Transcriptome Analysis of Ethylene Response in Chrysanthemum moriflolium Ramat. with an Emphasis on Flowering Delay
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Grown Conditions
2.2. Ethephon Treatment
2.3. RNA Extraction and Transcriptome Sequencing
2.4. De Novo Assembly, Gene Functional Annotation, and Differential Gene Expression Analysis
2.5. Quantitative RT-PCR (qRT-PCR)-Based Validation of DEGs
2.6. Floral Dip Transformation of Arabidopsis thaliana
2.7. Data Analysis
3. Results
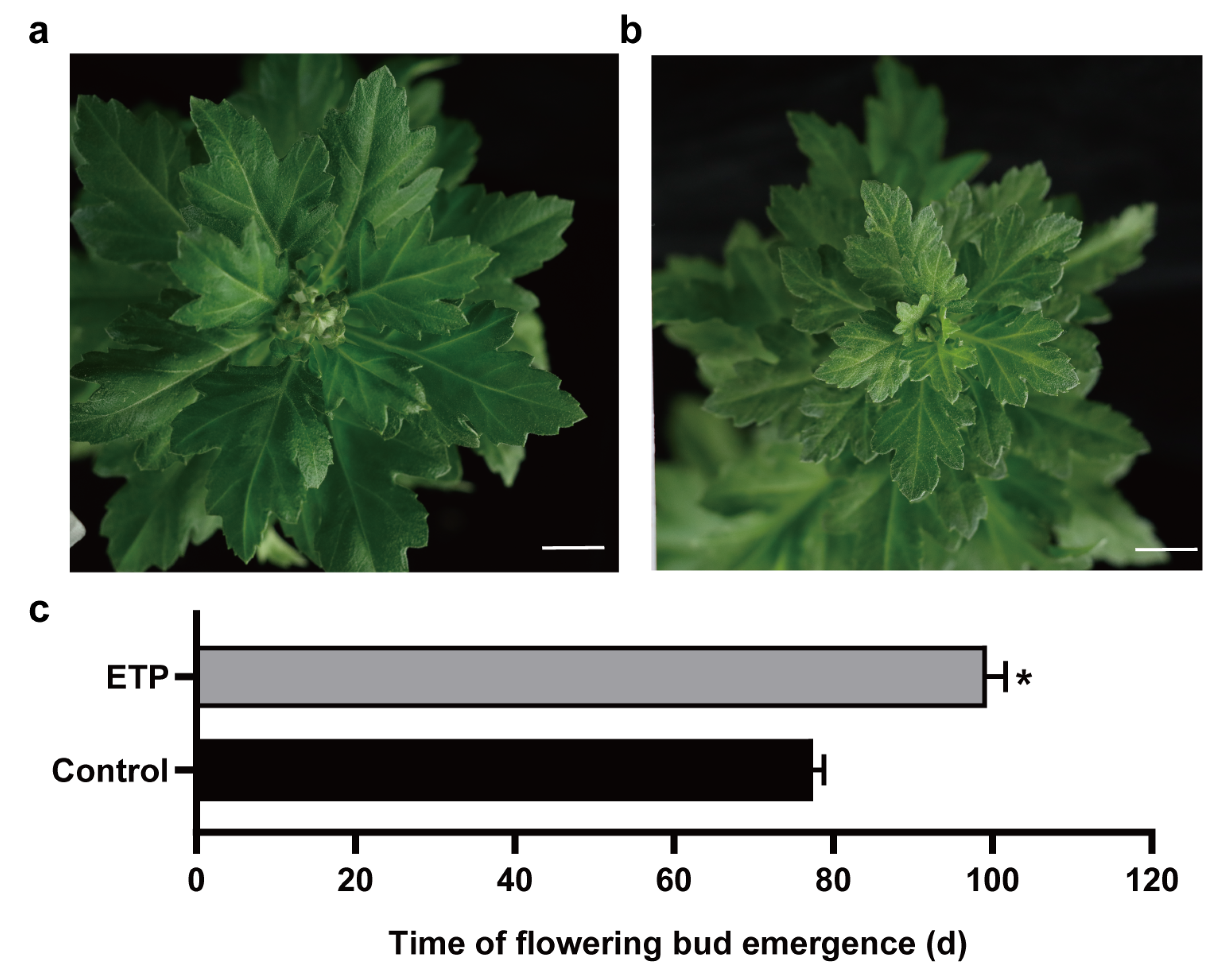
3.1. Ethylene Suppressed Flowering in Chrysanthemum ‘Jinba’
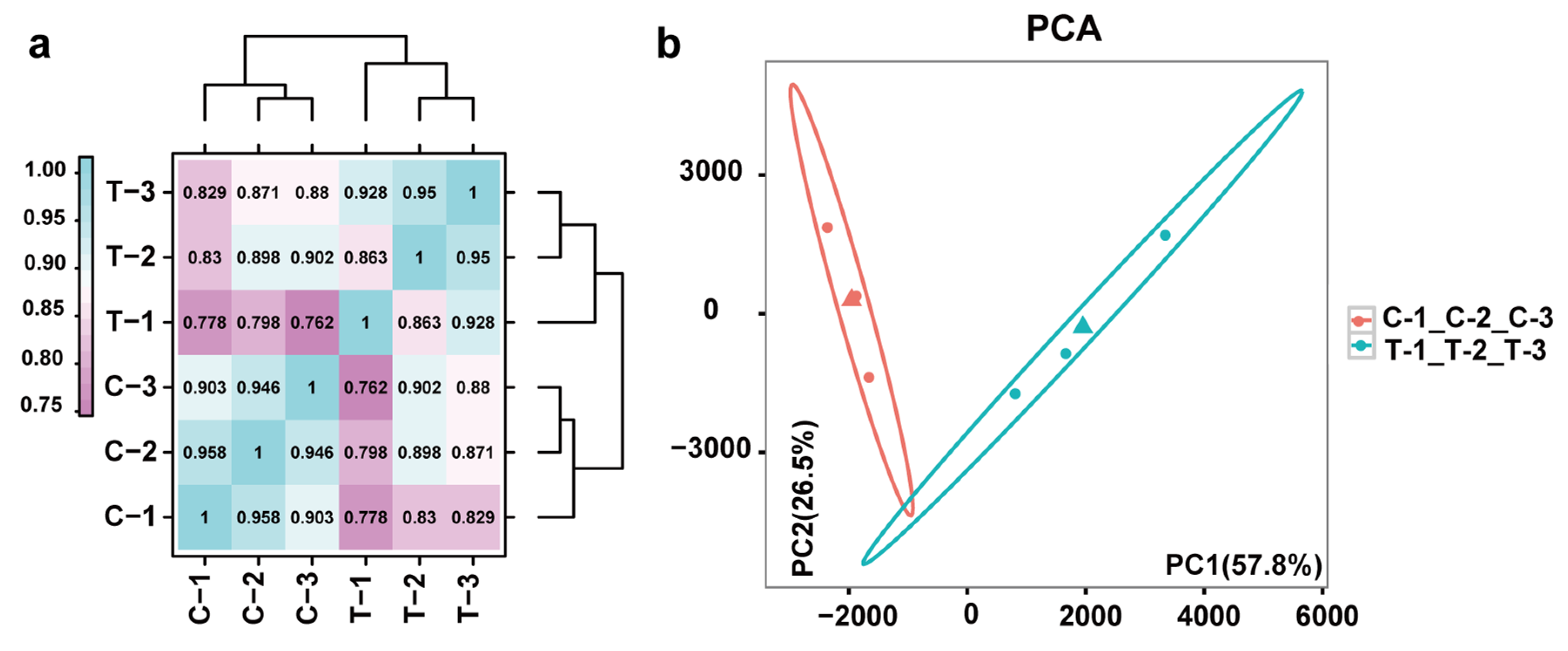
3.2. RNA-Seq after Ethylene Treatment
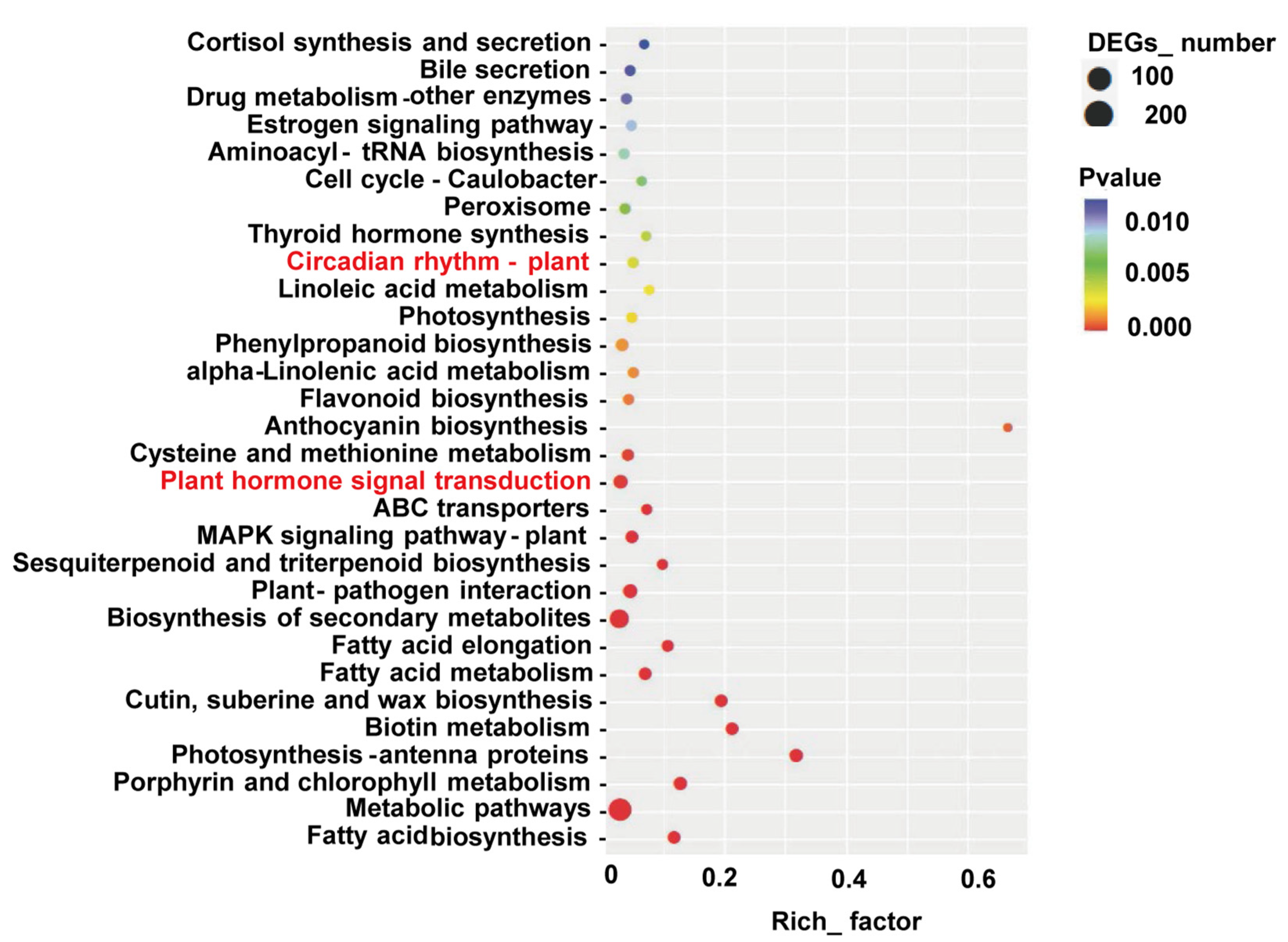
3.3. Differential Gene Expression Analysis
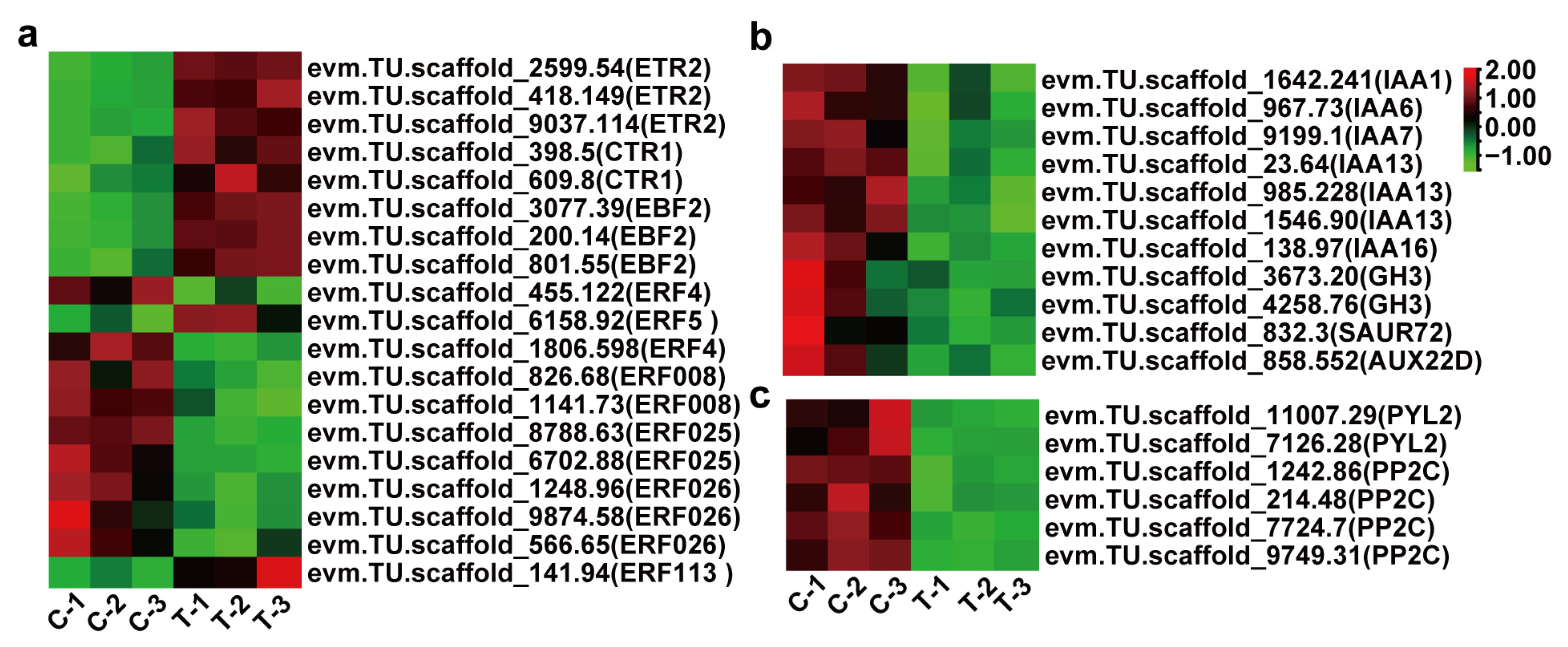
3.4. DEGs Related to Phytohormone Signaling and Circadian Rhythm
3.5. Floral Induction Genes and TFs Underlying in the Effect of Ethylene
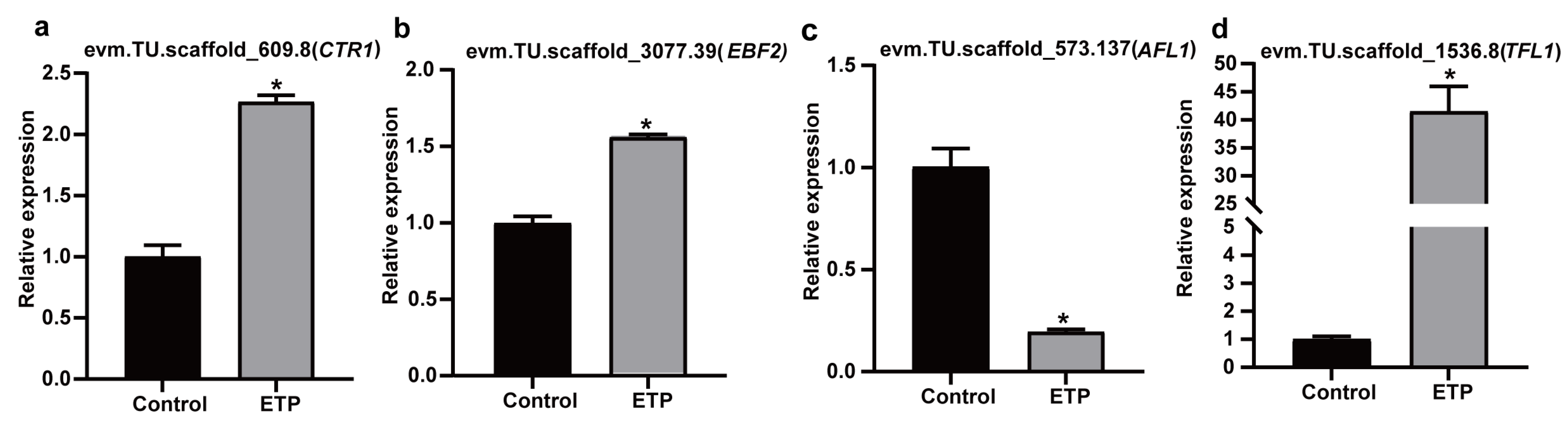
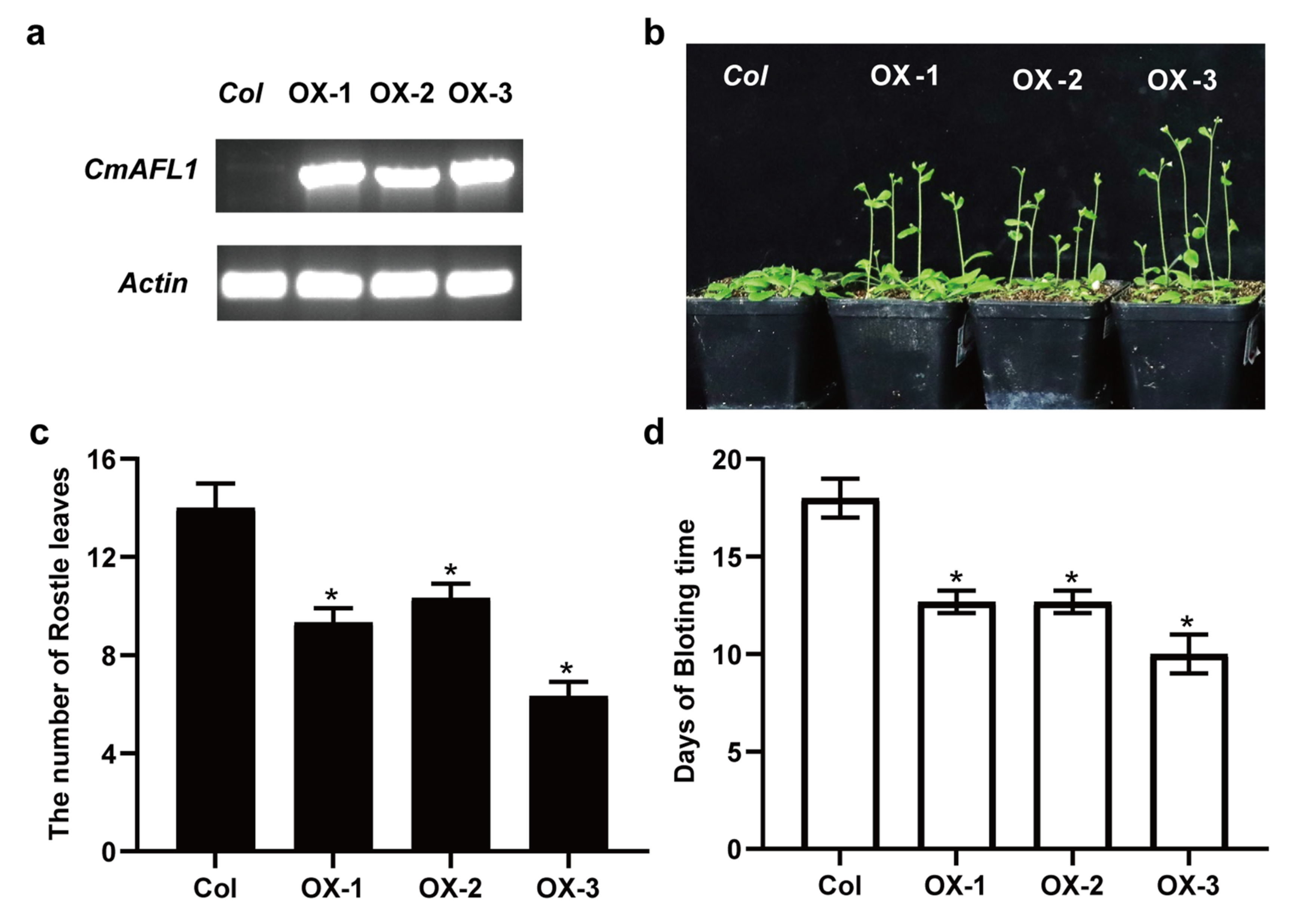
3.6. Validation of RNA-Seq Results Using qRT-PCR and Heterologous Expression in Arabidopsis thaliana
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boss, P.K.; Bastow, R.M.; Mylne, J.S.; Dean, C. Multiple pathways in the decision to flower: Enabling, promoting, and resetting. Plant Cell 2004, 16, S18–S31. [Google Scholar] [CrossRef] [PubMed]
- Putterill, J.; Laurie, R.; Macknight, R. It’s time to flower: The genetic control of flowering time. Bioessays 2004, 26, 363–373. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.; Ding, L.; Jiang, J.; Shentu, Y.; Zhao, W.; Zhao, K.; Zhang, X.; Song, A.; Chen, S.; Chen, F. Overexpression of the CmJAZ1-like gene delays flowering in Chrysanthemum morifolium. Hortic. Res. 2021, 8, 87. [Google Scholar] [CrossRef]
- Blümel, M.; Dally, N.; Jung, C. Flowering time regulation in crops—What did we learn from Arabidopsis? Curr. Opin. Biotechnol. 2015, 32, 121–129. [Google Scholar] [CrossRef]
- Golembeski, G.S.; Imaizumi, T. Photoperiodic regulation of florigen function in Arabidopsis thaliana. Arab. Book 2015, 13, e0178. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.Z.; Zhou, Y.P.; Lv, T.X.; Xie, C.P.; Tian, C.E. Research progress on the autonomous flowering time pathway in Arabidopsis. Physiol. Mol. Biol. Plants 2017, 23, 477–485. [Google Scholar] [CrossRef]
- Surkova, S.Y.; Samsonova, M.G. Mechanisms of vernalization-induced flowering in legumes. Int. J. Mol. Sci. 2022, 23, 9889. [Google Scholar] [CrossRef]
- Bao, S.; Hua, C.; Shen, L.; Yu, H. New insights into gibberellin signaling in regulating flowering in Arabidopsis. J. Integr. Plant Biol. 2020, 62, 118–131. [Google Scholar] [CrossRef]
- Qi, L.; Shi, Y.; Terzaghi, W.; Yang, S.; Li, J. Integration of light and temperature signaling pathways in plants. J. Integr. Plant Biol. 2022, 64, 393–411. [Google Scholar] [CrossRef]
- Wang, J.W. Regulation of flowering time by the miR156-mediated age pathway. J. Exp. Bot. 2014, 65, 4723–4730. [Google Scholar] [CrossRef] [PubMed]
- Pajoro, A.; Biewers, S.; Dougali, E.; Leal Valentim, F.; Mendes, M.A.; Porri, A.; Coupland, G.; Van de Peer, Y.; van Dijk, A.D.; Colombo, L.; et al. The (r)evolution of gene regulatory networks controlling Arabidopsis plant reproduction: A two-decade history. J. Exp. Bot. 2014, 65, 4731–4745. [Google Scholar] [CrossRef] [PubMed]
- Iqbal, N.; Khan, N.A.; Ferrante, A.; Trivellini, A.; Francini, A.; Khan, M.I.R. Ethylene role in plant growth, development and senescence: Interaction with other phytohormones. Front. Plant Sci. 2017, 8, 475. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Li, Z.; Lei, M.; Fu, Y.; Zhao, J.; Ao, M.; Xu, L. Integrated DNA methylome and transcriptome analysis reveals the ethylene-induced flowering pathway genes in pineapple. Sci. Rep. 2017, 7, 17167. [Google Scholar] [CrossRef] [PubMed]
- Ogawara, T.; Higashi, K.; Kamada, H.; Ezura, H. Ethylene advances the transition from vegetative growth to flowering in Arabidopsis thaliana. J. Plant Physiol. 2003, 160, 1335–1340. [Google Scholar] [CrossRef]
- Wuriyanghan, H.; Zhang, B.; Cao, W.H.; Ma, B.; Lei, G.; Liu, Y.F.; Wei, W.; Wu, H.J.; Chen, L.J.; Chen, H.W.; et al. The ethylene receptor ETR2 delays floral transition and affects starch accumulation in rice. Plant Cell 2009, 21, 1473–1494. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Zhang, W.; Yin, Z.; Wen, C.K. Rice Constitutive Triple-Response2 is involved in the ethylene-receptor signalling and regulation of various aspects of rice growth and development. J. Exp. Bot. 2013, 64, 4863–4875. [Google Scholar] [CrossRef]
- Achard, P.; Baghour, M.; Chapple, A.; Hedden, P.; Van Der Straeten, D.; Genschik, P.; Moritz, T.; Harberd, N.P. The plant stress hormone ethylene controls floral transition via DELLA-dependent regulation of floral meristem-identity genes. Proc. Natl. Acad. Sci. USA 2007, 104, 6484–6489. [Google Scholar] [CrossRef]
- Xu, M.; Li, X.; Xie, W.; Lin, C.; Wang, Q.; Tao, Z. ETHYLENE INSENSITIVE3/EIN3-LIKE1 modulate FLOWERING LOCUS C expression via histone demethylase interaction. Plant Physiol. 2023, kiad131. [Google Scholar] [CrossRef]
- Morita, S.; Murakoshi, Y.; Chisaka, K.; Harada, T.; Satoh, S.J.J.o.P.B. Early flowering and increased expression of a FLOWERING LOCUS T-like gene in chrysanthemum transformed with a mutated ethylene receptor gene mDG-ERS1(etr1-4). J. Plant Biol. 2012, 55, 398–405. [Google Scholar] [CrossRef]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yoshimi, O.; Nobutaka, M.; Masaru, N.; Tsuyoshi, N.; Shingo, N.; Ko, K.; Masaru, O.T. Novel vector systems to accelerate functional analysis of transcription factors using chimeric repressor gene-silencing technology (CRES-T). Plant Biotechnol. 2011, 28, 201–210. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Sumitomo, K.; Narumi, T.; Satoh, S.; Hisamatsu, T. Involvement of the ethylene response pathway in dormancy induction in chrysanthemum. J. Exp. Bot. 2008, 59, 4075–4082. [Google Scholar] [CrossRef]
- Son, K.; Yu, S.; Shin, W.; Han, K.; Kang, K. A simple guideline to assess the characteristics of RNA-seq data. Biomed Res. Int. 2018, 2906292. [Google Scholar] [CrossRef]
- Zhang, N.; Zhou, S.; Yang, D.; Fan, Z. Revealing shared and distinct genes responding to JA and SA signaling in Arabidopsis by Meta-analysis. Front. Plant Sci. 2020, 11, 908. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Serrano, G.; Herrera-Palau, R.; Romero, J.M.; Serrano, A.; Coupland, G.; Valverde, F. Chlamydomonas CONSTANS and the evolution of plant photoperiodic signaling. Curr. Biol. 2009, 19, 359–368. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, F.; Bai, J.; He, Y. BrpSPL9 (Brassica rapa ssp. pekinensis SPL9) controls the earliness of heading time in Chinese cabbage. Plant Biotechnol. J. 2014, 12, 312–321. [Google Scholar] [CrossRef]
- Matar, S.; Kumar, A.; Holtgräwe, D.; Weisshaar, B.; Melzer, S. The transition to flowering in winter rapeseed during vernalization. Plant Cell Environ. 2021, 44, 506–518. [Google Scholar] [CrossRef] [PubMed]
- Gou, J.; Tang, C.; Chen, N.; Wang, H.; Debnath, S.; Sun, L.; Flanagan, A.; Tang, Y.; Jiang, Q.; Allen, R.D.; et al. SPL7 and SPL8 represent a novel flowering regulation mechanism in switchgrass. New Phytol. 2019, 222, 1610–1623. [Google Scholar] [CrossRef] [PubMed]
- Nakano, Y.; Takase, T.; Takahashi, S.; Sumitomo, K.; Higuchi, Y.; Hisamatsu, T. Chrysanthemum requires short-day repeats for anthesis: Gradual CsFTL3 induction through a feedback loop under short-day conditions. Plant Sci. 2019, 283, 247–255. [Google Scholar] [CrossRef] [PubMed]
- Preston, J.C.; Hileman, L.C. SQUAMOSA-PROMOTER BINDING PROTEIN 1 initiates flowering in Antirrhinum majus through the activation of meristem identity genes. Plant J. 2010, 62, 704–712. [Google Scholar] [CrossRef]
- Gao, Y.; Gao, Y.; Wu, Z.; Bu, X.; Fan, M.; Zhang, Q. Characterization of TEMINAL FLOWER1 homologs CmTFL1c gene from Chrysanthemum morifolium. Plant Mol. Biol. 2019, 99, 587–601. [Google Scholar] [CrossRef]
- Chen, W.J.; Zhu, T. Networks of transcription factors with roles in environmental stress response. Trends Plant Sci. 2004, 9, 591–596. [Google Scholar] [CrossRef]
- Lai, X.; Stigliani, A.; Vachon, G.; Carles, C.; Smaczniak, C.; Zubieta, C.; Kaufmann, K.; Parcy, F. Building transcription factor binding site models to understand gene regulation in plants. Mol. Plant 2019, 12, 743–763. [Google Scholar] [CrossRef]
- Matías-Hernández, L.; Aguilar-Jaramillo, A.E.; Cigliano, R.A.; Sanseverino, W.; Pelaz, S. Flowering and trichome development share hormonal and transcription factor regulation. J. Exp. Bot. 2016, 67, 1209–1219. [Google Scholar] [CrossRef]
- Higuchi, Y.; Narumi, T.; Oda, A.; Nakano, Y.; Sumitomo, K.; Fukai, S.; Hisamatsu, T. The gated induction system of a systemic floral inhibitor, antiflorigen, determines obligate short-day flowering in chrysanthemums. Proc. Natl. Acad. Sci. USA 2013, 110, 17137–17142. [Google Scholar] [CrossRef] [PubMed]
- Cong, H.; Li, Z.; Xu, L. Characterizing developmental and inducible differentiation between juvenile and adult plants of Aechmea fasciata treated with ethylene by transcriptomic analysis. Plant Growth Regul. 2013, 69, 247–257. [Google Scholar] [CrossRef]
- Li, Z.; Wang, J.; Zhang, X.; Lei, M.; Fu, Y.; Zhang, J.; Wang, Z.; Xu, L. Transcriptome sequencing determined flowering pathway genes in Aechmea fasciata Treated with Ethylene. J. Plant Growth Regul. 2016, 35, 316–329. [Google Scholar] [CrossRef]
- KonishI, K.; Kajihara, S.; Kageyama, Y. Inducing rosette formation in chrysanthemum plants by ethephon treatment. J. Jpn. Soc. Hortic. Sci. 1985, 54, 87–93. [Google Scholar] [CrossRef]
- Zhu, C.; Huang, Z.; Sun, Z.; Feng, S.; Wang, S.; Wang, T.; Yuan, X.; Zhong, L.; Cheng, Y.; Bao, M.; et al. The mutual regulation between DcEBF1/2 and DcEIL3-1 is involved in ethylene induced petal senescence in carnation (Dianthus caryophyllus L.). Plant J. 2023. Epub ahead of print. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, Y.; Tu, Y.; Wang, Y.; Cheng, W.; Yang, Y. Overexpression of an EIN3-binding F-box protein2-like gene caused elongated fruit shape and delayed fruit development and ripening in tomato. Plant Sci. 2018, 272, 131–141. [Google Scholar] [CrossRef]
- Yang, Y.; Wu, Y.; Pirrello, J.; Regad, F.; Bouzayen, M.; Deng, W.; Li, Z. Silencing Sl-EBF1 and Sl-EBF2 expression causes constitutive ethylene response phenotype, accelerated plant senescence, and fruit ripening in tomato. J. Exp. Bot. 2010, 61, 697–708. [Google Scholar] [CrossRef]
- Upadhyay, R.K.; Soni, D.K.; Singh, R.; Dwivedi, U.N.; Pathre, U.V.; Nath, P.; Sane, A.P. SlERF36, an EAR-motif-containing ERF gene from tomato, alters stomatal density and modulates photosynthesis and growth. J. Exp. Bot. 2013, 64, 3237–3247. [Google Scholar] [CrossRef]
- Chen, Y.; Zhang, L.; Zhang, H.; Chen, L.; Yu, D. ERF1 delays flowering through direct inhibition of FLOWERING LOCUS T expression in Arabidopsis. J. Integr. Plant Biol. 2021, 63, 1712–1723. [Google Scholar] [CrossRef]
- Huang, Y.; Xing, X.; Tang, Y.; Jin, J.; Ding, L.; Song, A.; Chen, S.; Chen, F.; Jiang, J.; Fang, W. An ethylene-responsive transcription factor and a flowering locus KH domain homologue jointly modulate photoperiodic flowering in chrysanthemum. Plant Cell Environ. 2022, 45, 1442–1456. [Google Scholar] [CrossRef]
- Cheng, H.; Yu, Y.; Zhai, Y.; Wang, L.; Wang, L.; Chen, S.; Chen, F.; Jiang, J. An ethylene-responsive transcription factor and a B-box protein coordinate vegetative growth and photoperiodic flowering in chrysanthemum. Plant Cell Environ. 2023, 46, 440–450. [Google Scholar] [CrossRef]
- Oda, A.; Higuchi, Y.; Hisamatsu, T. Constitutive expression of CsGI alters critical night length for flowering by changing the photo-sensitive phase of anti-florigen induction in chrysanthemum. Plant Sci. 2020, 293, 110417. [Google Scholar] [CrossRef] [PubMed]
- Burg, S.P. Ethylene, plant senescence and abscission. Plant Physiol. 1968, 43, 1503–1511. [Google Scholar] [PubMed]
- Sehar, Z.; Iqbal, N.; Fatma, M.; Rather, B.A.; Albaqami, M.; Khan, N.A. Ethylene suppresses abscisic acid, modulates antioxidant system to counteract arsenic-inhibited photosynthetic performance in the presence of selenium in mustard. Front. Plant Sci. 2022, 13, 852704. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Niki, T.; Nishijima, T.; Douzono, M.; Hisamatsu, T.J.J.o.P.; Science, H. Roles of CmFL, CmAFL1, and CmSOC1 in the transition from vegetative to reproductive growth in Chrysanthemum morifolium Ramat. J. Pomol. Hortic. Sci. 2009, 84, 447–453. [Google Scholar]
- Oda, A.; Narumi, T.; Li, T.; Kando, T.; Higuchi, Y.; Sumitomo, K.; Fukai, S.; Hisamatsu, T. CsFTL3, a chrysanthemum FLOWERING LOCUS T-like gene, is a key regulator of photoperiodic flowering in chrysanthemums. J. Exp. Bot. 2012, 63, 1461–1477. [Google Scholar] [CrossRef]
- Xu, M.; Hu, T.; Zhao, J.; Park, M.Y.; Earley, K.W.; Wu, G.; Yang, L.; Poethig, R.S. Developmental Functions of miR156-Regulated SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) Genes in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006263. [Google Scholar] [CrossRef]
- Higuchi, Y.; Hisamatsu, T. CsTFL1, a constitutive local repressor of flowering, modulates floral initiation by antagonising florigen complex activity in chrysanthemum. Plant Sci. 2015, 237, 1–7. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, H.; Zhou, M.; Si, Y.; Li, W.; Wang, L.; Chen, S.; Chen, F.; Jiang, J. Transcriptome Analysis of Ethylene Response in Chrysanthemum moriflolium Ramat. with an Emphasis on Flowering Delay. Horticulturae 2023, 9, 428. https://doi.org/10.3390/horticulturae9040428
Cheng H, Zhou M, Si Y, Li W, Wang L, Chen S, Chen F, Jiang J. Transcriptome Analysis of Ethylene Response in Chrysanthemum moriflolium Ramat. with an Emphasis on Flowering Delay. Horticulturae. 2023; 9(4):428. https://doi.org/10.3390/horticulturae9040428
Chicago/Turabian StyleCheng, Hua, Min Zhou, Yuyang Si, Wenjie Li, Likai Wang, Sumei Chen, Fadi Chen, and Jiafu Jiang. 2023. "Transcriptome Analysis of Ethylene Response in Chrysanthemum moriflolium Ramat. with an Emphasis on Flowering Delay" Horticulturae 9, no. 4: 428. https://doi.org/10.3390/horticulturae9040428
APA StyleCheng, H., Zhou, M., Si, Y., Li, W., Wang, L., Chen, S., Chen, F., & Jiang, J. (2023). Transcriptome Analysis of Ethylene Response in Chrysanthemum moriflolium Ramat. with an Emphasis on Flowering Delay. Horticulturae, 9(4), 428. https://doi.org/10.3390/horticulturae9040428







