Genome-Wide Identification and Characterization of USP Gene Family in Grapes (Vitis vinifera L.)
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of USP Gene Family in Grape
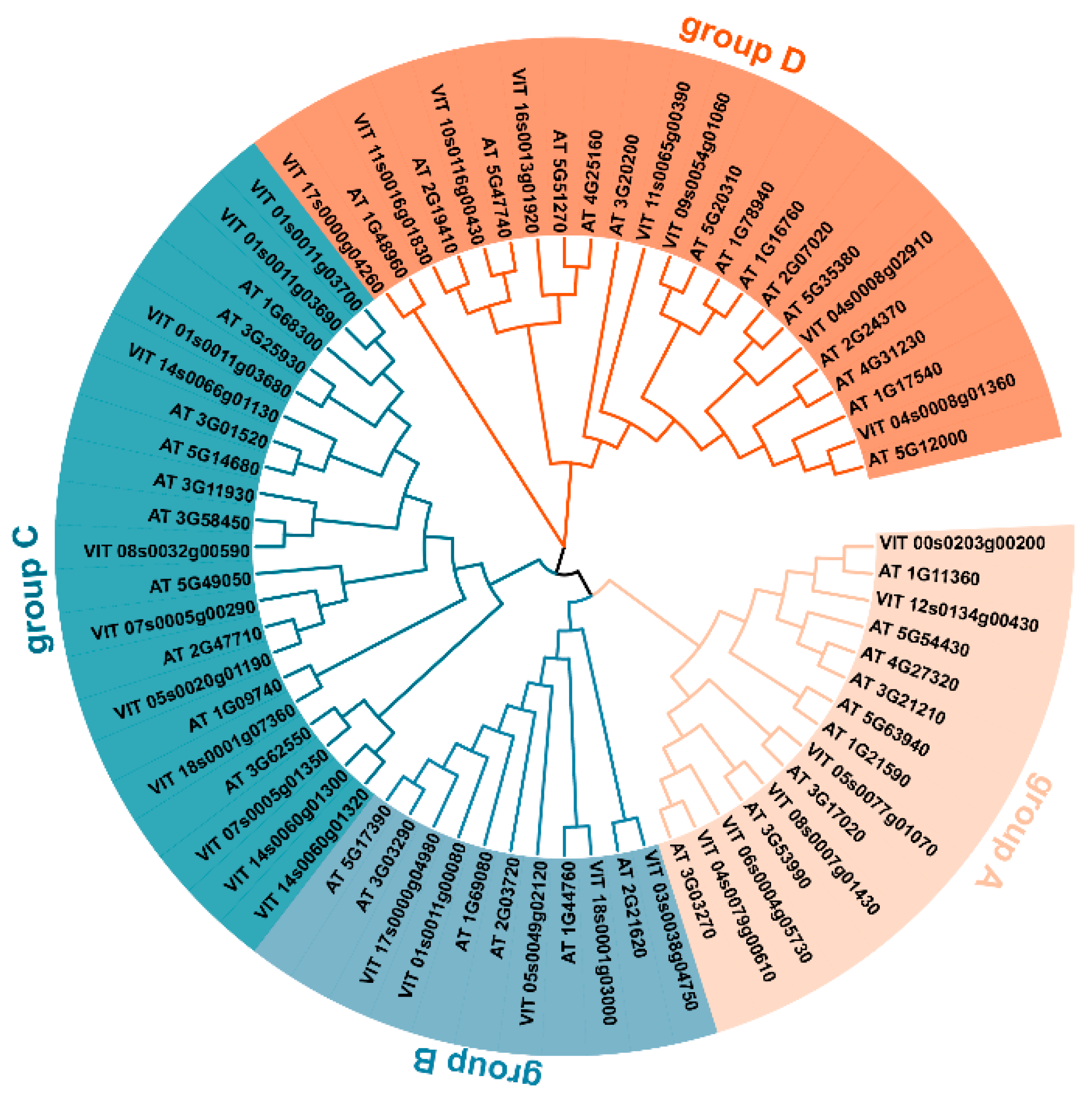
2.2. Phylogenetic Analysis, Gene Structure and Conserved Motif Analysis
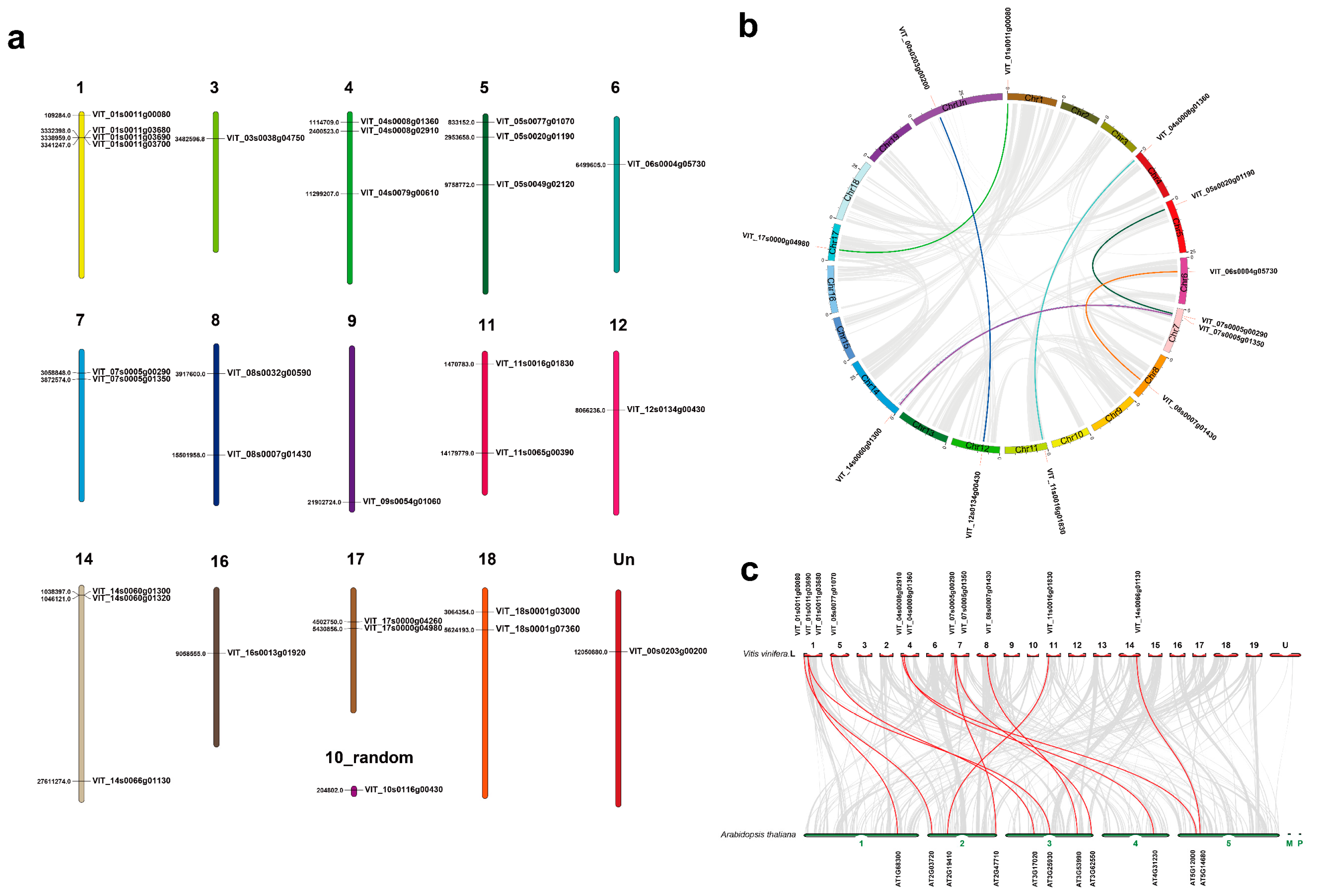
2.3. Chromosomal Location, Gene Duplication and Synteny Analysis
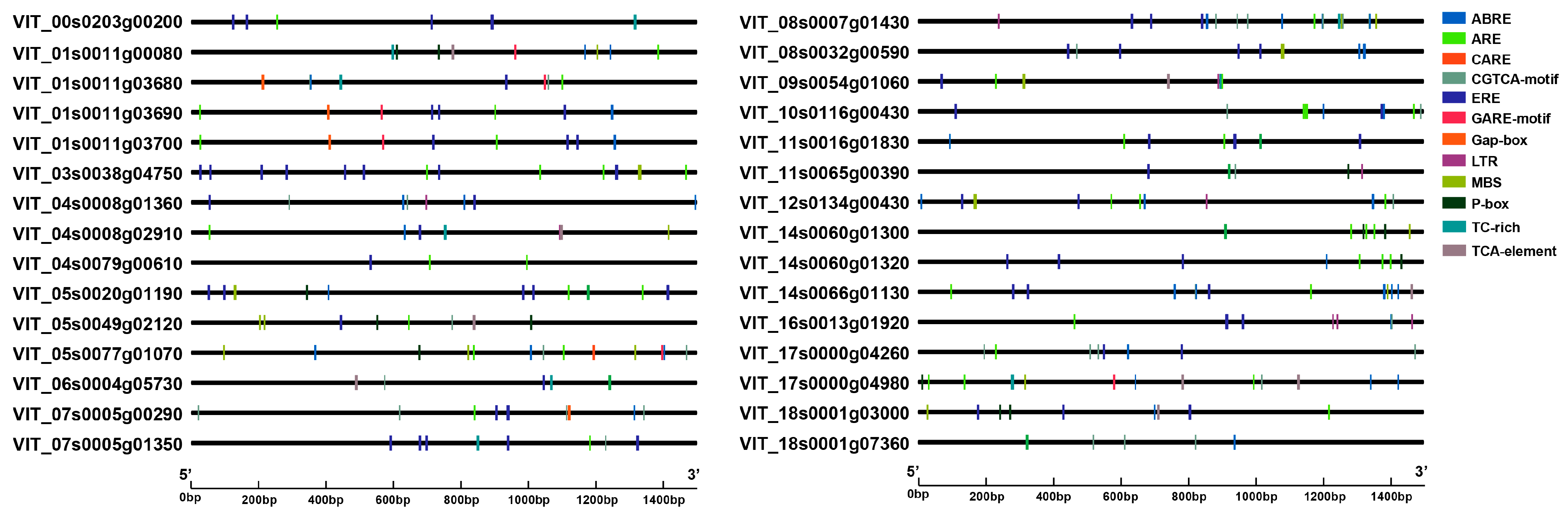
2.4. Cis-Acting Regulatory Element Analysis
2.5. Plant Materials and Treatments
2.6. RNA Extraction and qRT-PCR Analysis
3. Results
3.1. Identification and Analysis of VvUSPs
3.2. Gene Structure and Conserved Motifs
3.3. Chromosomal Location and Collinearity Analysis
3.4. Cis-Acting Elements Present in the Promoter Regions of VvUSP Family Genes
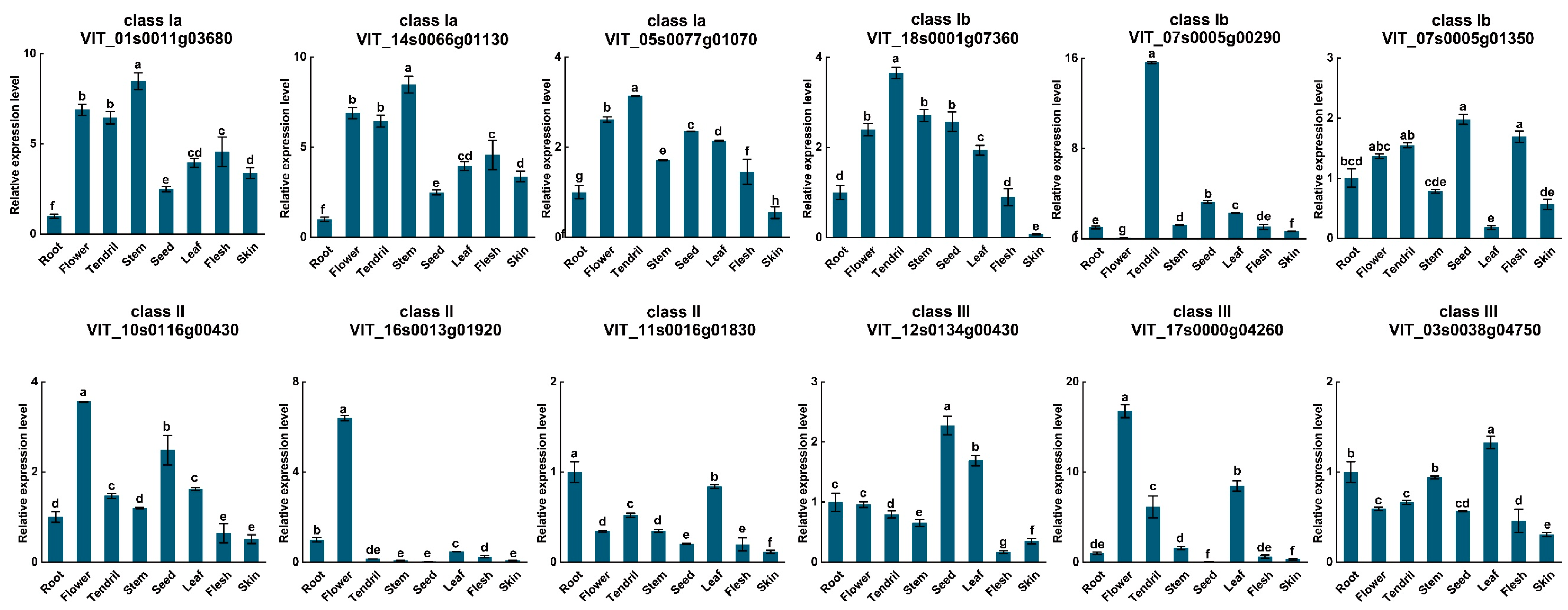
3.5. Tissue-Specific Analysis of VvUSPs
3.6. Expression Profiles of VvUSPs in Response to Diverse Hormone Treatments
3.7. Analysis of VvUSP Expression under Abiotic Stress Treatments
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yamori, N.; Levine, C.P.; Mattson, N.S.; Yamori, W. Optimum root zone temperature of photosynthesis and plant growth depends on air temperature in lettuce plants. Plant Mol. Biol. 2022. [Google Scholar] [CrossRef] [PubMed]
- Turner, L.B. The Effect of Water-stress on the Vegetative Growth of White Clover (Trifolium-repens L.)—Comparison of Long-term Water Deficit and A Short-term Development Water-stress. J. Exp. Bot. 1991, 42, 311–316. [Google Scholar] [CrossRef]
- Gao, J.; Liu, Z.; Zhao, B.; Dong, S.T.; Liu, P.; Zhang, J.W. Shade stress decreased maize grain yield, dry matter, and nitrogen accumulation. Agron. J. 2020, 112, 2768–2776. [Google Scholar] [CrossRef]
- Banakar, M.H.; Amiri, H.; Sarafraz Ardakani, M.R.; Ranjbar, G.H. Susceptibility and tolerance of fenugreek (Trigonella foenum-graceum L.) to salt stress: Physiological and biochemical inspections. Environ. Exp. Bot. 2022, 194, 104748. [Google Scholar] [CrossRef]
- George, I.S.; Haynes, P.A. Current perspectives in proteomic analysis of abiotic stress in Grapevines. Front. Plant Sci. 2014, 5, 686. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Abdelrahman, M.; Burritt, D.J.; Gupta, A.; Tsujimoto, H.; Tran, L.S.P. Heat stress effects on source sink relationships and metabolome dynamics in wheat. J. Exp. Bot. 2020, 71, 543–554. [Google Scholar] [CrossRef]
- Biswas, D.; Saha, S.C.; Dey, A. CRISPR-Cas genome-editing tool in plant abiotic stress-tolerance. Plant Gene 2021, 26, 100286. [Google Scholar] [CrossRef]
- Ranjan, A.; Sinha, R.; Bala, M.; Pareek, A.; Singla-Pareek, S.L.; Singh, A.K. Silicon-mediated abiotic and biotic stress mitigation in plants: Underlying mechanisms and potential for stress resilient agriculture. Plant Physiol. Biochem. 2021, 163, 15–25. [Google Scholar] [CrossRef]
- Islam, T.; Arabia, S.; Sami, A.A.; Akhter, S.; Sarker, R.H. Comprehensive in silico Characterization of Universal Stress Proteins in Rice (Oryza sativa L.) With Insight Into Their Stress-Specific Transcriptional Modulation. Front. Plant Sci. 2021, 12, 712607. [Google Scholar] [CrossRef]
- Bhuria, M.; Goel, P.; Kumar, S.; Singh, A.K. Genome-wide identification and expression profiling of genes encoding universal stress proteins (USP) identify multi-stress responsive USP genes in Arabidopsis thaliana. Plant Physiol. Rep. 2019, 24, 434–445. [Google Scholar] [CrossRef]
- Kvint, K.; Nachin, L.; Diez, A.; Nystrm, T. The bacterial universal stress protein: Function and regulation. Curr. Opin. Microbiol. 2003, 6, 140–145. [Google Scholar] [CrossRef]
- Nachin, L.; Nannmark, U.; Nystrom, T. Differential Roles of the Universal Stress Proteins of Escherichia coli in Oxidative Stress Resistance, Adhesion, and Motility. J. Bacteriol. 2005, 187, 6265–6272. [Google Scholar] [CrossRef] [PubMed]
- Cui, X.Y.; Zhang, P.Y.; Hu, Y.F.; Chen, C.C.; Liu, Q.Y.; Guan, P.Y.; Zhang, J.X. Genome-wide analysis of the Universal stress protein A gene family in Vitis and expression in response to abiotic stress. Plant Physiol. Biochem. 2021, 165, 57–70. [Google Scholar] [CrossRef] [PubMed]
- Pushpika, U.; Jha, R.K.; Dinkar, S.; Avinash, M.; Bhavanath, J. Overexpression of a Cytosolic Abiotic Stress Responsive Universal Stress Protein (SbUSP) Mitigates Salt and Osmotic Stress in Transgenic Tobacco Plants. Front. Plant Sci. 2016, 7, 518. [Google Scholar] [CrossRef]
- Chi, Y.H.; Koo, S.S.; Oh, H.T.; Lee, E.S.; Park, J.H.; Phan, K.A.T.; Wi, S.D.; Bae, S.B.; Paeng, S.K.; Chae, H.B. The Physiological Functions of Universal Stress Proteins and Their Molecular Mechanism to Protect Plants From Environmental Stresses. Front. Plant Sci. 2019, 10, 750. [Google Scholar] [CrossRef]
- Gorshkova, D.S.; Pojidaeva, E.S. Members of the Universal Stress Protein Family are Indirectly Involved in Gibberellin-Dependent Regulation of Germination and Post-Germination Growth. Russ. J. Plant Physiol. 2021, 68, 451–462. [Google Scholar] [CrossRef]
- Galant, C.; Marchandise, J.; Stoenoiu, M.S.; Ducreux, J.; Groof, A.D.; Pirenne, S.; Benoit, V.; Houssiau, F.A.; Lauwerys, B.R. Overexpression of ubiquitin-specific peptidase 15 in systemic sclerosis fibroblasts increases response to transforming growth factor β. Rheumatology 2019, 58, 708–718. [Google Scholar] [CrossRef]
- Francois, S.; Rene, A.; Georg, H.; Sylvain, F.; Kiyokazu, A. Phylogenomics Reveals an Anomalous Distribution of USP Genes in Metazoans. Mol. Biol. Evol. 2011, 28, 153–161. [Google Scholar] [CrossRef]
- Basters, A.; Knobeloch, K.-P.; Fritz, G. USP18—A multifunctional component in the Interferon response. Biosci. Rep. 2018, 38, BSR20180250. [Google Scholar] [CrossRef]
- Rainer Waadt, C.A.S.; Hsu, P.-K.; Takahashi, Y.; Munemasa, S.; Schroeder, J.I. Plant hormone regulation of abiotic stress responses. Nat. Rev. Mol. Cell Biol. 2022, 23, 680–694. [Google Scholar] [CrossRef]
- Kosakivska, I.V.; Vedenicheva, N.P.; Babenko, L.M.; Voytenko, L.V.; Romanenko, K.O.; Vasyuk, V.A. Exogenous phytohormones in the regulation of growth and development of cereals under abiotic stresses. Mol. Biol. Rep. 2022, 49, 617–628. [Google Scholar] [CrossRef] [PubMed]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Prakash, V.; Singh, V.P.; Tripathi, D.K.; Sharma, S.; Corpas, F.J. Nitric oxide (NO) and salicylic acid (SA): A framework of their relationship in plant development under abiotic stress conditions. Plant Biol. 2021, 23, 104–119. [Google Scholar] [CrossRef] [PubMed]
- Nadarajah, K.; Hamid, N.; Rahman, N. SA-Mediated Regulation and Control of Abiotic Stress Tolerance in Rice. Int. J. Mol. Sci. 2021, 22, 5591. [Google Scholar] [CrossRef]
- Wu, Y.; Li, B.; Li, X.; Wang, L.; Zhang, W.; Duan, S.; Wang, S. Regulatory effect of root restriction on aroma quality of Red Alexandria grape. Food Chem. 2022, 372, 131118. [Google Scholar] [CrossRef]
- Pengfei, W.; Ling, S.; Huanhuan, G.; Xilong, J.; Xinying, W.; Yi, L.; Qianqian, Z.; Yongmei, W.; Fengshan, R. Genome-Wide Characterization of bHLH Genes in Grape and Analysis of their Potential Relevance to Abiotic Stress Tolerance and Secondary Metabolite Biosynthesis. Front. Plant Sci. 2018, 9, 64. [Google Scholar] [CrossRef]
- Hao, X.; Jiao, B.; Liu, Z.; Wang, X.; Wang, Q.C. Crosstalk between grapevine leafroll-associate virus-3 (GLRaV-3) and NaCl-induced salt stress in in vitro cultures of the red grape ‘Cabernet Sauvignon’. Plant Cell Tissue Organ Cult. 2021, 144, 73. [Google Scholar] [CrossRef]
- Liu, G.; Chai, F.; Wang, Y.; Jiang, J.; Duan, W.; Wang, Y.; Wang, F.; Li, S.; Wang, L. Genome-wide Identification and Classification of HSF Family in Grape, and Their Transcriptional Analysis under Heat Acclimation and Heat Stress. Hortic. Plant J. 2018, 4, 133–143. [Google Scholar] [CrossRef]
- Chen, T.; Xu, T.; Zhang, T.; Liu, T.; Shen, L.; Chen, Z.; Wu, Y.; Yang, J. Genome-Wide Identification and Characterization of DnaJ Gene Family in Grape (Vitis vinifera L.). Horticulturae 2021, 7, 589. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Li, J.; Zhao, X.-Q.; Wang, J.; Wong, G.K.-S.; Yu, J. KaKs_Calculator: Calculating Ka and Ks Through Model Selection and Model Averaging. Genom. Proteom. Bioinform. 2006, 4, 259–263. [Google Scholar] [CrossRef]
- Koch, M.A.; Haubold, B.; Mitchell-Olds, T. Comparative evolutionary analysis of chalcone synthase and alcohol dehydrogenase loci in Arabidopsis, Arabis, and related genera (Brassicaceae). Mol. Biol. Evol. 2000, 17, 1483–1498. [Google Scholar] [CrossRef]
- Lescot, M.; Déhais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van De Peer, Y.; Rouzé, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Guo, B.-B.; Li, J.-M.; Liu, X.; Qiao, X.; Musana, R.F.; Wang, P.; Zhang, S.-L.; Wu, J.-Y. Identification and expression analysis of the PbrMLO gene family in pear, and functional verification of PbrMLO23. J. Integr. Agric. 2021, 20, 2410–2423. [Google Scholar] [CrossRef]
- Liu, Z.; Fu, M.; Li, H.; Chen, Y.; Wang, L.; Liu, R. Systematic analysis of NAC transcription factors in Gossypium barbadense uncovers their roles in response to Verticillium wilt. PeerJ 2019, 7, e7995. [Google Scholar] [CrossRef]
- Hu, Z.; Song, H.; Feng, J.; Zhou, C.; Yang, M.-J.; Shi, P.; Yu, Z.-L.; Li, Y.-R.; Guo, Y.-J.; Li, H.-Z.; et al. Genome-wide analysis of the hard clam mitogen-activated protein kinase kinase gene family and their transcriptional profiles under abiotic stress. Mar. Environ. Res. 2022, 176, 105606. [Google Scholar] [CrossRef]
- Feng, T.; Sun, J.; Song, S.; Wang, H.; Yao, L.; Sun, M.; Wang, K.; Chen, D. Geographical differentiation of Molixiang table grapes grown in China based on volatile compounds analysis by HS-GC-IMS coupled with PCA and sensory evaluation of the grapes. Food Chem. X 2022, 15, 100423. [Google Scholar] [CrossRef]
- Zahra, N.; Wahid, A.; Hafeez, M.B.; Ullah, A.; Siddique, K.H.M.; Farooq, M. Grain development in wheat under combined heat and drought stress: Plant responses and management. Environ. Exp. Bot. 2021, 188, 104517. [Google Scholar] [CrossRef]
- Zhou, T.; Fan, M.; Irfan, M.; Wang, H.; Wang, D.; Wang, L.; Zhang, C.; Feng, L. Phylogenetic analysis of STK gene family and Usp domain in maize. Mol. Biol. Rep. 2014, 41, 8273–8284. [Google Scholar] [CrossRef] [PubMed]
- Xiao-Fan, W.; Jiao, S.; Na, Y.; Hui, Z.; Xiao-Yan, C.; Jie-Fang, K. Functional Characterization of Selected Universal Stress Protein from Salvia miltiorrhiza (SmUSP) in Escherichia coli. Genes 2017, 8, 224. [Google Scholar] [CrossRef]
- Merkouropoulos, G.; Tsaftaris, A.S. Differential Expression of Gossypium hirsutum USP-Related Genes, GhUSP1 and GhUSP2, During Development and upon Salt Stress. Plant Mol. Biol. Report. 2013, 31, 1539–1547. [Google Scholar] [CrossRef]
- Nazmul Hasan, M.; Islam, S.; Bhuiyan, F.H.; Arefin, S.; Hoque, H.; Azad Jewel, N.; Ghosh, A.; Prodhan, S.H. Genome wide analysis of the heavy-metal-associated (HMA) gene family in tomato and expression profiles under different stresses. Gene 2022, 835, 146664. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Shi, C.; Zhang, Y.; Hou, X. Genome-Wide Identification and Characterization of TCP Family Genes in Pak-Choi [ Brassica campestris (syn. Brassica rapa) ssp. chinensis var. communis]. Front. Plant Sci. 2022, 13, 854171. [Google Scholar] [CrossRef] [PubMed]
- Ruoqiu, W.; Peng, Z.; Nana, K.; Ruize, L.; Yue, P.; Chenxi, H.; Haoli, M.; Qin, C. Genome-Wide Identification and Characterization of the Potato bHLH Transcription Factor Family. Genes 2018, 9, 54. [Google Scholar] [CrossRef]
- Mostofa, M.G.; Li, W.Q.; Nguyen, K.H.; Fujita, M.; Tran, L.S.P. Strigolactones in plant adaptation to abiotic stresses: An emerging avenue of plant research. Curr. Opin. Plant Biol. 2018, 41, 2227–2243. [Google Scholar] [CrossRef]
- Peleg, Z.; Blumwald, E. Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 2011, 14, 290–295. [Google Scholar] [CrossRef]
- Pan, R.; Ding, M.; Feng, Z.; Zeng, F.; Medison, M.B.; Hu, H.; Han, Y.; Xu, L.; Li, C.; Zhang, W. HvGST4 enhances tolerance to multiple abiotic stresses in barley: Evidence from integrated meta-analysis to functional verification. Plant Physiol. Biochem. 2022, 188, 47–59. [Google Scholar] [CrossRef]
- Long, S.; Liu, Q.; Guo, H.; Li, X.; You, X.; Liu, B.; Gao, S.; Wen, S.; Liu, T.-Y.; Xu, Y. Integrated physiological and transcriptomic analyses reveal differential photosynthetic responses to low-light stress in tall fescue cultivars. Sci. Hortic. 2022, 304, 111343. [Google Scholar] [CrossRef]
- Li, X.; Wang, S.; Chen, X.; Cong, Y.; Cui, J.; Shi, Q.; Liu, H.; Diao, M. The positive effects of exogenous sodium nitroprusside on the plant growth, photosystem II efficiency and Calvin cycle of tomato seedlings under salt stress. Sci. Hortic. 2022, 299, 111016. [Google Scholar] [CrossRef]

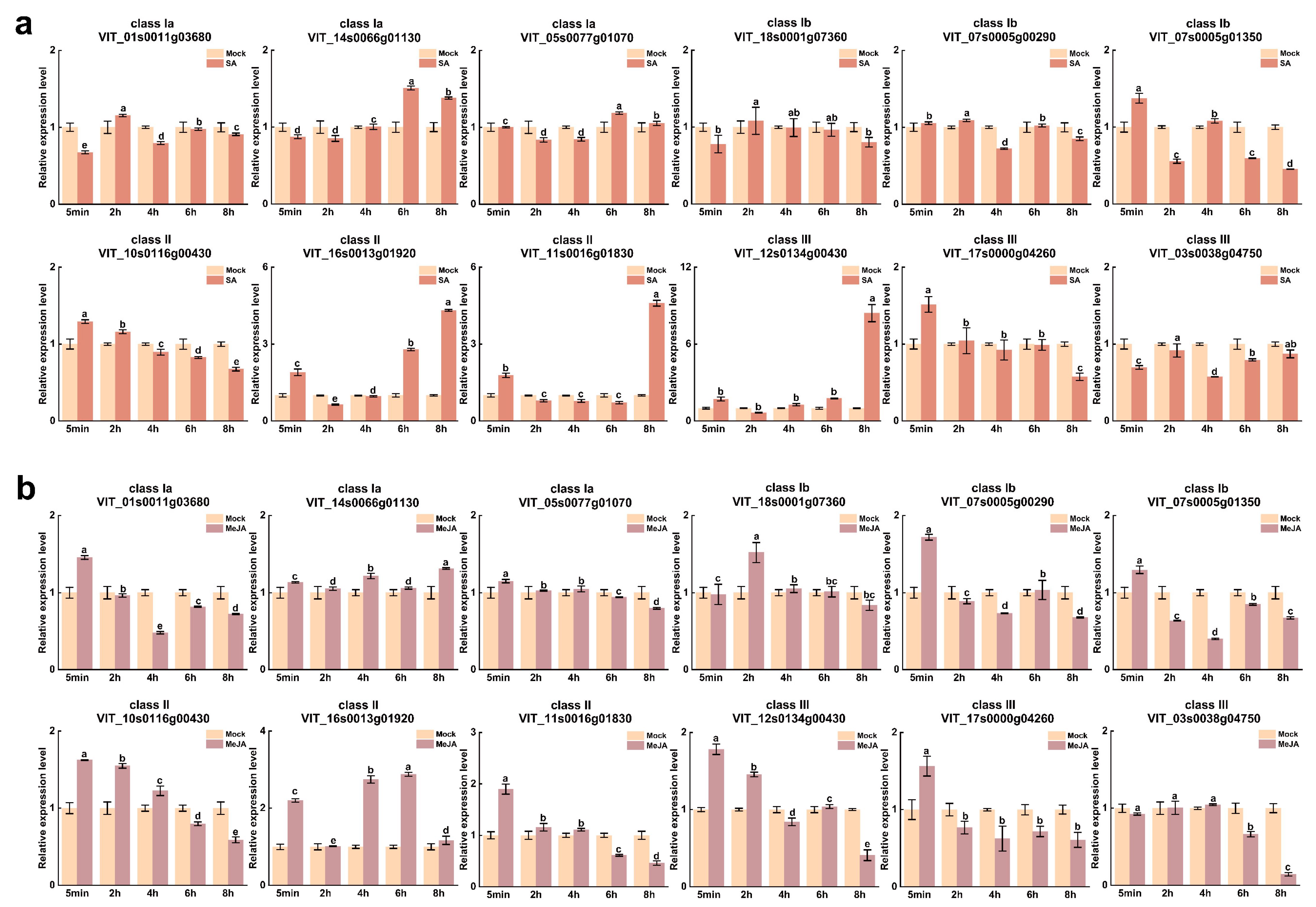
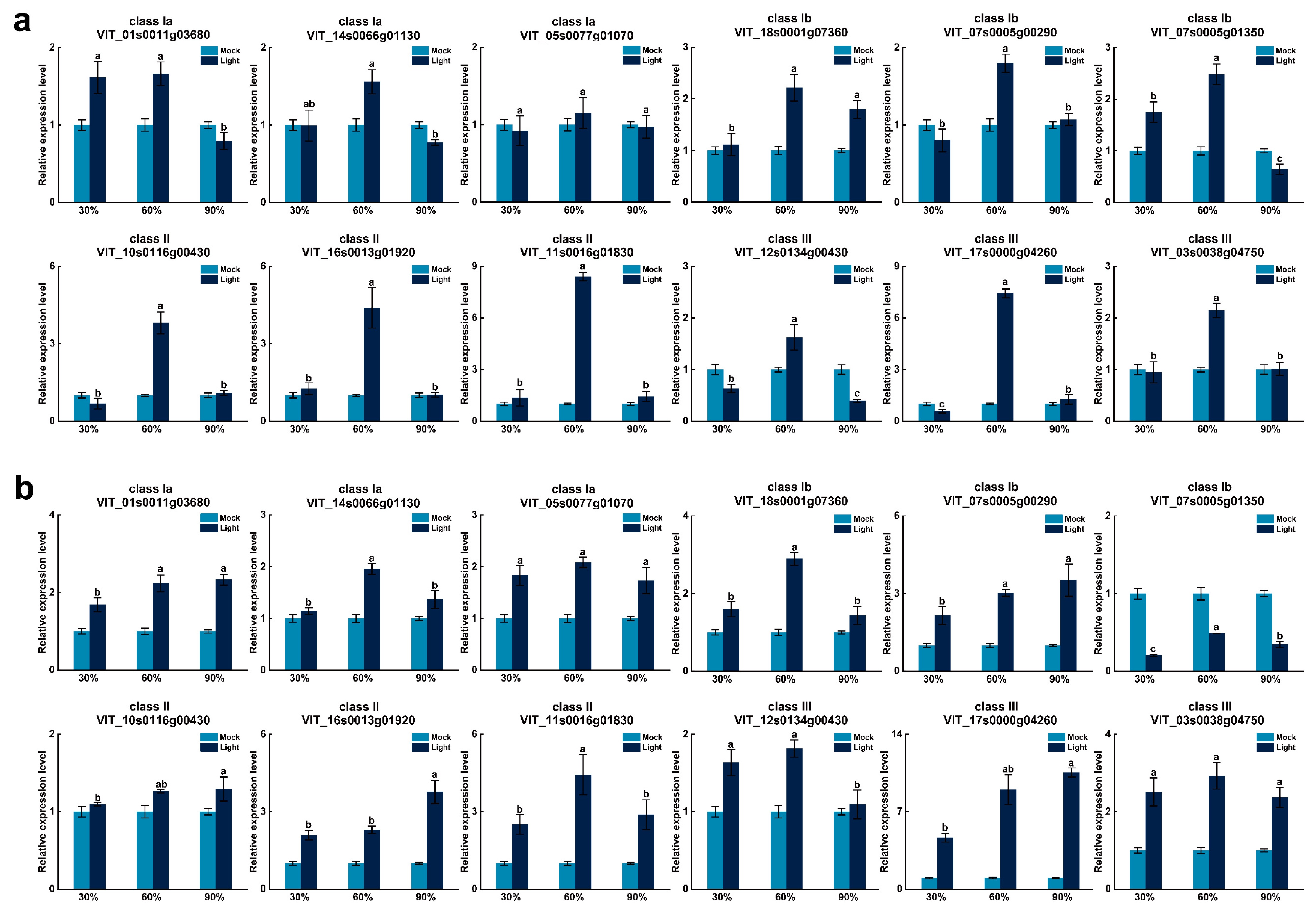
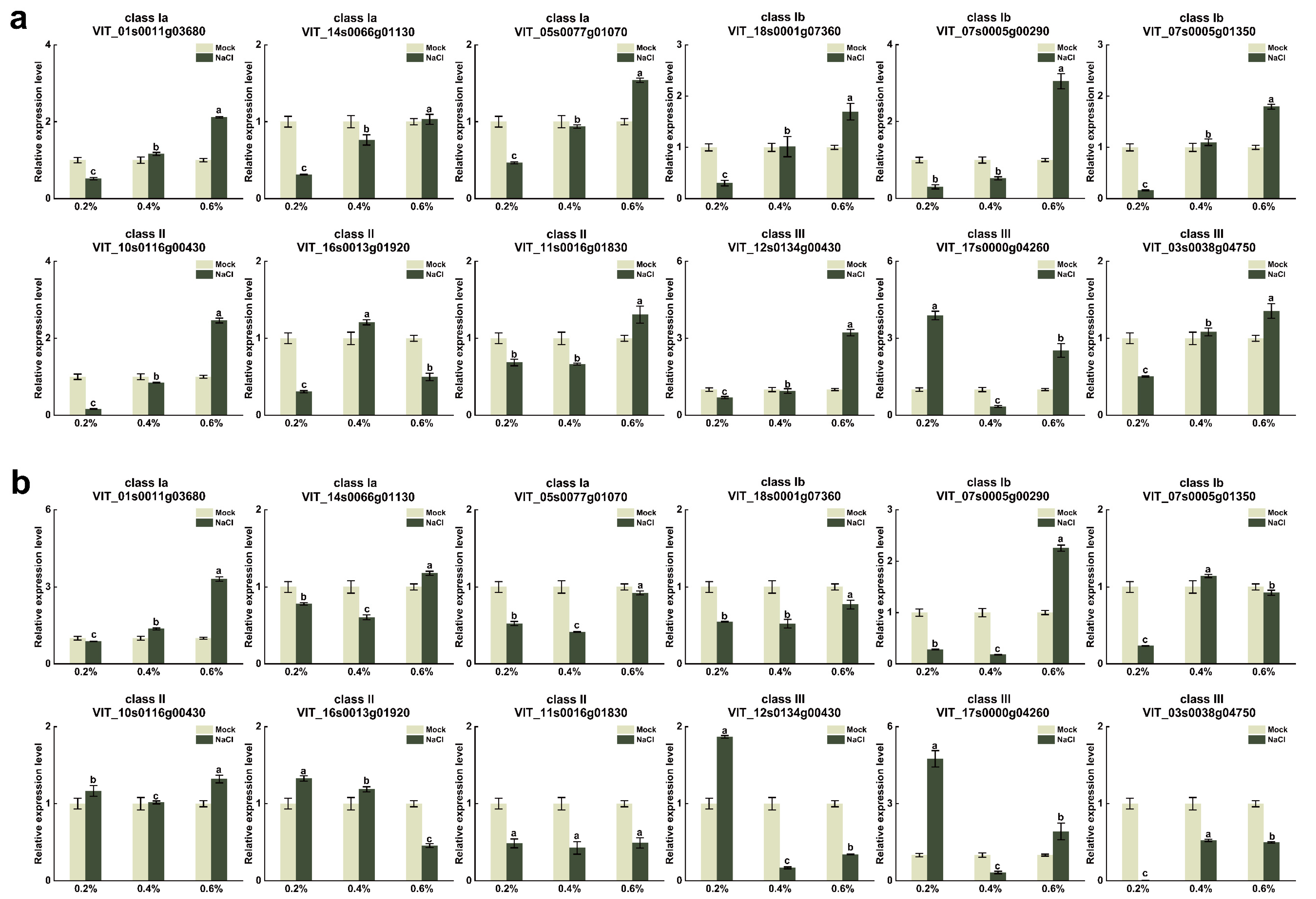
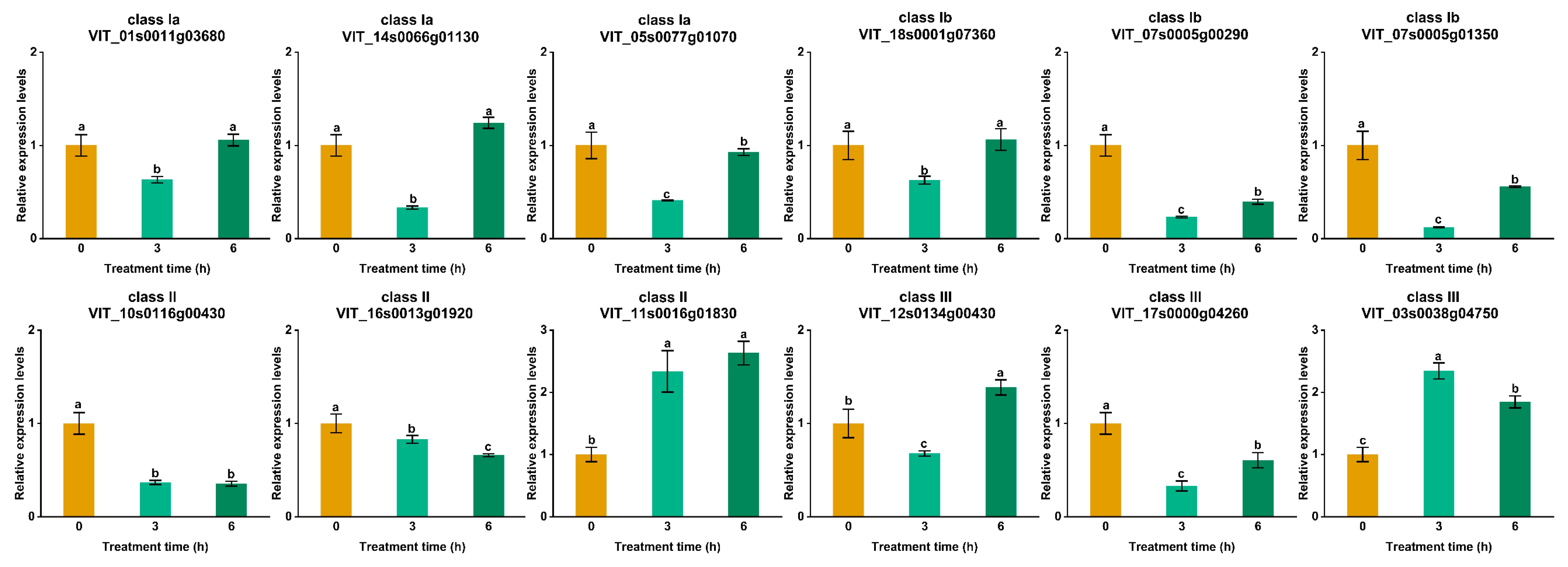
| Gene Name | CDS Length | Protein Size | Theoretical pI | Molecular Weight | Subcellular Localization |
| VIT_01s0011g00080 | 678 | 225 | 10.39 | 25,021.2 | Mitochondrial |
| VIT_01s0011g03680 | 513 | 170 | 8.6 | 18,218.9 | Extracellular |
| VIT_01s0011g03690 | 495 | 164 | 7.01 | 18,070.7 | Plasma membrane |
| VIT_01s0011g03700 | 1092 | 363 | 8.16 | 40,109.7 | Plasma membrane |
| VIT_03s0038g04750 | 522 | 173 | 5.64 | 18,899.7 | Cytoplasmic |
| VIT_04s0008g01360 | 2199 | 732 | 8.67 | 81,742.4 | Golgi |
| VIT_04s0008g02910 | 2394 | 797 | 7.27 | 89,051.2 | Cytoplasmic |
| VIT_04s0079g00610 | 495 | 164 | 7.29 | 17,816.4 | Mitochondrial |
| VIT_05s0020g01190 | 477 | 158 | 8.02 | 17,088.6 | Cytoplasmic |
| VIT_05s0049g02120 | 600 | 199 | 10.43 | 21,761 | Mitochondrial |
| VIT_05s0077g01070 | 498 | 165 | 5.82 | 18,055.7 | Cytoplasmic |
| VIT_06s0004g05730 | 495 | 164 | 6.79 | 18,014.6 | Mitochondrial |
| VIT_07s0005g00290 | 1014 | 337 | 7.9 | 36,033.5 | Cytoplasmic |
| VIT_07s0005g01350 | 432 | 143 | 8.3 | 15,577 | Cytoplasmic |
| VIT_08s0007g01430 | 495 | 164 | 7.51 | 18,238.8 | Mitochondrial |
| VIT_08s0032g00590 | 390 | 129 | 8.65 | 14,197.5 | Extracellular |
| VIT_09s0054g01060 | 2340 | 779 | 7.26 | 87,164.3 | Mitochondrial |
| VIT_10s0116g00430 | 828 | 275 | 5.11 | 30,278.2 | Cytoplasmic |
| VIT_11s0016g01830 | 2310 | 769 | 7.06 | 86,112.4 | Mitochondrial |
| VIT_11s0065g00390 | 2226 | 741 | 8.1 | 82,191 | Golgi |
| VIT_12s0134g00430 | 762 | 253 | 5.18 | 27,630.7 | Nuclear |
| VIT_14s0060g01300 | 513 | 170 | 7.41 | 18,301.1 | Mitochondrial |
| VIT_14s0060g01320 | 612 | 203 | 7.25 | 22,214.7 | Cytoplasmic |
| VIT_14s0066g01130 | 528 | 175 | 6.62 | 19,891.7 | Cytoplasmic |
| VIT_16s0013g01920 | 2355 | 784 | 7 | 87,702.1 | Cytoplasmic |
| VIT_17s0000g04260 | 633 | 210 | 10.19 | 23,398.6 | Nuclear |
| VIT_17s0000g04980 | 696 | 231 | 10.93 | 25,818.9 | Mitochondrial |
| VIT_18s0001g03000 | 660 | 219 | 9.94 | 23,863.5 | Mitochondrial |
| VIT_18s0001g07360 | 507 | 168 | 7.11 | 17,991.6 | Mitochondrial |
| VIT_00s0203g00200 | 465 | 154 | 4.71 | 16,917 | Nuclear |
| Duplicated Gene Pairs | Ka | Ks | Ka/Ks | Duplicated Type | Divergence Time (Mya) |
| VIT_17s0000g04980/VIT_01s0011g00080 | 0.33 | 1.30 | 0.25 | Segmental | 43.44 |
| VIT_08s0007g01430/VIT_06s0004g05730 | 0.25 | 1.93 | 0.13 | Segmental | 64.28 |
| VIT_14s0060g01300/VIT_07s0005g01350 | 0.98 | 1.06 | 0.92 | Segmental | 35.43 |
| VIT_04s0008g01360/VIT_11s0016g01830 | 0.94 | 0.69 | 1.37 | Segmental | 22.97 |
| VIT_12s0134g00430/VIT_00s0203g00200 | 0.12 | 4.43 | 0.03 | Segmental | 147.80 |
| VIT_17s0000g04980/VIT_01s0011g00080 | 0.33 | 1.30 | 0.25 | Segmental | 43.44 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, T.; Chen, T.; Zhang, T.; Shen, L.; Chen, Z.; Xu, Y.; Wu, Y.; Yang, J. Genome-Wide Identification and Characterization of USP Gene Family in Grapes (Vitis vinifera L.). Horticulturae 2022, 8, 1024. https://doi.org/10.3390/horticulturae8111024
Xu T, Chen T, Zhang T, Shen L, Chen Z, Xu Y, Wu Y, Yang J. Genome-Wide Identification and Characterization of USP Gene Family in Grapes (Vitis vinifera L.). Horticulturae. 2022; 8(11):1024. https://doi.org/10.3390/horticulturae8111024
Chicago/Turabian StyleXu, Tao, Tianchi Chen, Tianye Zhang, Leyi Shen, Zhe Chen, Yue Xu, Yueyan Wu, and Jian Yang. 2022. "Genome-Wide Identification and Characterization of USP Gene Family in Grapes (Vitis vinifera L.)" Horticulturae 8, no. 11: 1024. https://doi.org/10.3390/horticulturae8111024
APA StyleXu, T., Chen, T., Zhang, T., Shen, L., Chen, Z., Xu, Y., Wu, Y., & Yang, J. (2022). Genome-Wide Identification and Characterization of USP Gene Family in Grapes (Vitis vinifera L.). Horticulturae, 8(11), 1024. https://doi.org/10.3390/horticulturae8111024







