cucurbita genomes. Mol. Plant 2017, 10, 1293–1306. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Lozano, R.; Hamblin, M.T.; Prochnik, S.; Jannink, J.L. Identification and distribution of the NBS-LRR gene family in the Cassava genome. BMC Genom. 2015, 16, 360. [Google Scholar] [CrossRef]
- Artimo, P.; Jonnalagedda, M.; Arnold, K.; Baratin, D.; Csardi, G.; De Castro, E.; Duvaud, S.; Flegel, V.; Fortier, A.; Gasteiger, E. ExPASy: SIB bioinformatics resource portal. Nucleic Acids Res. 2012, 40, W597–W603. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree Of Life (iTOL) v4: Recent updates and new developments. Nucleic Acids Res. 2019, 47, W256–W259. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Freeling, M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu. Rev. Plant Biol. 2009, 60, 433–453. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.I.; Schein, J.E.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Rombauts, S.; Dehais, P.; Van Montagu, M.; Rouze, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Gan, X.; Stegle, O.; Behr, J.; Steffen, J.G.; Drewe, P.; Hildebrand, K.L.; Lyngsoe, R.; Schultheiss, S.J.; Osborne, E.J.; Sreedharan, V.T.; et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature 2011, 477, 419–423. [Google Scholar] [CrossRef]
- Li, Z.; Zhang, Z.; Yan, P.; Huang, S.; Fei, Z.; Lin, K. RNA-Seq improves annotation of protein-coding genes in the cucumber genome. BMC Genom. 2011, 12, 540. [Google Scholar] [CrossRef] [PubMed]
- Sato, S.; Tabata, S.; Hirakawa, H.; Asamizu, E.; Shirasawa, K.; Isobe, S.; Kaneko, T.; Nakamura, Y.; Shibata, D.; Aoki, K.; et al. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635–641. [Google Scholar]
- Jiang, B.; Xie, D.; Liu, W.; Peng, Q.; He, X. De novo assembly and characterization of the transcriptome, and development of SSR markers in wax gourd (Benicasa hispida). PLoS ONE 2013, 8, e71054. [Google Scholar] [CrossRef] [PubMed]
- Grassi, S.; Piro, G.; Lee, J.M.; Zheng, Y.; Fei, Z.; Dalessandro, G.; Giovannoni, J.J.; Lenucci, M.S. Comparative genomics reveals candidate carotenoid pathway regulators of ripening watermelon fruit. BMC Genom. 2013, 14, 781. [Google Scholar] [CrossRef] [PubMed]
- Rhee, S.J.; Seo, M.; Jang, Y.J.; Cho, S.; Lee, G.P. Transcriptome profiling of differentially expressed genes in floral buds and flowers of male sterile and fertile lines in watermelon. BMC Genom. 2015, 16, 914. [Google Scholar] [CrossRef]
- Rhee, S.J.; Kwon, T.; Seo, M.; Jang, Y.J.; Sim, T.Y.; Cho, S.; Han, S.W.; Lee, G.P. De novo-based transcriptome profiling of male-sterile and fertile watermelon lines. PLoS ONE 2017, 12, e0187147. [Google Scholar] [CrossRef]
- Yano, R.; Nonaka, S.; Ezura, H. Melonet-DB, a grand RNA-Seq gene expression atlas in melon (Cucumis melo L.). Plant Cell Physiol. 2018, 59, e4. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Mengarelli, D.A.; Zanor, M.I. Genome-wide characterization and analysis of the CCT motif family genes in soybean (Glycine max). Planta 2021, 253, 15. [Google Scholar] [CrossRef] [PubMed]
- Haga, N.; Kobayashi, K.; Suzuki, T.; Maeo, K.; Kubo, M.; Ohtani, M.; Mitsuda, N.; Demura, T.; Nakamura, K.; Jurgens, G.; et al. Mutations in MYB3R1 and MYB3R4 cause pleiotropic developmental defects and preferential down-regulation of multiple G2/M-specific genes in Arabidopsis. Plant Physiol. 2011, 157, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Lang, L.; Pettko-Szandtner, A.; Tuncay Elbasi, H.; Takatsuka, H.; Nomoto, Y.; Zaki, A.; Dorokhov, S.; De Jaeger, G.; Eeckhout, D.; Ito, M.; et al. The DREAM complex represses growth in response to DNA damage in Arabidopsis. Life Sci. Alliance 2021, 4, e202101141. [Google Scholar] [CrossRef]
- Cordelia Bolle, C.K.; Chua, N.-H. PAT1, a new member of the GRAS family, is involved in phytochrome a signal transduction. Genes Dev. 2000, 14, 1269–1278. [Google Scholar] [CrossRef]
- Greb, T.; Clarenz, O.; Schafer, E.; Muller, D.; Herrero, R.; Schmitz, G.; Theres, K. Molecular analysis of the LATERAL SUPPRESSOR gene in Arabidopsis reveals a conserved control mechanism for axillary meristem formation. Genes Dev. 2003, 17, 1175–1187. [Google Scholar] [CrossRef] [PubMed]
- Tyler, L.; Thomas, S.G.; Hu, J.; Dill, A.; Alonso, J.M.; Ecker, J.R.; Sun, T.P. Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 2004, 135, 1008–1019. [Google Scholar] [CrossRef]
- Fode, B.; Siemsen, T.; Thurow, C.; Weigel, R.; Gatz, C. The Arabidopsis GRAS protein SCL14 interacts with class II TGA transcription factors and is essential for the activation of stress-inducible promoters. Plant Cell 2008, 20, 3122–3135. [Google Scholar] [CrossRef]
- Heo, J.O.; Chang, K.S.; Kim, I.A.; Lee, M.H.; Lee, S.A.; Song, S.K.; Lee, M.M.; Lim, J. Funneling of gibberellin signaling by the GRAS transcription regulator scarecrow-like 3 in the Arabidopsis root. Proc. Natl. Acad. Sci. USA 2011, 108, 2166–2171. [Google Scholar] [CrossRef]
- Della Rovere, F.; Fattorini, L.; D’Angeli, S.; Veloccia, A.; Del Duca, S.; Cai, G.; Falasca, G.; Altamura, M.M. Arabidopsis SHR and SCR transcription factors and AUX1 auxin influx carrier control the switch between adventitious rooting and xylogenesis in planta and in in vitro cultured thin cell layers. Ann. Bot. 2015, 115, 617–628. [Google Scholar] [CrossRef]
- Li, X.; Qian, Q.; Fu, Z.; Wang, Y.; Xiong, G.; Zeng, D.; Wang, X.; Liu, X.; Teng, S.; Hiroshi, F.; et al. Control of tillering in rice. Nature 2003, 422, 618–621. [Google Scholar] [CrossRef]
- Xu, K.; Chen, S.; Li, T.; Ma, X.; Liang, X.; Ding, X.; Liu, H.; Luo, L. OsGRAS23, a rice GRAS transcription factor gene, is involved in drought stress response through regulating expression of stress-responsive genes. BMC Plant Biol. 2015, 15, 141. [Google Scholar] [CrossRef]
- Tanabe, S.; Onodera, H.; Hara, N.; Ishii-Minami, N.; Day, B.; Fujisawa, Y.; Hagio, T.; Toki, S.; Shibuya, N.; Nishizawa, Y.; et al. The elicitor-responsive gene for a GRAS family protein, CIGR2, suppresses cell death in rice inoculated with rice blast fungus via activation of a heat shock transcription factor, OsHsf23. Biosci. Biotechnol. Biochem. 2016, 80, 145–151. [Google Scholar] [CrossRef]
- Liu, Y.; Huang, W.; Xian, Z.; Hu, N.; Lin, D.; Ren, H.; Chen, J.; Su, D.; Li, Z. Overexpression of SlGRAS40 in tomato enhances tolerance to abiotic stresses and influences auxin and gibberellin signaling. Front. Plant Sci. 2017, 8, 1659. [Google Scholar] [CrossRef]
- Huang, W.; Peng, S.; Xian, Z.; Lin, D.; Hu, G.; Yang, L.; Ren, M.; Li, Z. Overexpression of a tomato miR171 target gene SlGRAS24 impacts multiple agronomical traits via regulating gibberellin and auxin homeostasis. Plant Biotechnol. J. 2017, 15, 472–488. [Google Scholar] [CrossRef]
- Zhou, S.; Hu, Z.; Li, F.; Yu, X.; Naeem, M.; Zhang, Y.; Chen, G. Manipulation of plant architecture and flowering time by down-regulation of the GRAS transcription factor SlGRAS26 in Solanum lycopersicum. Plant Sci. 2018, 271, 81–93. [Google Scholar] [CrossRef]
- Liu, Y.; Shi, Y.; Zhu, N.; Zhong, S.; Bouzayen, M.; Li, Z. SlGRAS4 mediates a novel regulatory pathway promoting chilling tolerance in tomato. Plant Biotechnol. J. 2020, 18, 1620–1633. [Google Scholar] [CrossRef]
- Ho-Plagaro, T.; Molinero-Rosales, N.; Farina Flores, D.; Villena Diaz, M.; Garcia-Garrido, J.M. Identification and expression analysis of GRAS transcription factor genes involved in the control of arbuscular mycorrhizal development in tomato. Front. Plant Sci. 2019, 10, 268. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).