Transcriptomic Analysis of Salicylic Acid Promoting Seed Germination of Melon under Salt Stress
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Germination Treatments
2.2. Determination of SOD, CAT, POD Activities, and Proline (Pro) Content
2.3. RNA-Seq Library Preparation and Sequencing
2.4. De Novo Assembly, Functional Annotation, and Enrichment Analysis of Differentially Expressed Genes
2.5. qRT-PCR Analysis
2.6. Statistical Analysis
3. Results
3.1. Effect of Different Treatments on Germination of Melon Seeds
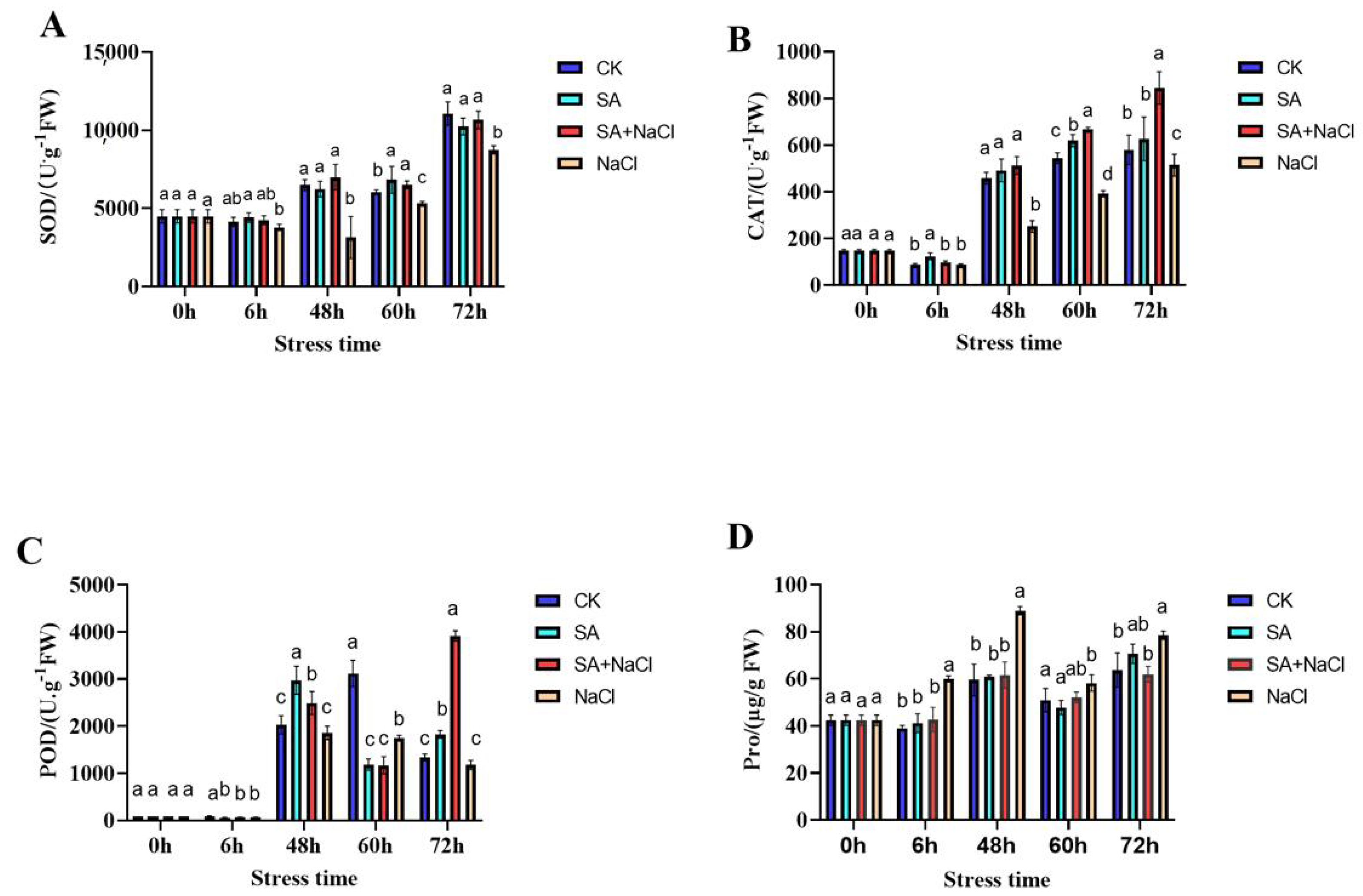
3.2. Salicylic Acid (SA)-Mediated Physiological Response to Salt Stress
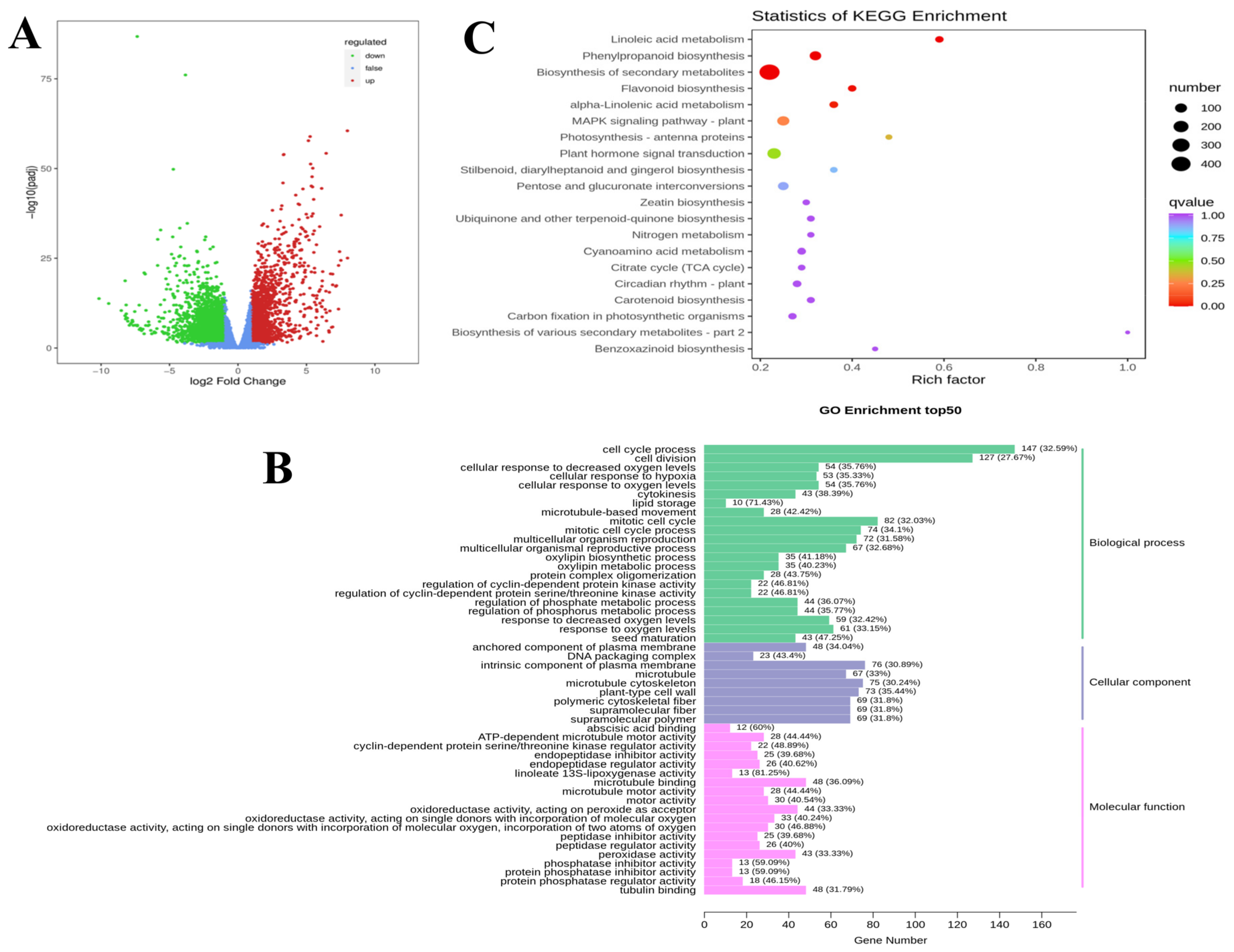
3.3. Transcriptome Data Analysis
3.4. Differential Gene Expression Analysis
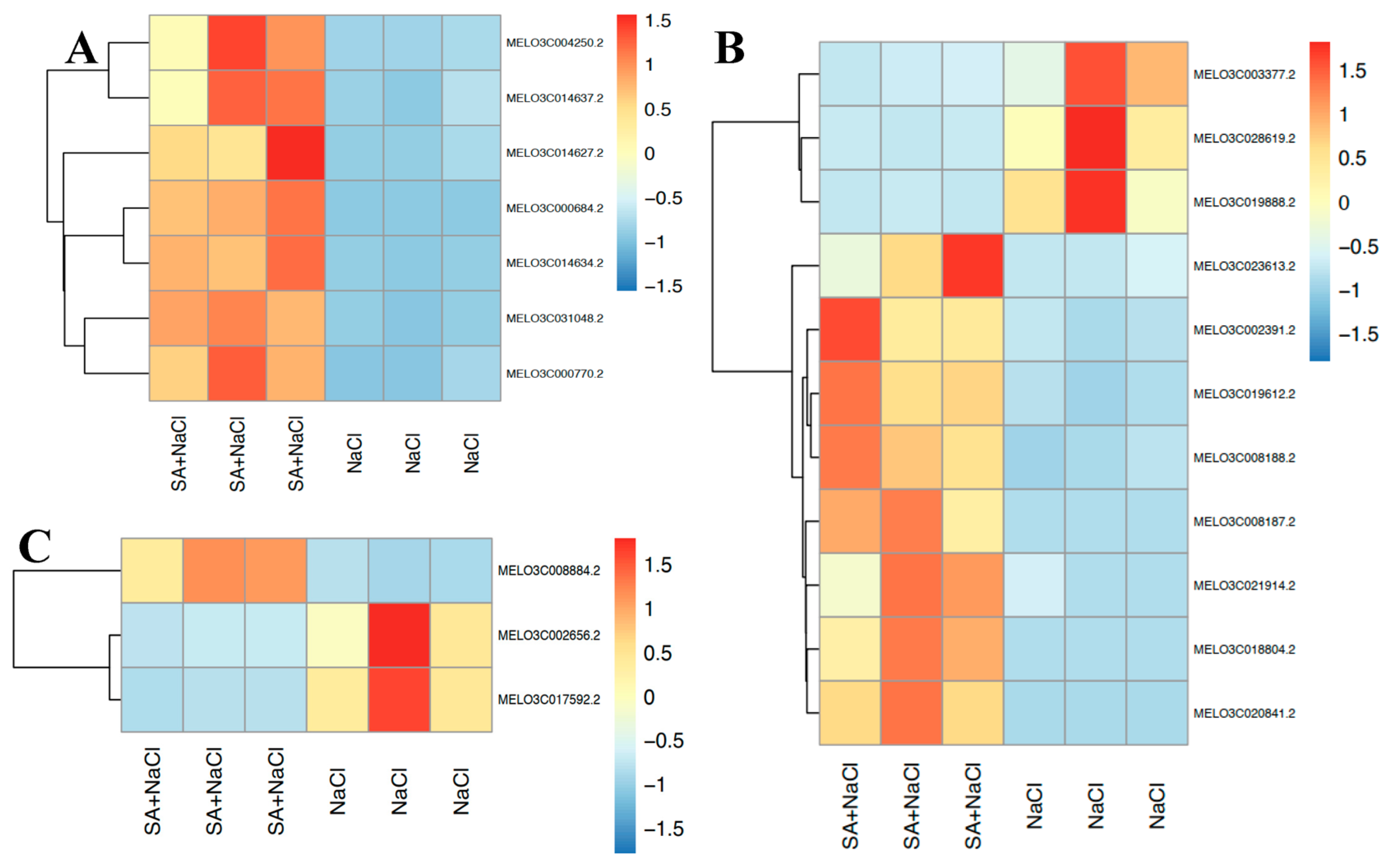
3.5. DEGs Related to Lipid Metabolism
3.6. DEGs Related to Biosynthesis of Secondary Metabolites
3.7. DEGs Related to Signal Transduction
3.8. Validation of Gene Expression Using qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Hazell, P.; Wood, S. Drivers of change in global agriculture. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008, 363, 495–515. [Google Scholar] [CrossRef] [PubMed]
- Qadir, M.; Quillérou, E.; Nangia, V.; Murtaza, G.; Singh, M.; Thomas, R.J. Economics of salt-induced land degradation and restoration. Nat. Resour. Forum 2014, 38, 282–295. [Google Scholar] [CrossRef]
- Jia, X.-M.; Wang, H.; Svetla, S.; Zhu, Y.-F.; Hu, Y.; Cheng, L.; Zhao, T.; Wang, Y.-X. Comparative physiological responses and adaptive strategies of apple Malus halliana to salt, alkali and saline-alkali stress. Sci. Hortic. 2019, 245, 154–162. [Google Scholar] [CrossRef]
- Hamann, T. The plant cell wall integrity maintenance mechanism—Concepts for organization and mode of action. Plant Cell Physiol. 2015, 56, 215–223. [Google Scholar] [CrossRef] [PubMed]
- Manishankar, P.; Wang, N.; Köster, P.; Alatar, A.A.; Kudla, J. Calcium signaling during salt stress and in the regulation of ion homeostasis. J. Exp. Bot. 2018, 69, 4215–4226. [Google Scholar] [CrossRef]
- Van Zelm, E.; Zhang, Y.; Testerink, C. Salt tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Zhang, N.; Zhao, B.; Zhang, H.-J.; Weeda, S.; Yang, C.; Yang, Z.-C.; Ren, S.; Guo, Y.-D. Melatonin promotes water-stress tolerance, lateral root formation, and seed germination in cucumber (Cucumis sativus L.). J. Pineal Res. 2013, 54, 15–23. [Google Scholar] [CrossRef]
- Uniyal, R.C.; Nautiyal, A.R. Seed germination and seedling extension growth in Ougeinia dalbergioides Benth under water and salinity stress. New For. 1998, 16, 265–272. [Google Scholar] [CrossRef]
- Liu, Z.; Ma, C.; Hou, L.; Wu, X.; Wang, D.; Zhang, L.; Liu, P. Exogenous SA affects rice seed germination under salt stress by regulating Na+/K+ balance and endogenous GAs and ABA homeostasis. Int. J. Mol. Sci. 2022, 23, 3293. [Google Scholar] [CrossRef]
- Luo, X.; Dai, Y.; Zheng, C.; Yang, Y.; Chen, W.; Wang, Q.; Chandrasekaran, U.; Du, J.; Liu, W.; Shu, K. The ABI4-RbohD/VTC2 regulatory module promotes reactive oxygen species (ROS) accumulation to decrease seed germination under salinity stress. New Phytol. 2021, 229, 950–962. [Google Scholar] [CrossRef]
- Guzmán-Murillo, M.A.; Ascencio, F.; Larrinaga-Mayoral, J.A. Guzmán-Murillo, M.A.; Ascencio, F.; Larrinaga-Mayoral, J.A. Germination and ROS detoxification in bell pepper (Capsicum annuum L.) under NaCl stress and treatment with microalgae extracts. Protoplasma 2013, 250, 33–42. [Google Scholar] [CrossRef]
- Khan, M.N.; Li, Y.; Khan, Z.; Chen, L.; Liu, J.; Hu, J.; Wu, H.; Li, Z. Nanoceria seed priming enhanced salt tolerance in rapeseed through modulating ROS homeostasis and α-amylase activities. J. Nanobiotechnol. 2021, 19, 276. [Google Scholar] [CrossRef]
- Taşgın, E.; Atıcı, Ö.; Nalbantoğlu, B.; Popova, L.P. Effects of salicylic acid and cold treatments on protein levels and on the activities of antioxidant enzymes in the apoplast of winter wheat leaves. Phytochemistry 2006, 67, 710–715. [Google Scholar] [CrossRef]
- Senaratna, T.; Touchell, D.; Bunn, E.; Dixon, K. Acetyl salicylic acid (Aspirin) and salicylic acid induce multiple stress tolerance in bean and tomato plants. Plant Growth Regul. 2000, 30, 157–161. [Google Scholar] [CrossRef]
- Bharti, A.; Garg, N. SA and AM symbiosis modulate antioxidant defense mechanisms and asada pathway in chickpea genotypes under salt stress. Ecotoxicol. Environ. Saf. 2019, 178, 66–78. [Google Scholar] [CrossRef]
- Garg, N.; Bharti, A. Salicylic acid improves arbuscular mycorrhizal symbiosis, and chickpea growth and yield by modulating carbohydrate metabolism under salt stress. Mycorrhiza 2018, 28, 727–746. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Li, L.; Yuan, F.; Chen, M. Exogenous salicylic acid improves the germination of Limonium bicolor seeds under salt stress. Plant Signal. Behav. 2019, 14, e1644595. [Google Scholar] [CrossRef]
- Wang, L.; Wang, Y.; Wang, X.; Li, Y.; Peng, F.; Wang, L. Regulation of POD activity by pelargonidin during vegetative growth in radish (Raphanus sativus L.). Sci Hortic. 2014, 174, 105–111. [Google Scholar] [CrossRef]
- Burger, Y.; Sa’ar, U.; Paris, H.S.; Lewinsohn, E.; Katzir, N.; Tadmor, Y.; Schaffer, A. Genetic variability for valuable fruit quality traits in Cucumis melo. Isr. J. Plant Sci. 2006, 54, 233–242. [Google Scholar] [CrossRef]
- Nunez-Palenius, H.G.; Gomez-Lim, M.; Ochoa-Alejo, N.; Grumet, R.; Lester, G.; Cantliffe, D.J. Melon fruits: Genetic diversity, physiology, and biotechnology features. Crit. Rev. Biotechnol. 2008, 28, 13–55. [Google Scholar] [CrossRef] [PubMed]
- Liang, S.X.; Gao, N.; Li, X.; Xi, X. Toxic effects of antimony on the seed germination and seedlings accumulation in Raphanus sativus L. radish and Brassica napus L. Mol. Biol. Rep. 2018, 45, 2609–2614. [Google Scholar] [CrossRef]
- Chen, L.; Lu, B.; Liu, L.; Duan, W.; Jiang, D.; Li, J.; Zhang, K.; Sun, H.; Zhang, Y.; Li, C.; et al. Melatonin promotes seed germination under salt stress by regulating ABA and GA3 in cotton (Gossypium hirsutum L.). Plant Physiol Biochem. 2021, 162, 506–516. [Google Scholar] [CrossRef]
- Yin, Y.J.; Chen, C.J.; Guo, S.W.; Li, K.M.; Ma, Y.N.; Sun, W.M.; Xu, F.R.; Cheng, Y.X.; Dong, X. The Fight Against Panax notoginseng Root-Rot Disease Using Zingiberaceae Essential Oils as Potential Weapons. Front. Plant Sci. 2018, 9, 1346. [Google Scholar] [CrossRef]
- Reuveni, R. Peroxidase activity as a biochemical marker for resistance of muskmelon (Cucumis melo) to pseudoperonospora cubensis. Phytopathology 1992, 82, 749–753. [Google Scholar] [CrossRef]
- Vieira, S.M.; Silva, T.M.; Glória, M.B.A. Influence of processing on the levels of amines and proline and onthe physico-chemical characteristics of concentrated orange juice. Food Chem. 2010, 119, 7–11. [Google Scholar] [CrossRef]
- Spitz, D.R.; Oberley, L.W. An assay for superoxide dismutase activity in mammalian tissue homogenates. Anal. Biochem. 1989, 179, 8–18. [Google Scholar] [CrossRef]
- Zhu, Q.; Chen, L.; Chen, T.; Xu, Q.; He, T.; Wang, Y.; Deng, X.; Zhang, S.; Pan, Y.; Jin, A. Integrated transcriptome and metabolome analyses of biochar-induced pathways in response to Fusarium wilt infestation in pepper. Genomics 2021, 113, 2085–2095. [Google Scholar] [CrossRef] [PubMed]
- Kong, Q.; Yuan, J.; Niu, P.; Xie, J.; Jiang, W.; Huang, Y.; Bie, Z. Screening suitable reference genes for normalization in reverse transcription quantitative real-time PCR analysis in melon. PLoS ONE 2014, 9, e87197. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Arif, Y.; Sami, F.; Siddiqui, H.; Bajguz, A.; Hayat, S. Salicylic acid in relation to other phytohormones in plant: A study towards physiology and signal transduction under challenging environment. Environ. Exp. Bot. 2020, 175, 104040. [Google Scholar] [CrossRef]
- Hasan, F.; Mohaddeseh, S.S. Effects of foliar application of salicylic acid on vegetative growth of maize under saline conditions. Afr. J. Plant Sci. 2011, 5, 575–578. [Google Scholar] [CrossRef]
- Liu, S.; Dong, Y.; Xu, L.; Kong, J. Effects of foliar applications of nitric oxide and salicylic acid on salt-induced changes in photosynthesis and antioxidative metabolism of cotton seedlings. Plant Growth Regul. 2014, 73, 67–78. [Google Scholar] [CrossRef]
- Dong, C.J.; Wang, X.L.; Shang, Q.M. Salicylic acid regulates sugar metabolism that confers tolerance to salinity stress in cucumber seedlings. Sci. Hortic. 2011, 129, 629–636. [Google Scholar] [CrossRef]
- Horváth, E.; Csiszár, J.; Gallé, Á.; Poór, P.; Szepesi, Á.; Tari, I. Hardening with salicylic acid induces concentration-dependent changes in abscisic acid biosynthesis of tomato under salt stress. J. Plant Physiol. 2015, 183, 54–63. [Google Scholar] [CrossRef]
- Farooq, M.; Hussain, M.; Wahid, A.; Siddique, K.H.M. Drought stress in plants: An overview. In Plant Responses Drought Stress; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–33. [Google Scholar] [CrossRef]
- Chapin, F.S. Integrated responses of plants to stress. Bioscience 1991, 41, 29–36. [Google Scholar] [CrossRef]
- Meloni, D.A.; Oliva, M.A.; Martinez, C.A.; Cambraia, J. Photosynthesis and activity of superoxide dismutase, peroxidase and glutathione reductase in cotton under salt stress. Environ. Exp. Bot. 2003, 49, 69–76. [Google Scholar] [CrossRef]
- Torun, H. Time-course analysis of salicylic acid effects on ROS regulation and antioxidant defense in roots of hulled and hulless barley under combined stress of drought, heat and salinity. Physiol. Plant. 2019, 165, 169–182. [Google Scholar] [CrossRef]
- Flores, A.; Grau, A.; Laurich, F.; Drffling, K. Effect of new terpenoid analogues of abscisic acid on chilling and freezing resistance. J. Plant Physiol. 1988, 132, 362–369. [Google Scholar] [CrossRef]
- Upadhyaya, A.; Davis, T.D.; Walser, R.H.; Galbraith, A.B.; Sankhla, N. Uniconazole-induced alleviation of low-temperature damage in relation to antioxidant activity. HortScience 1989, 24, 955–957. [Google Scholar] [CrossRef]
- Feng, Z.; Guo, A.; Feng, Z. Amelioration of chilling stress by triadimefon in cucumber seedlings. Plant Growth Regul. 2003, 39, 277–283. [Google Scholar] [CrossRef]
- Grechkin, A. Recent developments in biochemistry of the plant lipoxygenase pathway. Prog. Lipid Res. 1998, 37, 317–352. [Google Scholar] [CrossRef] [PubMed]
- Ding, H.; Lai, J.; Wu, Q.; Zhang, S.; Chen, L.; Dai, Y.-S.; Wang, C.; Du, J.; Xiao, S.; Yang, C. Jasmonate complements the function of Arabidopsis lipoxygenase3 in salinity stress response. Plant Sci. 2016, 244, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Lim, C.W.; Han, S.W.; Hwang, I.S.; Kim, D.S.; Hwang, B.K.; Lee, S.C. The pepper lipoxygenase CaLOX1 plays a role in osmotic, drought and high salinity stress response. Plant Cell Physiol. 2015, 56, 930–942. [Google Scholar] [CrossRef] [PubMed]
- Boerjan, W.; Ralph, J.; Baucher, M. Lignin biosynthesis. Annu. Rev. Plant Biol. 2003, 54, 519–546. [Google Scholar] [CrossRef]
- Goubet, F.; Misrahi, A.; Park, S.K.; Zhang, Z.; Twell, D.; Dupree, P. AtCSLA7, a cellulose synthase-like putative glycosyltransferase, is important for pollen tube growth and embryogenesis in Arabidopsis. Plant Physiol. 2003, 131, 547–557. [Google Scholar] [CrossRef]
- Zhu, J.; Lee, B.-H.; Dellinger, M.; Cui, X.; Zhang, C.; Wu, S.; Nothnagel, E.A.; Zhu, J.-K. A cellulose synthase-like protein is required for osmotic stress tolerance in Arabidopsis. Plant J. 2010, 63, 128–140. [Google Scholar] [CrossRef]
- Parrotta, L.; Guerriero, G.; Sergeant, K.; Cai, G.; Hausman, J.F. Target or barrier? The cell wall of early-and later-diverging plants vs. cadmium toxicity: Differences in the response mechanisms. Front. Plant Sci. 2015, 6, 133. [Google Scholar] [CrossRef]
- Kim, H.J.; Triplett, B. Involvement of extracellular Cu/Zn superoxide dismutase in cotton fiber primary and secondary cell wall biosynthesis. Plant Signal. Behav. 2008, 3, 1119–1121. [Google Scholar] [CrossRef]
- Somerville, C.; Youngs, H.; Taylor, C.; Davis, S.C.; Long, S.P. Feedstocks for lignocellulosic biofuels. Science 2010, 329, 790–792. [Google Scholar] [CrossRef]
- Levin, D.E. Regulation of cell wall biogenesis in Saccharomyces cerevisiae: The cell wall integrity signaling pathway. Genetics 2011, 189, 1145–1175. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.; Zhang, Z.; Zhang, L.; Wang, J.; Wu, C. Glycosyltransferase GT1 family: Phylogenetic distribution, substrates coverage, and representative structural features. Comput. Struct. Biotechnol. J. 2020, 18, 1383–1390. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Li, Y.; Wang, Y.; Liu, H.; Lei, L.; Yang, H.; Liu, G.; Ren, D. Activation of MAPK kinase 9 induces ethylene and camalexin biosynthesis and enhances sensitivity to salt stress in Arabidopsis. J. Biol. Chem. 2008, 283, 26996–27006. [Google Scholar] [CrossRef]
- Kawa, D.; Meyer, A.J.; Dekker, H.L.; Abd-El-Haliem, A.M.; Gevaert, K.; Van De Slijke, E.; Testerink, C. SnRK2 protein kinases and mRNA decapping machinery control root development and response to salt. Plant Physiol. 2020, 182, 361–377. [Google Scholar] [CrossRef]
- Takahashi, Y.; Zhang, J.; Hsu, P.K.; Ceciliato, P.H.; Zhang, L.; Dubeaux, G.; Schroeder, J.I. MAP3Kinase-dependent SnRK2-kinase activation is required for abscisic acid signal transduction and rapid osmotic stress response. Nat. Commun. 2020, 11, 12. [Google Scholar] [CrossRef] [PubMed]
- De Zélicourt, A.; Colcombet, J.; Hirt, H. The role of MAPK modules and ABA during abiotic stress signaling. Trends Plant Sci. 2016, 21, 677–685. [Google Scholar] [CrossRef]
- Yoshida, T.; Fujita, Y.; Maruyama, K.; Mogami, J.; Todaka, D.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef] [PubMed]
| Treatment | Germination Potential/% | Germination Rate/% | Fresh Weight/g | Seedling Length/cm |
|---|---|---|---|---|
| CK | 91.75 ± 0.04 ab | 99.75 ± 0.00 a | 4.39 ± 0.08 a | 8.91 ± 0.29 a |
| SA | 97.00 ± 0.00 a | 99.25 ± 0.04 a | 4.35 ± 0.05 a | 8.31 ± 0.29 a |
| SA + NaCl | 89.50 ± 0.01 b | 93.75 ± 0.01 a | 3.80 ± 0.16 b | 6.75 ± 0.37 b |
| NaCl | 44.25 ± 0.02 c | 56.50 ± 0.03 b | 1.55 ± 0.03 c | 0.96 ± 0.17 c |
| Sample | Raw Reads | Clean Reads | Q30 Content | GC Content (%) |
|---|---|---|---|---|
| NaCl | 50,782,597 | 46,390,802 | 90.85 | 43.52 |
| SA + NaCl | 61,157,746 | 56,421,838 | 92.10 | 43.58 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, M.; Mao, J.; Wu, T.; Xiong, T.; Huang, Q.; Wu, H.; Hu, G. Transcriptomic Analysis of Salicylic Acid Promoting Seed Germination of Melon under Salt Stress. Horticulturae 2023, 9, 375. https://doi.org/10.3390/horticulturae9030375
Yan M, Mao J, Wu T, Xiong T, Huang Q, Wu H, Hu G. Transcriptomic Analysis of Salicylic Acid Promoting Seed Germination of Melon under Salt Stress. Horticulturae. 2023; 9(3):375. https://doi.org/10.3390/horticulturae9030375
Chicago/Turabian StyleYan, Miao, Jiancai Mao, Ting Wu, Tao Xiong, Quansheng Huang, Haibo Wu, and Guozhi Hu. 2023. "Transcriptomic Analysis of Salicylic Acid Promoting Seed Germination of Melon under Salt Stress" Horticulturae 9, no. 3: 375. https://doi.org/10.3390/horticulturae9030375
APA StyleYan, M., Mao, J., Wu, T., Xiong, T., Huang, Q., Wu, H., & Hu, G. (2023). Transcriptomic Analysis of Salicylic Acid Promoting Seed Germination of Melon under Salt Stress. Horticulturae, 9(3), 375. https://doi.org/10.3390/horticulturae9030375




