Genome-Wide Identification and Expression Analysis of the fw2.2-like Gene Family in Pear
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of the fw2.2-like Family in Pear
2.2. Analysis of fw2.2-like Gene Structure and Conserved Motifs
2.3. Analysis of Cis-Acting Elements in fw2.2-like Gene Promoters
2.4. Chromosomal Localization and Gene Duplication
2.5. Phylogenetic Analysis of fw2.2-like Proteins
2.6. Plant Material
2.7. Measuring the Characteristics of Fruit at Early Stages of Pear Fruit Development
2.8. Measurements of Receptacle/Fruit Cell Size and Number
2.9. RNA Isolation and qRT-PCR
3. Results
3.1. Identification and Sequence Analysis of fw2.2-likes in Pear
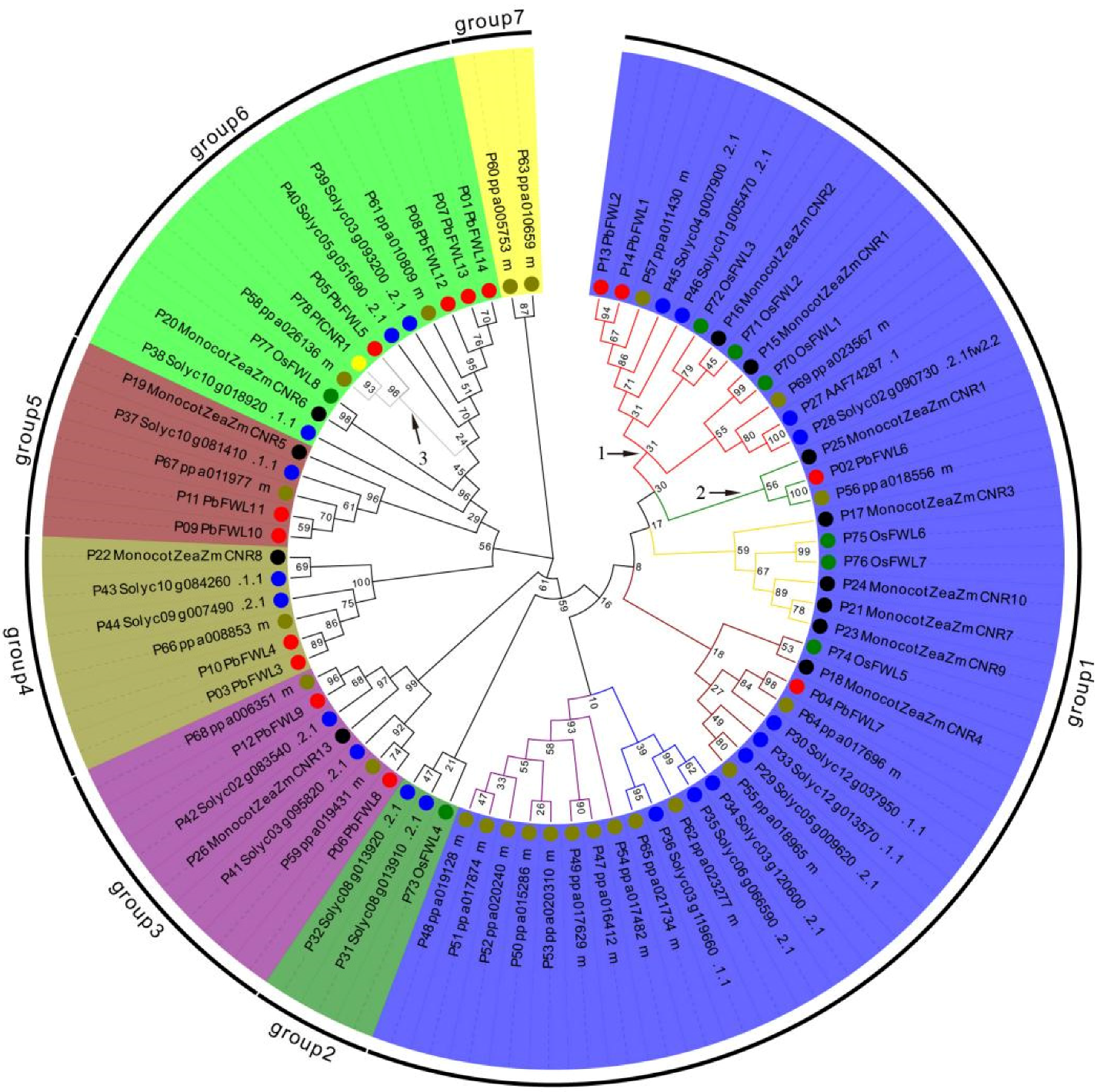
3.2. Phylogenetic Analyses of PbFWL Gene Family Members
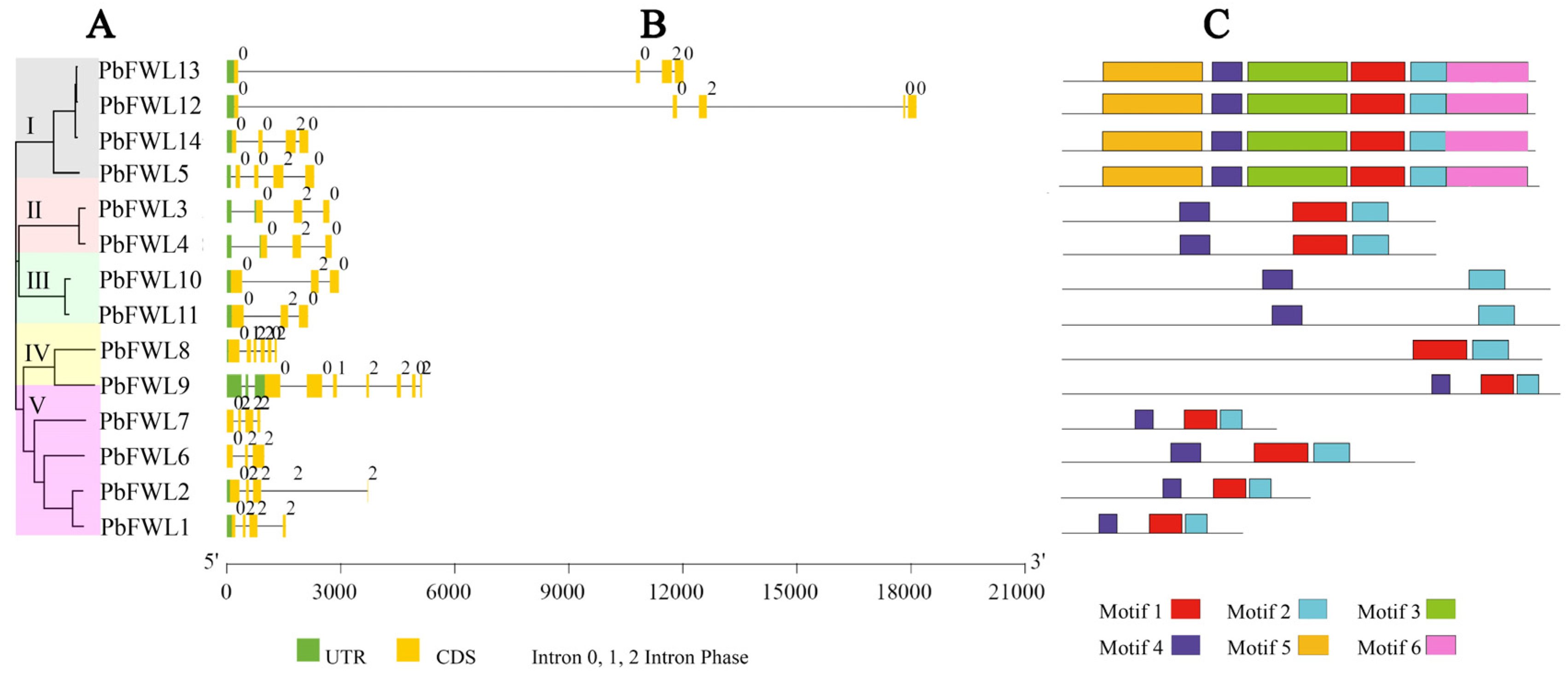
3.3. Gene Structure and Motif Analysis
3.4. Chromosomal Distribution and Synteny Analysis of PbFWLs in Pyrus Bretschneideri
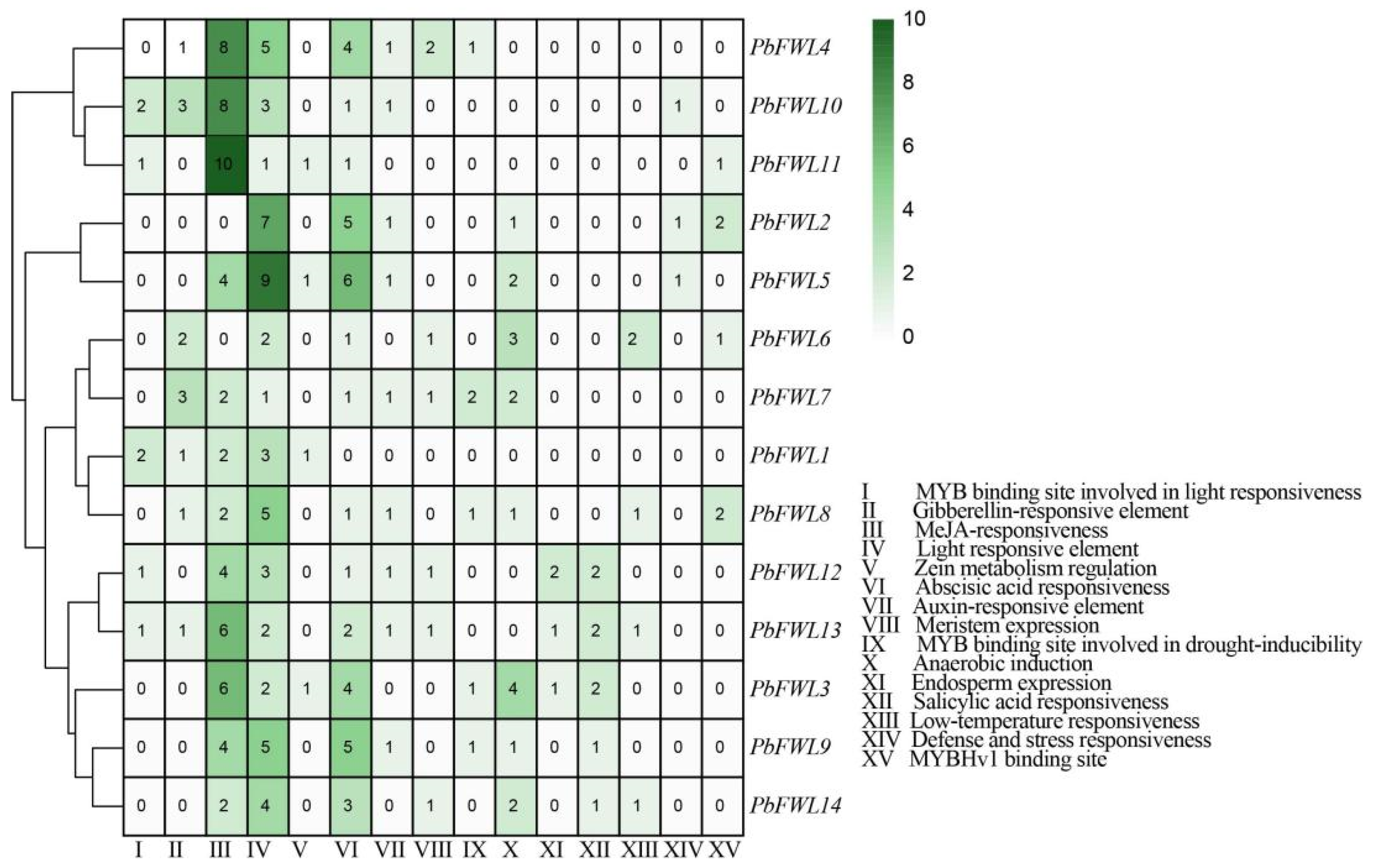
3.5. Analysis of the fw2.2-like Gene Family Promoter Elements in Pear
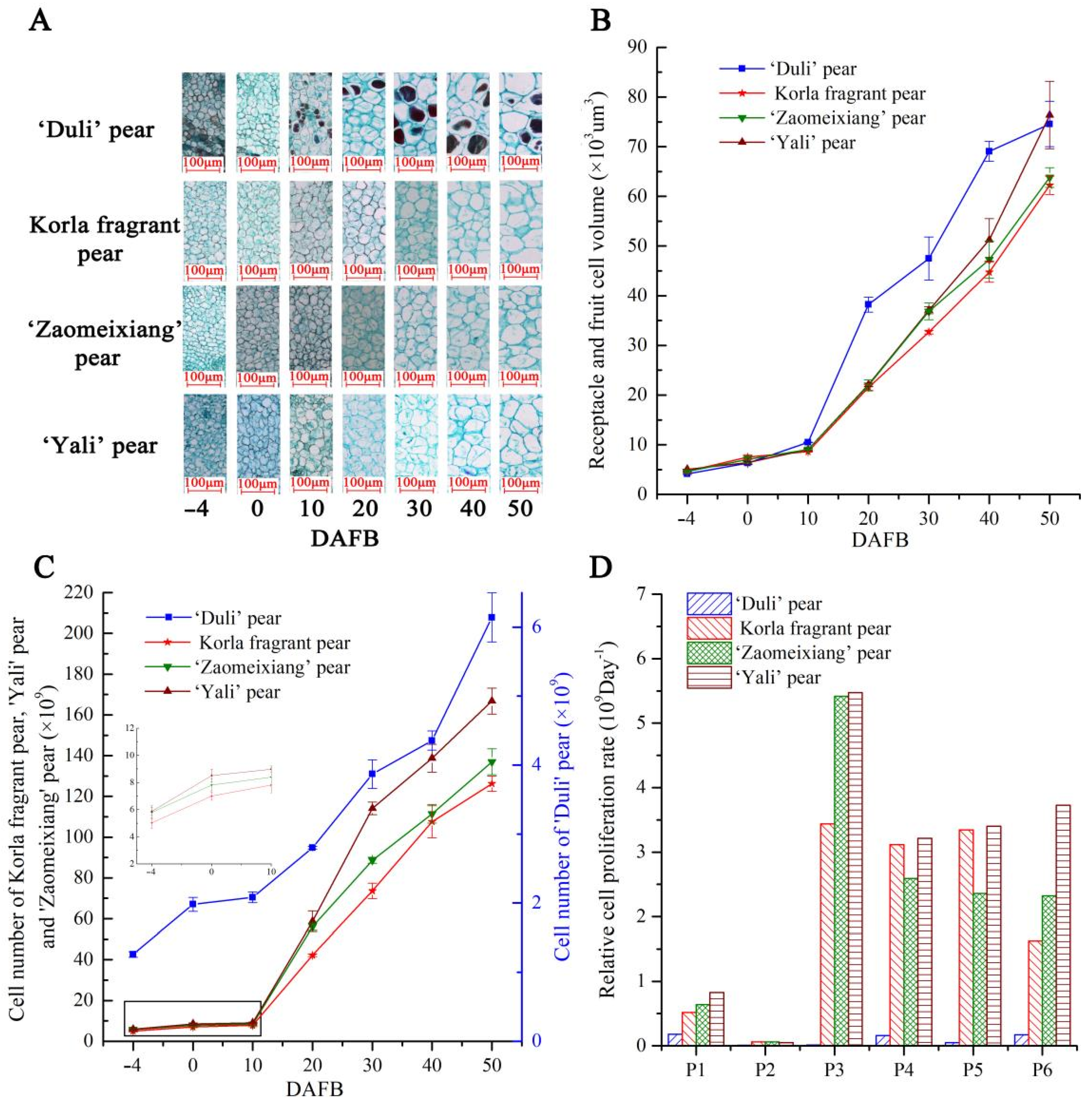
3.6. Differential Expression of the PbFWL Genes during Early Fruit Development among Pear Varieties with Different Fruit Sizes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gillaspy, G.; Gruissem, B.D. Fruits: A develomental perspective. Plant Cell. 1993, 5, 1439–1451. [Google Scholar] [CrossRef] [PubMed]
- Monforte, A.J.; Díaz, A.; Caño-Delgado, A.; van der Knaap, E. The genetic basis of fruit morphology in horticultural crops: Lessons from tomato and melon. J. Exp. Bot. 2013, 65, 4625–4637. [Google Scholar] [CrossRef] [PubMed]
- Frary, A.; Nesbitt, T.C.; Frary, A.; Grandillo, S.; van der Knaap, E.; Cong, B.; Liu, J.; Meller, J.; Elber, R.; Alpert, K.B.; et al. fw2.2: A quantitative trait locus key to the evolution of tomato fruit size. Science 2000, 289, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, N.; Beemster, G.T.; Inzé, D. David and goliath: What can the tiny weed arabidopsis teach us to improve biomass production in crops? Curr. Opin. Plant Biol. 2009, 12, 157–164. [Google Scholar] [CrossRef] [PubMed]
- Hussain, Q.; Shi, J.; Scheben, A.; Zhan, J.; Wang, X.; Liu, G.; Yan, G.; King, G.J.; Edwards, D.; Wang, H. Genetic and signalling pathways of dry fruit size: Targets for genome editing-based crop improvement. Plant Biotechnol. J. 2019, 18, 1124–1140. [Google Scholar] [CrossRef]
- Pan, Y.; Wang, Y.; McGregor, C.; Liu, S.; Luan, F.; Gao, M.; Weng, Y. Genetic architecture of fruit size and shape variation in cucurbits: A comparative perspective. Theor. Appl. Genet. 2019, 133, 1–21. [Google Scholar] [CrossRef]
- Tanksley, S.D. The genetic, developmental, and molecular bases of fruit size and shape variation in tomato. Plant Cell. 2004, 16, S181–S189. [Google Scholar] [CrossRef]
- Alpert, K.B.; Tanksley, S.D. High-resolution mapping and isolation of a yeast artificial chromosome contig containing fw2.2: A major fruit weight quantitative trait locus in tomato. Proc. Natl. Acad. Sci. USA 1996, 93, 15503–15507. [Google Scholar] [CrossRef]
- Guo, M.; Rupe, M.A.; Dieter, J.A.; Zou, J.; Spielbauer, D.; Duncan, K.E.; Howard, R.J.; Hou, Z.; Simmons, C.R. Cell number regulator1 affects plant and organ size in maize: Implications for crop yield enhancement and heterosis. Plant Cell. 2010, 22, 1057–1073. [Google Scholar] [CrossRef]
- Thibivilliers, S.; Farmer, A.; Libault, M. Biological and cellular functions of the microdomain-associated FWL/CNR protein family in plants. Plants 2020, 9, 377. [Google Scholar] [CrossRef]
- Arthur, B.; Frédéric, G.; Nathalie, G.; Christian, C. In search of the still unknown function of FW2.2/CELL NUMBER REGU-LATOR, a major regulator of fruit size in tomato. J. Exp. Bot. 2021, 72, 5300–5311. [Google Scholar] [CrossRef]
- Li, Z.; He, C. Physalis floridana Cell Number Regulator1 encodes a cell membrane-anchored modulator of cell cycle and neg-atively controls fruit size. J. Exp. Bot. 2014, 66, 257–270. [Google Scholar] [CrossRef]
- De Franceschi, P.; Stegmeir, T.; Cabrera, A.; van der Knaap, E.; Rosyara, U.R.; Sebolt, A.M.; Dondini, L.; Dirlewanger, E.; Quero-Garcia, J.; Campoy, J.A.; et al. Cell number regulator genes in Prunus provide candidate genes for the control of fruit size in sweet and sour cherry. Mol. Breed. 2013, 32, 311–326. [Google Scholar] [CrossRef]
- Dahan, Y.; Rosenfeld, R.; Zadiranov, V.; Irihimovitch, V. A proposed conserved role for an avocado fw2.2-like gene as a negative regulator of fruit cell division. Planta 2010, 232, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Zhang, L.; Jiang, Y.; Huang, T.; Chen, X.; Liu, Y.; Wu, J.; Yang, X.; Lin, S. EjFWLs are repressors of cell division during early fruit morphogenesis of loquat. Sci. Hortic. 2021, 287, 110261. [Google Scholar] [CrossRef]
- Kuang, C.; Li, J.; Liu, H.; Sun, X.; Zhu, X.; Hua, W. Genome-wide identification and evolutionary analysis of the fruit-weight 2.2-like gene family in polyploid oilseed rape (Brassica napus L.). DNA Cell Biol. 2020, 39, 766–782. [Google Scholar] [CrossRef]
- Xu, J.; Xiong, W.; Cao, B.; Yan, T.; Luo, T.; Fan, T.; Luo, M. Molecular characterization and functional analysis of “fruit-weight 2.2-like” gene family in rice. Planta 2013, 238, 643–655. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhang, T.; Yuan, Z. Pomegranate PLAC8 family. Acta Hortic. 2019, 1245, 35–40. [Google Scholar] [CrossRef]
- Li, J.; Zhang, M.; Li, X.; Khan, A.; Kumar, S.; Allan, A.C.; Lin-Wang, K.; Espley, R.V.; Wang, C.; Wang, R.; et al. Pear genetics: Recent advances, new prospects, and a roadmap for the future. Hortic. Res. 2021, 9, uhab040. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H.; et al. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef]
- Zhang, F.; Jiang, Z.Q.; Li, S.Q.; He, T.M. The development factors of Pyrus sinkiangensis Korla fragrant pear industry. Chin. Agric. Sci. Bull. 2021, 37, 159–164. (In Chinese) [Google Scholar]
- Zhang, R.-P.; Wu, J.; Li, X.-G.; Khan, M.A.; Chen, H.; Korban, S.S.; Zhang, S.-L. An AFLP, SRAP, and SSR genetic linkage map and identification of QTLs for fruit traits in Pear (Pyrus L.). Plant Mol. Biol. Rep. 2012, 31, 678–687. [Google Scholar] [CrossRef]
- Yamamoto, T.; Terakami, S.; Takada, N.; Nishio, S.; Onoue, N.; Nishitani, C.; Kunihisa, M.; Inoue, E.; Iwata, H.; Hayashi, T.; et al. Identification of QTLs controlling harvest time and fruit skin color in Japanese pear (Pyrus pyrifolia Nakai). Breed. Sci. 2014, 64, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Marchler-Bauer, A.; Bo, Y.; Han, L.; He, J.; Lanczycki, C.J.; Lu, S.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; et al. CDD/SPARCLE: Functional classification of proteins via subfamily domain architectures. Nucleic Acids Res. 2017, 45, D200–D203. [Google Scholar] [CrossRef] [PubMed]
- Finn, R.D.; Coggill, P.; Eberhardt, R.Y.; Eddy, S.R.; Mistry, J.; Mitchell, A.L.; Potter, S.C.; Punta, M.; Qureshi, M.; Sangrador-Vegas, A.; et al. The Pfam protein families database: Towards a more sustainable future. Nucleic Acids Res. 2016, 44, D279–D285. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A top-down strategy to augment the power for predicting plant protein subcellular lo-calization. PLoS ONE 2010, 28, e11335. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Gu, Z.; Cavalcanti, A.; Chen, F.-C.; Bouman, P.; Li, W.-H. Extent of gene duplication in the genomes of drosophila, nematode, and yeast. Mol. Biol. Evol. 2002, 19, 256–262. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, X.; Yue, J.-X.; Tian, D.; Chen, J.-Q. Recent duplications dominate NBS-encoding gene expansion in two woody species. Mol. Genet. Genom. 2008, 280, 187–198. [Google Scholar] [CrossRef]
- Wang, L.; Guo, K.; Li, Y.; Tu, Y.; Hu, H.; Wang, B.; Cui, X.; Peng, L. Expression profiling and integrative analysis of the CE-SA/CSL superfamily in rice. BMC Plant Biol. 2010, 10, 282. [Google Scholar] [CrossRef]
- Huang, Z.; Duan, W.; Song, X.; Tang, J.; Peng, W.; Bei, Z.; Hou, X. Retention, molecular evolution, and expression divergence of the auxin/indole acetic acid and auxin response factor gene families inbrassica rapashed light on their evolution patterns in plants. Genome Biol. Evol. 2015, 8, 302–316. [Google Scholar] [CrossRef] [PubMed]
- Nesbitt, T.C.; Tanksley, S.D. Comparative sequencing in the genus lycopersicon: Implications for the evolution of fruit size in the domestication of cultivated tomatoes. Genetics 2002, 162, 365–379. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 337, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.Z.; Ma, X.T. Comparison of several common methods to measure the volume of fruit. China Fruits 1995, 2, 44–45. (In Chinese) [Google Scholar]
- Zhang, C.; Tanabe, K.; Wang, S.; Tamura, F.; Yoshida, A.; Matsumoto, K. The impact of cell division and cell enlargement on the evolution of fruit size in Pyrus pyrifolia. Ann. Bot. 2006, 98, 537–543. [Google Scholar] [CrossRef]
- Cong, B.; Tanksley, S.D. FW2.2 and cell cycle control in developing tomato fruit: A possible example of gene co-option in the evolution of a novel organ. Plant Mol. Biol. 2006, 62, 867–880. [Google Scholar] [CrossRef]
- Tian, J.; Zeng, B.; Luo, S.-P.; Li, X.-G.; Wu, B.; Li, J. Cloning, localization and expression analysis of two fw2.2-like genes in small- and large-fruited pear species. J. Integr. Agric. 2016, 15, 282–294. [Google Scholar] [CrossRef]
- Libault, M.; Stacey, G. Evolution of FW2.2-like (FWL) and PLAC8 genes in eukaryotes. Plant Signal. Behav. 2010, 5, 1226–1228. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Wang, L.; Yan, L.; Li, Q.; Yong, B.; Zhu, W. Evolutionary developmental mechanisms underlying the origin and diversification of the fruits. Sci. Sin. Vitae 2019, 49, 301–319. [Google Scholar] [CrossRef]
- Azzi, L. Etude du rôle de FW2.2 dans le développement du fruit de tomate. In Pour Obtenir le Grade de Docteur; Université Sciences et Technologies-Bordeaux I: Bordeaux, France, 2013; Available online: https://theses.hal.science/tel-01089718 (accessed on 16 December 2013).
- Schnable, P.S.; Ware, D.; Fulton, R.S.; Stein, J.C.; Wei, F.; Pasternak, S.; Liang, C.; Zhang, J.; Fulton, L.; Graves, T.A.; et al. The B73 maize genome: Complexity, diversity, and dynamics. Science 2009, 326, 1112–1115. [Google Scholar] [CrossRef] [PubMed]
- Cabreira-Cagliari, C.; Dias, N.D.C.; Bohn, B.; Fagundes, D.G.D.S.; Margis-Pinheiro, M.; Bodanese-Zanettini, M.H.; Cagliari, A. Revising the PLAC8 gene family: From a central role in differentiation, proliferation, and apoptosis in mammals to a mul-tifunctional role in plants. Genome 2018, 61, 857–865. [Google Scholar] [CrossRef] [PubMed]
- Song, W.Y.; Choi, Y.I.; Shim, D.; Kim, D.Y.; Noh, E.W.; Martinoia, E.; Lee, Y. Transgenic poplar for phytoremediation. In Biotechnology and Sustainable Agriculture 2006 and Beyond; Springer: Berlin, Germany, 2007; pp. 265–271. [Google Scholar] [CrossRef]
- Arnoys, E.J.; Wang, J.L. Dual localization: Proteins in extracellular and intracellular compartments. Acta Histochem. 2007, 109, 89–110. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Fujita, M.; Satoh, R.; Maruyama, K.; Parvez, M.M.; Seki, M.; Hiratsu, K.; Ohme-Takagi, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in arabidopsis. Plant Cell 2005, 17, 3470–3488. [Google Scholar] [CrossRef]
- Robert-Seilaniantz, A.; Grant, M.; Jones, J.D.G. Hormone crosstalk in plant disease and defense: More than just JASMONATE-SALICYLATE antagonism. Annu. Rev. Phytopathol. 2011, 49, 317–343. [Google Scholar] [CrossRef]
- Cong, B.; Liu, J.; Tanksley, S.D. Natural alleles at a tomato fruit size quantitative trait locus differ by heterochronic regulatory mutations. Proc. Natl. Acad. Sci. USA 2002, 99, 13606–13611. [Google Scholar] [CrossRef]
- Liu, J.; Cong, B.; Tanksley, S.D.; Mergaert, P.; Nikovics, K.; Kelemen, Z.; Maunoury, N.; Vaubert, D.; Kondorosi, A.; Kondorosi, E. Generation and analysis of an artificial gene dosage series in tomato to study the mechanisms by which the cloned quantitative trait locus fw2.2 controls fruit size. Plant Physiol. 2003, 132, 292–299. [Google Scholar] [CrossRef]
- Song, W.Y.; Martinoia, E.; Lee, J.; Kim, D.; Kim, D.Y.; Vogt, E.; Shim, D.; Choi, K.S.; Hwang, I.; Lee, Y. A novel family of cys-rich membrane proteins mediates cadmium resistance in Arabidopsis. Plant Physiol. 2004, 135, 1027–1039. [Google Scholar] [CrossRef]



| Gene Name | Gene ID | Chr | Gene Location (bp) | AA | MW (kDa) | pI | Instability Index(Ⅱ) | GRAVY | cys | S.L. |
|---|---|---|---|---|---|---|---|---|---|---|
| PbFWL1 | rna5458 | 16 | 312,566(-)314,118 | 151 | 16.99 | 7.41 | 54.00 | −0.124 | 16 | Cell membrane |
| PbFWL2 | rna5456 | 16 | 310,397(-)314,118 | 208 | 23.08 | 5.67 | 54.46 | −0.333 | 15 | Cell membrane |
| PbFWL3 | rna12156 | 4 | 11,872,621(-)11,875,857 | 246 | 26.72 | 5.29 | 56.18 | −0.363 | 16 | Cell membrane |
| PbFWL4 | rna4422 | 12 | 12,008,588(-)12,011,010 | 251 | 27.19 | 4.93 | 56.13 | −0.445 | 15 | Cell membrane |
| PbFWL5 | rna27717 | 7 | 6,529,212(-)6,531,603 | 242 | 26.91 | 4.79 | 56.34 | −0.314 | 17 | Cell membrane |
| PbFWL6 | rna1124 | 3 | 23,944,140(-)23,945,131 | 178 | 19.54 | 7.79 | 53.62 | 0.022 | 20 | Cell membrane |
| PbFWL7 | rna4877 | 6 | 18,544,328(-)18,545,220 | 179 | 19.33 | 6.04 | 50.31 | −0.232 | 16 | Cell membrane |
| PbFWL8 | rna23574 | 7 | 17,632,755(-)17,634,097 | 242 | 27.16 | 5.81 | 45.29 | −0.135 | 19 | Cell membrane Nucleus |
| PbFWL9 | rna17541 | 15 | 14,880,874(-)14,886,180 | 415 | 47.51 | 6.58 | 49.00 | −0.484 | 18 | Cell membrane Nucleus |
| PbFWL10 | rna19452 | 12 | 1,210,378(-)1,213,360 | 180 | 19.42 | 4.87 | 60.41 | 0.239 | 17 | Cell membrane |
| PbFWL11 | rna6937 | 14 | 433,409(-)436,373 | 188 | 20.36 | 4.9 | 59.62 | 0.134 | 17 | Cell membrane |
| PbFWL12 | rna24884 | 7 | 19,812,175(-)19,824,369 | 239 | 26.55 | 5.28 | 47.81 | −0.365 | 17 | Cell membrane |
| PbFWL13 | rna27387 | 7 | 18,336,597(-)18,354,923 | 239 | 26.53 | 5.27 | 45.69 | −0.365 | 17 | Cell membrane |
| PbFWL14 | rna41755 | 1 | 5,027,259(-)5,029,618 | 239 | 26.47 | 5.28 | 41.77 | −0.318 | 17 | Cell membrane |
| Cell Division (C) | Expression | |||
|---|---|---|---|---|
| PbFWL1 | PbFWL2 | PbFWL3 | ||
| ‘Duli’ pear | 0.036 | 27.179 | 19.683 | 198.781 |
| Korla fragrant pear | 0.126 | 12.127 | 5.962 | 41.572 |
| ‘Zaomeixiang’ pear | 0.124 | 9.432 | 5.305 | 28.070 |
| ‘Yali’ pear | 0.132 | 4.067 | 3.410 | 15.589 |
| Correlation | r = −0.958 | r = −0.996 | r = −0.996 | |
| P = 0.042 | P = 0.004 | P = 0.004 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pu, X.; Tian, J.; Li, J.; Wen, Y. Genome-Wide Identification and Expression Analysis of the fw2.2-like Gene Family in Pear. Horticulturae 2023, 9, 429. https://doi.org/10.3390/horticulturae9040429
Pu X, Tian J, Li J, Wen Y. Genome-Wide Identification and Expression Analysis of the fw2.2-like Gene Family in Pear. Horticulturae. 2023; 9(4):429. https://doi.org/10.3390/horticulturae9040429
Chicago/Turabian StylePu, Xiaoqiu, Jia Tian, Jiang Li, and Yue Wen. 2023. "Genome-Wide Identification and Expression Analysis of the fw2.2-like Gene Family in Pear" Horticulturae 9, no. 4: 429. https://doi.org/10.3390/horticulturae9040429
APA StylePu, X., Tian, J., Li, J., & Wen, Y. (2023). Genome-Wide Identification and Expression Analysis of the fw2.2-like Gene Family in Pear. Horticulturae, 9(4), 429. https://doi.org/10.3390/horticulturae9040429




