Investigation of Diversity by Analyzing the Polymorphism of SSR Markers and the Composition of Leaf and Fruit Essential Oils of 72 Mandarins (Citrus reticulata Blanco)
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Genotyping
2.3. Sampling, Peel and Leaf Essential Oils
2.4. EO Analysis
2.4.1. Gas Chromatography (GC) Analysis
2.4.2. Mass Spectrometry
2.4.3. NMR Analysis
2.4.4. Identification of Individual Components
2.5. Data Analysis
3. Results
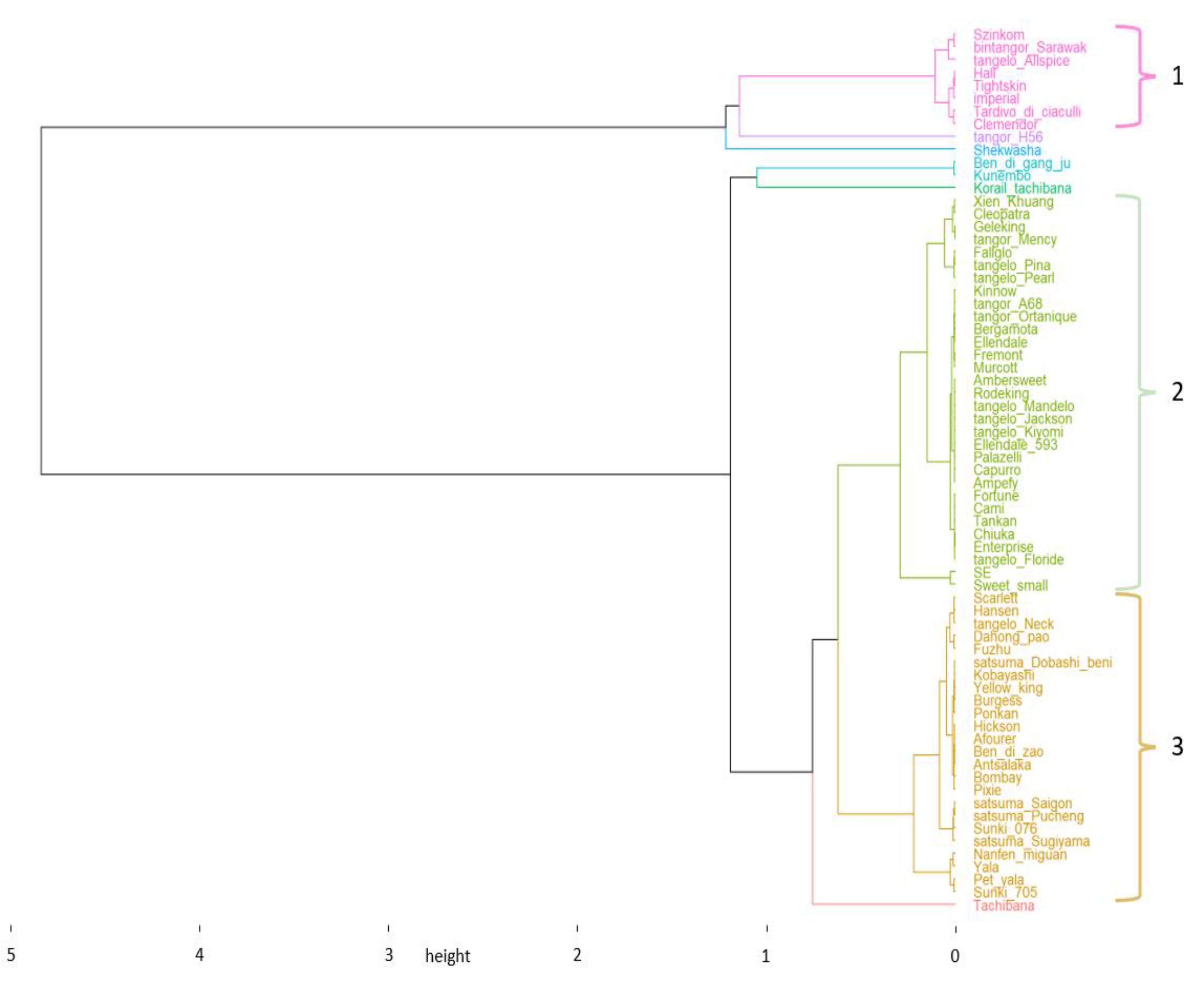
3.1. Genetic Diversity in Mandarins
3.2. Variation in PEO Yield
3.3. Diversity of Mandarin PEO Composition
3.4. Diversity of Mandarin LEO Composition
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Curk, F.; Ollitrault, F.; Garcia-Lor, A.; Luro, F.; Navarro, L.; Ollitrault, P. Phylogenetic origin of limes and lemons revealed by cytoplasmic and nuclear markers. Ann. Bot. 2016, 117, 565–583. [Google Scholar] [CrossRef]
- Wu, G.A.; Prochnik, S.; Jenkins, J.; Salse, J.; Hellsten, U.; Murat, F.; Perrier, X.; Ruiz, M.; Scalabrin, S.; Terol, J.; et al. Sequencing of diverse mandarin, pummelo and orange genomes reveals complex history of admixture during citrus domestication. Na. Biotech. 2014, 32, 656–662. [Google Scholar] [CrossRef]
- Wu, G.A.; Terol, J.; Ibanez, V.; López-García, A.; Pérez-Román, E.; Borredá, C.; Domingo, C.; Tadeo, F.R.; Carbonell-Caballero, J.; Alonso, R.; et al. Genomics of the Origin and Evolution of Citrus. Nature 2018, 554, 311–316. [Google Scholar] [CrossRef]
- Wang, L.; He, F.; Huang, Y.; He, J.; Yang, S.; Zeng, J.; Deng, C.; Jiang, X.; Fang, Y.; Wen, S.; et al. Genome of wild mandarin and domestication history of Mandarin. Mol. Plant 2018, 11, 1024–1037. [Google Scholar] [CrossRef] [PubMed]
- Ollitrault, P.; Curk, F.; Krueger, R. Citrus taxonomy. In The Genus Citrus; Talon, M., Carruso, M., Gmitter, G., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 57–81. [Google Scholar]
- Oueslati, A.; Salhi-Hannachi, A.; Luro, F.; Vignes, H.; Mournet, P.; Ollitrault, P. Genotyping by sequencing reveals the interspecific C. maxima/C. reticulata admixture along the genomes of modern citrus varieties of mandarins, tangors, tangelos, orangelos and grapefruits. PLoS ONE 2017, 12, e0185618. [Google Scholar] [CrossRef]
- Wu, G.A.; Sugimoto, C.; Kinjo, H.; Azama, C.; Mitsube, F.; Talon, M.; Gmitter, F.G.; Rokhsar, D.S. Diversification of mandarin citrus by hybrid speciation and apomixis. Nat. Commun. 2021, 12, 4377. [Google Scholar] [CrossRef]
- Wang, X.; Xu, Y.; Zhang, S.; Cao, L.; Huang, Y.; Cheng, J.; Wu, G.; Tian, S.; Chen, C.; Liu, Y.; et al. Genomic analyses of primitive, wild and cultivated citrus provide insights into asexual reproduction. Nat. Genet. 2017, 49, 765–772. [Google Scholar] [CrossRef]
- Shimada, T.; Endo, T.; Fujii, H.; Nakano, M.; Sugiyama, A.; Daido, G.; Ohta, S.; Yoshioka, T.; Omura, M. MITE insertion-dependent expression of CitRKD1 with a RWP-RK domain regulates somatic embryogenesis in citrus nucellar tissues. BMC Plant Biol. 2018, 18, 166. [Google Scholar] [CrossRef]
- Swingle, W.T.; Reece, P.C. The botany of Citrus and its wild relatives. In The Citrus Industry; Reuther, W., Webber, H.J., Batchelor, L.D., Eds.; University of California Press: Berkeley, CA, USA, 1967; pp. 190–430. [Google Scholar]
- Webber, H.J. Cultivated varieties of citrus. In The Citrus Industry. History, World Distribution, Botany and Varieties; Reuther, W., Webber, H.J., Batchelor, L.D., Eds.; University of California Press: Berkeley, CA, USA, 1943; pp. 475–668. [Google Scholar]
- Caruso, M.; Smith, M.W.; Froelicher, Y.; Russo, G.; Gmitter, F.G., Jr. Traditional breeding. In The Genus Citrus; Talon, M., Caruso, M., Gmitter, F.G., Jr., Eds.; Elsevier: Duxford, UK, 2020; pp. 129–148. [Google Scholar]
- Jacquemond, C.; Curk, F.; Froelicher, Y.; Luro, F. Variétés et porte-greffes: Création, description et sélection. In Les Clémentiniers et Autres Petits Agrumes; Jacquemond, C., Curk, F., Heuzet, M., Eds.; Quae Editions Collection Savoir-Faire: Versailles, France, 2013; pp. 37–108. [Google Scholar]
- Garcia-Lor, A.; Luro, F.; Ollitrault, P.; Navarro, L. Genetic diversity and population-structure analysis of mandarin germplasm by nuclear, chloroplastic and mitochondrial markers. Tree Genet. Genomes 2015, 72, 123. [Google Scholar] [CrossRef]
- Lota, M.L.; de Rocca Serra, D.; Tomi, F.; Casanova, J. Chemical variability of peel and leaf essential oils of Mandarins from Citrus reticulata Blanco species. Biochem. Syst. Ecol. 2000, 28, 61–78. [Google Scholar] [CrossRef]
- Lota, M.-L.; de Rocca Serra, D.; Tomi, F.; Casanova, J. Chemical variability of peel and leaf essential oils of 15 species of mandarins. Biochem. Syst. Ecol. 2001, 29, 77–104. [Google Scholar] [CrossRef] [PubMed]
- Fanciullino, A.-L.; Tomi, F.; Luro, F.; Desjobert, J.M.; Casanova, J. Chemical variability of peel and leaf oils of mandarins. Flavour Fragr. J. 2006, 21, 359–367. [Google Scholar] [CrossRef]
- Ollitrault, P.; Terol, J.; Garcia-Lor, A.; Bérard, A.; Chauveau, A.; Froelicher, Y.; Belzile, C.; Morillon, R.; Navarro, L.; Brunel, D.; et al. SNP mining in C. clementina BAC end sequences; transferability in the Citrus genus (Rutaceae), phylogenetic inferences and perspectives for genetic mapping. BMC Genom. 2012, 13, 13. [Google Scholar] [CrossRef] [PubMed]
- González-Mas, M.C.; Rambla, J.L.; López-Gresa, M.P.; Blázquez, M.A.; Granell, A. Volatile Compounds in Citrus Essential Oils: A Comprehensive Review. Front. Plant Sci. 2019, 10, 12. [Google Scholar] [CrossRef] [PubMed]
- Luro, F.; Garcia Neves, C.; Costantino, G.; da Silva Gesteira, A.; Paoli, M.; Ollitrault, P.; Tomi, F.; Micheli, F.; Gibernau, M. Effect of Environmental Conditions on the Yield of Peel and Composition of Essential Oils from Citrus Cultivated in Bahia (Brazil) and Corsica (France). Agronomy 2020, 10, 1256. [Google Scholar] [CrossRef]
- Ferrer, V.; Paymal, N.; Quinton, C.; Tomi, F.; Luro, F. Investigations of the Chemical Composition and Aromatic Properties of Peel Essential Oils throughout the Complete Phase of Fruit Development for Two Cultivars of Sweet Orange (Citrus sinensis (L.) Osb.). Plants 2022, 11, 2747. [Google Scholar] [CrossRef]
- Benjamin, G.; Tietel, Z.; Porat, R. Effects of Rootstock/Scion Combinations on the Flavor of Citrus Fruit. J. Agric. Food Chem. 2013, 61, 11286–11294. [Google Scholar] [CrossRef]
- Ferrer, V.; Paymal, N.; Quinton, C.; Costantino, G.; Paoli, M.; Froelicher, Y.; Ollitrault, P.; Tomi, F.; Luro, F. Influence of the Rootstock and the Ploidy Level of the Scion and the Rootstock on Sweet Orange (Citrus sinensis) Peel Essential Oil Yield, Composition and Aromatic Properties. Agriculture 2022, 12, 214. [Google Scholar] [CrossRef]
- Ferhat, M.A.; Boukhatem, M.N.; Hazzit, M.; Meklati, B.Y.; Chemat, F. Cold Pressing, Hydrodistillation and Microwave Dry Distillation of Citrus Essential Oil from Algeria: A Comparative Study. Electron. J. Biol. 2016, 1, 30–41. [Google Scholar]
- Sun, X.; Yang, H.; Zhao, W.; Bourcier, E.; Baldwin, E.A.; Plotto, A.; Irey, M.; Bai, J. Huanglongbing and Foliar Spray Programs Affect the Chemical Profile of “Valencia” Orange Peel Oil. Front. Plant Sci. 2021, 12, 611449. [Google Scholar] [CrossRef]
- Zouaghi, G.; Najar, A.; Aydi, A.; Claumann, C.A.; Wüst Zibetti, A.; Ben Mahmoud, K.; Jemmali, A.; Bleton, J.; Moussa, F.; Abderrabba, M.; et al. Essential oil components of Citrus cultivar ‘MALTAISE DEMI SANGUINE’ (Citrus sinensis) as affected by the effects of rootstocks and viroid infection. Int. J. Food Prop. 2019, 22, 438–448. [Google Scholar] [CrossRef]
- Gioffrè, G.; Ursino, D.; Concetta Labate, M.L.C.; Giuffrè, A.M. The peel essential oil composition of bergamot fruit (Citrus bergamia, Risso) of Reggio Calabria (Italy): A review. Emir. J. Food Agric. 2020, 32, 835–845. [Google Scholar] [CrossRef]
- Krueger, R.R.; Navarro, L. Citrus germplasm resources. In Citrus Genetics, Breeding and Biotechnology; Khan, I., Ed.; CABI Publishing: Wallington, NJ, USA, 2007; pp. 45–140. [Google Scholar]
- Krueger, R.R.; Roose, M.L. Use of molecular markers in the management of citrus germplasm resources. J. Am. Soc. Hortic. Sci 2003, 128, 827–837. [Google Scholar] [CrossRef]
- Luro, F.; Bloquel, E.; Tomu, B.; Costantino, G.; Tur, I.; Riolacci, S.; Varamo, F.; Ollitrault, P.; Froelicher, Y.; Curk, F.; et al. The INRACIRAD Citrus Germplasm Collection of San Giuliano, Corsica; Publications du Centre Jean Bérard: Naples, Italy, 2017; pp. 243–261. [Google Scholar]
- Ollitrault, P.; Terol, J.; Chen, C.; Federici, C.T.; Lotfy, S.; Hippolyte, I.; Ollitrault, F.; Bérard, A.; Chauveau, A.; Cuenca, J.; et al. A reference genetic map of C. clementina hort. ex Tan.; citrus evolution inferences from comparative mapping. BMC Genom. 2012, 13, 593. [Google Scholar] [CrossRef]
- Luro, F.; Costantino, G.; Argout, X.; Froelicher, Y.; Terol, J.; Talon, M.; Wincker, P.; Ollitrault, P.; Morillon, R. Transferability of the EST-SSRs developed on Nules clementine (Citrus clementina Hort ex Tan) to other Citrus species and their effectiveness for genetic mapping. BMC Genom. 2008, 9, 287. [Google Scholar] [CrossRef]
- Ferrer, V.; Costantino, G.; Paoli, M.; Paymal, N.; Quinton, C.; Ollitrault, P.; Tomi, F.; Luro, F. Intercultivar Diversity of Sour Orange (Citrus aurantium L.) Based on Genetic Markers, Phenotypic Characteristics, Aromatic Compounds and Sensorial Analysis. Agronomy 2021, 11, 1084. [Google Scholar] [CrossRef]
- Perrier, X.; Jacquemoud-Collet, J.P. DARwin Software: Dissimilarity Analysis and Representation for Windows. Website 2006. Available online: http://darwin.cirad.fr/darwin (accessed on 21 February 2023).
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Shimizu, T.; Kitajima, A.; Nonaka, K.; Yoshioka, T.; Ohta, S.; Goto, S.; Toyoda, A.; Fujiyama, A.; Mochizuki, T.; Nagasaki, H.; et al. Hybrid Origins of Citrus Varieties Inferred from DNA Marker Analysis of Nuclear and Organelle Genomes. PLoS ONE 2016, 11, e0166969. [Google Scholar] [CrossRef]
- Demarcq, B.; Cavailles, M.; Lambert, L.; Schippa, C.; Ollitrault, P.; Luro, F. Characterization of Odor-Active Compounds of Ichang Lemon (Citrus wilsonii Tan.) and Identification of Its Genetic Interspecific Origin by DNA Genotyping. J. Agric. Food Chem. 2021, 69, 3175–3188. [Google Scholar] [CrossRef]
- Luro, F.; Baccati, C.; Paoli, M.; Marchi, E.; Costantino, G.; Gibernau, M.; Ollitrault, P.; Tomi, F. Phylogenetic and taxonomic status of Citrus halimii B.C. Stone determined by genotyping complemented by chemical analysis of leaf and fruit rind essential oils. Sci. Hortic. 2022, 299, 111018. [Google Scholar] [CrossRef]
- Gaff, M.; Esteban-Decloux, M.; Giampaoli, P. Bitter Orange Peel Essential Oil: A Review of the Different Factors and Chemical Reactions Influencing Its Composition. Flavour Fragr. J. 2020, 35, 247–269. [Google Scholar] [CrossRef]
- Rowshan, V.; Najafian, S. Changes of Peel Essential Oil Composition of Citrus aurantium L. During Fruit Maturation in Iran. J. Essent. Oil Bear. Plants 2015, 18, 1006–1012. [Google Scholar] [CrossRef]
- Froelicher, Y.; Dambier, D.; Costantino, G.; Lotfy, S.; Didout, C.; Beaumont, V.; Brottier, P.; Risterucci, A.M.; Luro, F.; Ollitrault, P. Characterization of microsatellite markers in Citrus reticulata Blanco. Mol. Eco. Notes 2008, 8, 119–122. [Google Scholar]
- Garcia-Lor, A.; Curk, F.; Snoussi-Trifa, H.; Morillon, R.; Ancillo, G.; Luro, F.; Navarro, L.; Ollitrault, P. A nuclear phylogeny: SNPs, indels and SSRs deliver new insights into the relationships in the ’true citrus fruit trees group’ (Citrinae, Rutaceae) and the origin of cultivated species. Ann. Bot. 2012, 111, 1–19. [Google Scholar] [CrossRef]
- Kijas, J.M.H.; Thomas, M.R.; Fowler, J.C.S.; Roose, M.L. Integration of trinucleotide microsatellites into a linkage map of Citrus. Theor. Appl. Genet. 1997, 94, 701–706. [Google Scholar] [CrossRef]
- Cuenca, J.; Froelicher, Y.; Aleza, P.; Juarez, J.; Navarro, L.; Ollitrault, P. Multilocus half-tetrad analysis and centromere mapping in citrus: Evidence of SDR mechanism for 2n megagametophyte production and partial chiasma interference in mandarin cv ‘Fortune’. Heredity 2011, 107, 462–470. [Google Scholar] [CrossRef]
- Luro, F.; Viglietti, G.; Marchi, E.; Costantino, G.; Scarpa, M.G.; Tomi, F.; Paoli, M.; Curk, F.; Ollitrault, P. Genetic, morphological and chemical investigations reveal the genetic origin of Pompia (C. medica tuberosa Risso&Poiteau)—An old endemic Sardinian citrus fruit. Phytochemistry 2019, 168, 112083. [Google Scholar] [CrossRef]
- Kamiri, M.; Stift, M.; Srairi, I.; Costantino, G.; El Moussadik, A.; Hmyene, A.; Bakry, F.; Ollitrault, P.; Froelicher, Y. Evidence for non-disomic inheritance in a Citrus interspecific tetraploid somatic hybrid between C. reticulata and C. limon using SSR markers and cytogenetic analysis. Plant Cell Rep. 2011, 30, 1415–1425. [Google Scholar] [CrossRef]
- Aleza, P.; Froelicher, Y.; Schwarz, S.; Hernández, M.; Juárez, J.; Morillon, R.; Navarro, L.; Ollitrault, P. Tetraploidization events by chromosome doubling of nucellar cells are frequent in apomictic citrus and are dependent on genotype and environment. Ann. Bot. 2011, 108, 37–50. [Google Scholar] [CrossRef]





| Genotype | Cultivar | Tanaka’s System | ICVN |
|---|---|---|---|
| 1 | Clemendor | C. deliciosa | 0100658 |
| 1 | Avana Apireno | C. deliciosa | 0100527 |
| 1 | Hall | C. reticulata | 0100356 |
| 1 | Thighskin | C. reticulata | 0100466 |
| 1 | Tardivo di Ciaculli | C. deliciosa | 0100704 |
| 1 | Willow Leaf | C. deliciosa | 0100131 |
| 2 | Sweet small | C. tangerina | 0100826 |
| 2 | Cleopatra | C. reshni | 0100591 |
| 2 | Xien Khuang | C. reticulata | 0100601 |
| 3 | Bombay | C. reticulata | 0100518 |
| 3 | Antsalaka Diego | C. reticulata | 0100495 |
| 3 | Ponkan | C. reticulata | 0100584 |
| 4 | Yala | C. reticulata | 0100674 |
| 4 | Pet Yala | C. suhuiensis | 0100273 |
| 5 | Dobashi Beni | C. unshiu | 0100342 |
| 5 | Pusheng | C. unshiu | 0100657 |
| 5 | Saigon | C. unshiu | 0100225 |
| 5 | Sugiyama | C. unshiu | 0100446 |
| 6 | Ellendale | Tangor | 0100454 |
| 6 | Ellendale | Tangor | 0100455 |
| 7 | Scarlett | C. tangerina | 0100681 |
| 7 | Hansen | C. reticulata | 0100587 |
| 8 | Sarawak | Bintangor | 0100582 |
| 8 | Szinkom | C. suhuiensis | 0100597 |
| 9 | Ben di gang Ju | C. unshiu | 0100433 |
| 9 | Kunembo | C. nobilis | 0100326 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luro, F.; Paoli, M.; Marchi, E.; Costantino, G.; Tomi, F. Investigation of Diversity by Analyzing the Polymorphism of SSR Markers and the Composition of Leaf and Fruit Essential Oils of 72 Mandarins (Citrus reticulata Blanco). Horticulturae 2023, 9, 577. https://doi.org/10.3390/horticulturae9050577
Luro F, Paoli M, Marchi E, Costantino G, Tomi F. Investigation of Diversity by Analyzing the Polymorphism of SSR Markers and the Composition of Leaf and Fruit Essential Oils of 72 Mandarins (Citrus reticulata Blanco). Horticulturae. 2023; 9(5):577. https://doi.org/10.3390/horticulturae9050577
Chicago/Turabian StyleLuro, François, Mathieu Paoli, Elodie Marchi, Gilles Costantino, and Félix Tomi. 2023. "Investigation of Diversity by Analyzing the Polymorphism of SSR Markers and the Composition of Leaf and Fruit Essential Oils of 72 Mandarins (Citrus reticulata Blanco)" Horticulturae 9, no. 5: 577. https://doi.org/10.3390/horticulturae9050577
APA StyleLuro, F., Paoli, M., Marchi, E., Costantino, G., & Tomi, F. (2023). Investigation of Diversity by Analyzing the Polymorphism of SSR Markers and the Composition of Leaf and Fruit Essential Oils of 72 Mandarins (Citrus reticulata Blanco). Horticulturae, 9(5), 577. https://doi.org/10.3390/horticulturae9050577







