Comparative Transcriptome Analysis Reveals Potential Genes Conferring Resistance or Susceptibility to Bacterial Canker in Tomato
Abstract
1. Introduction
2. Results
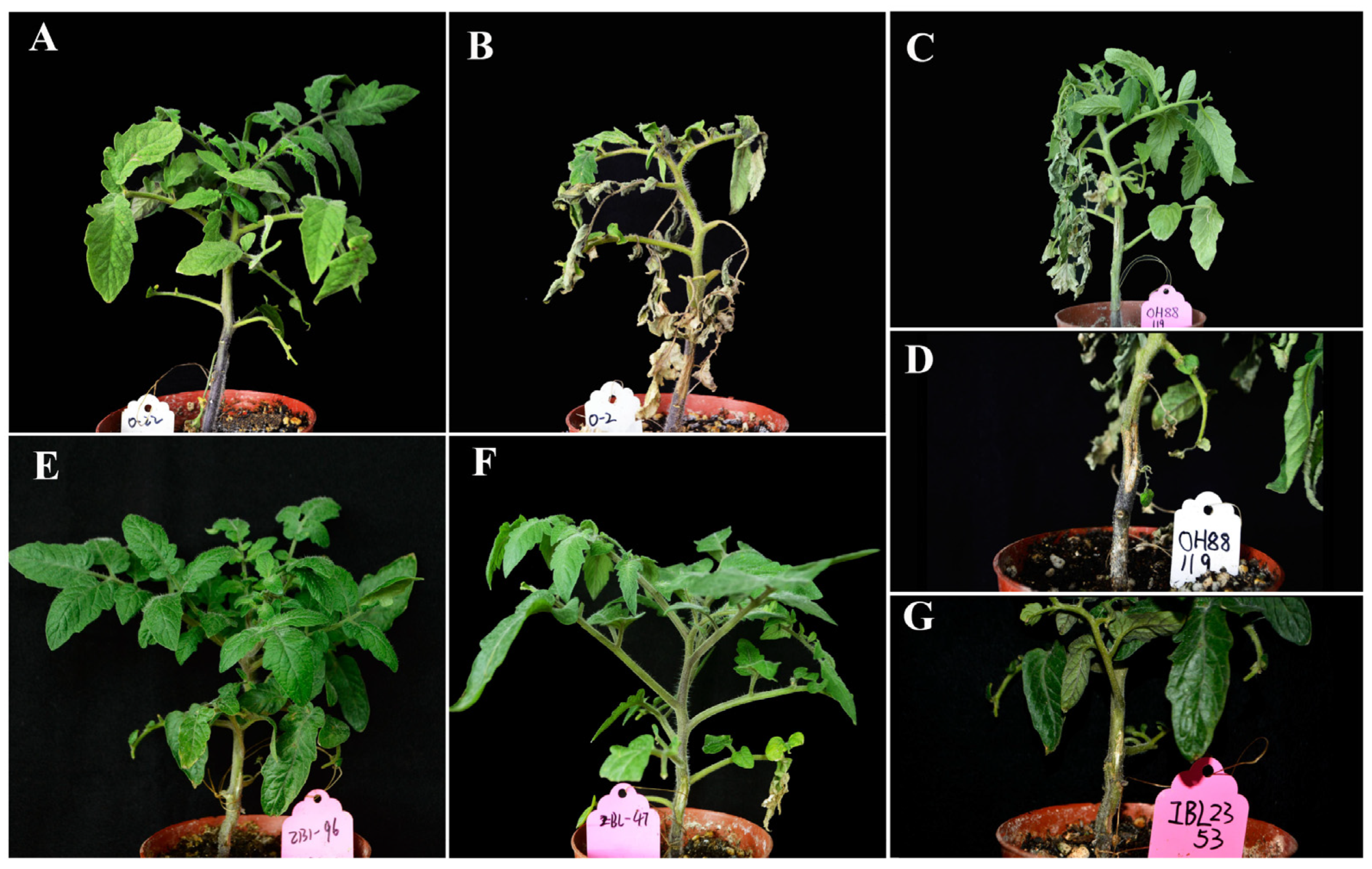
2.1. Phenotype Response of IBL2353 and Ohio88119 to Cm Infection
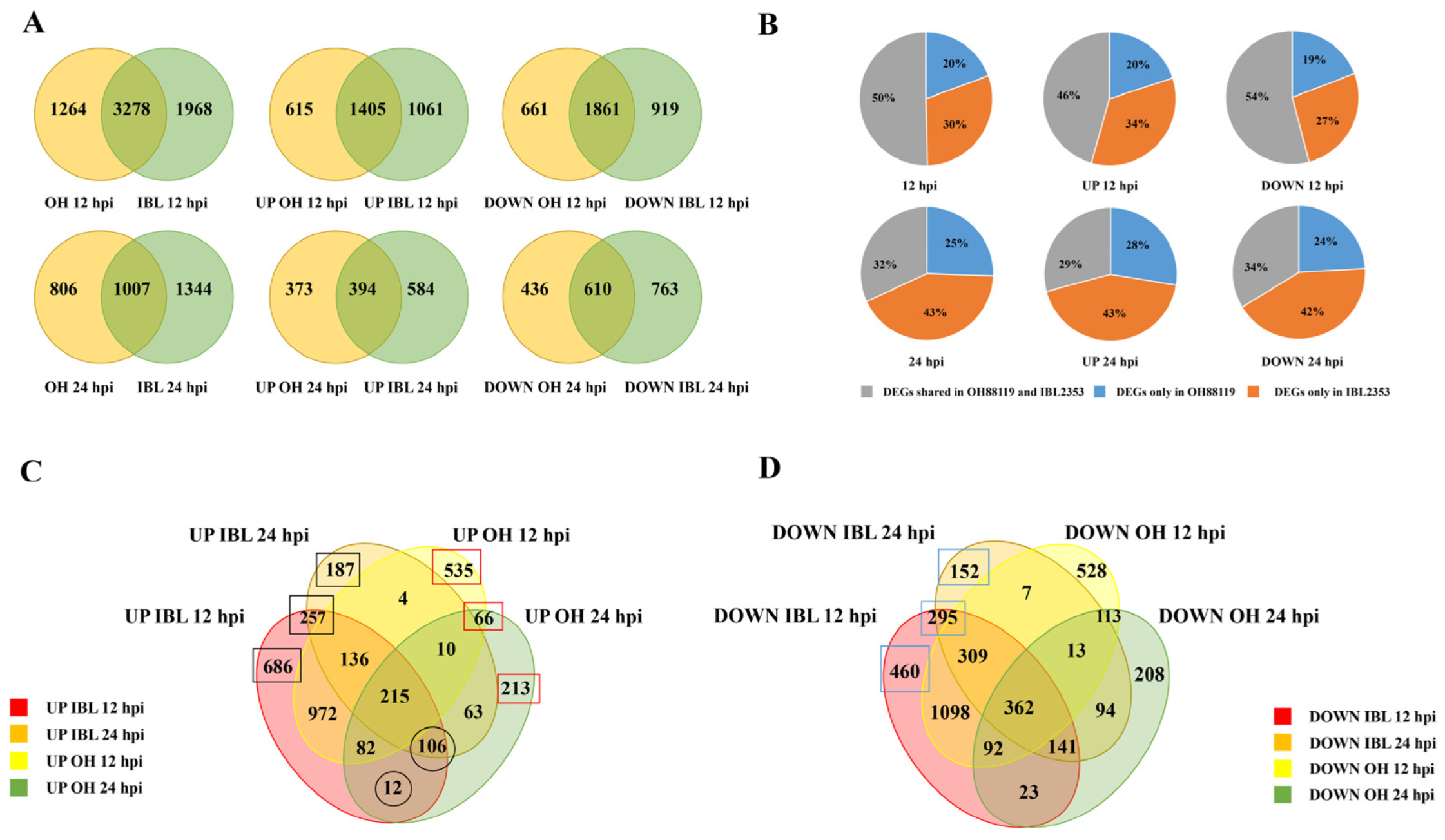
2.2. Cm-Induced Differentially Expressed Genes
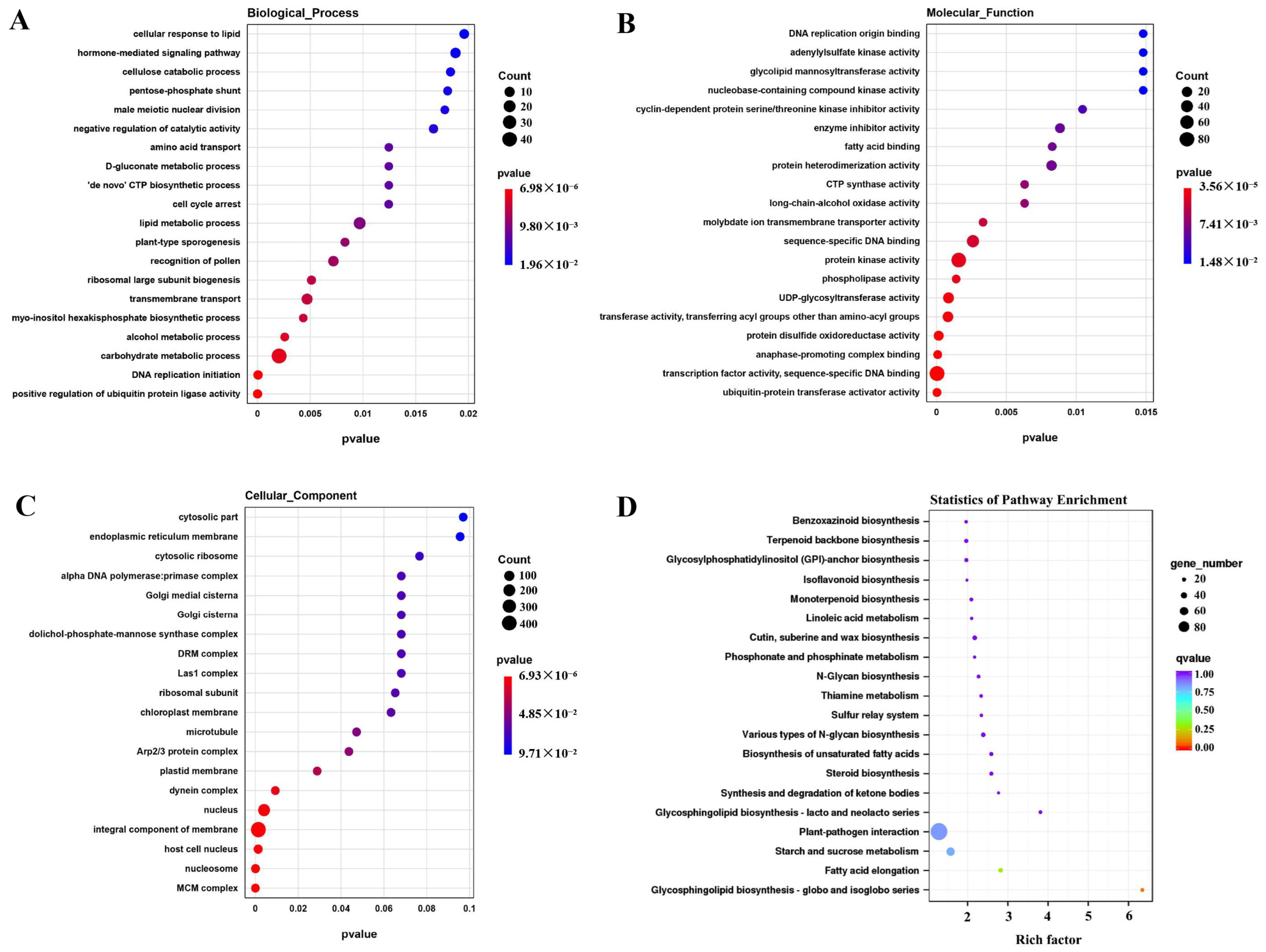
2.3. GO Term Enrichment and KEGG Pathway Analysis
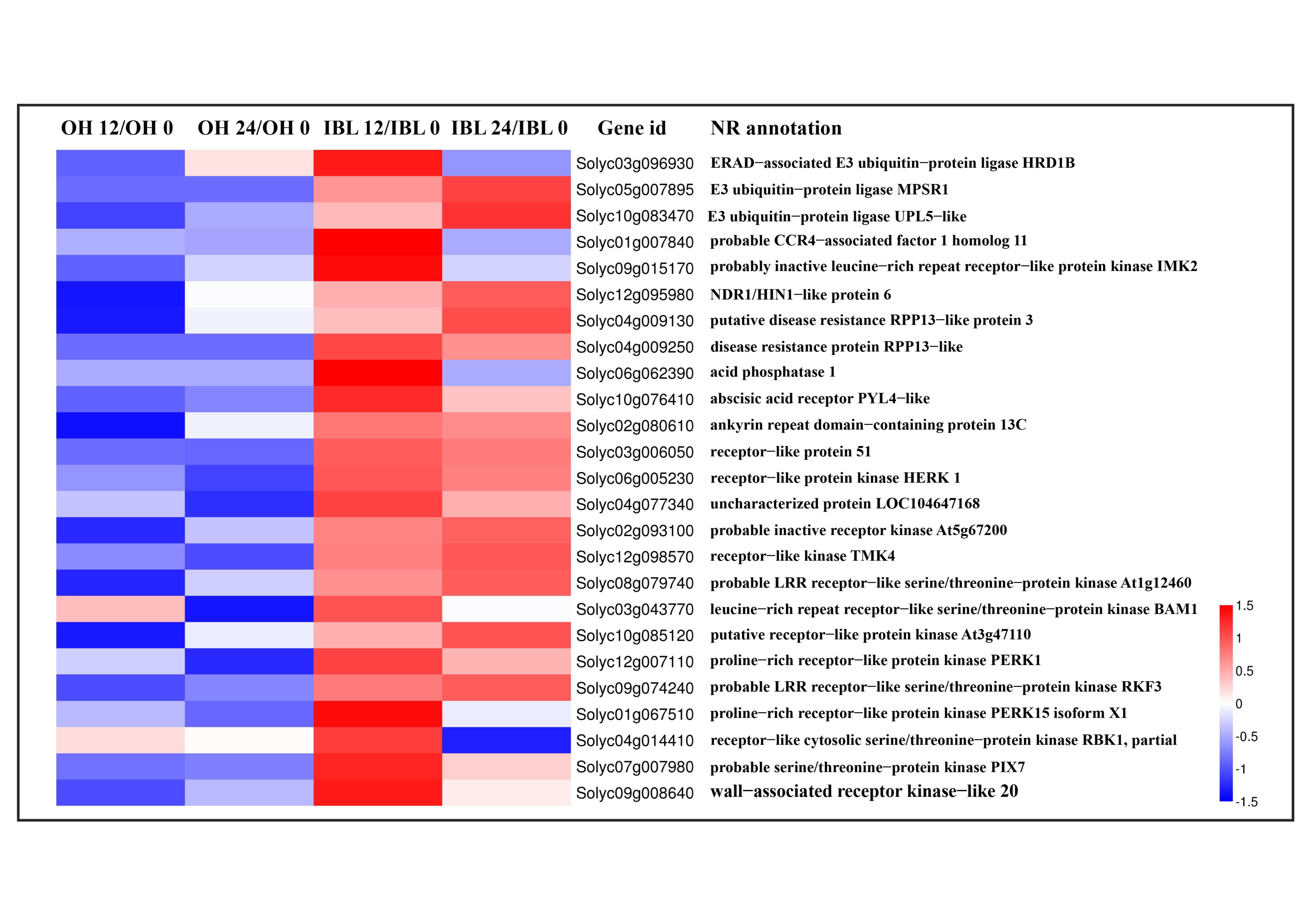
2.4. Genes Associated with the Defense Response against Cm Infection
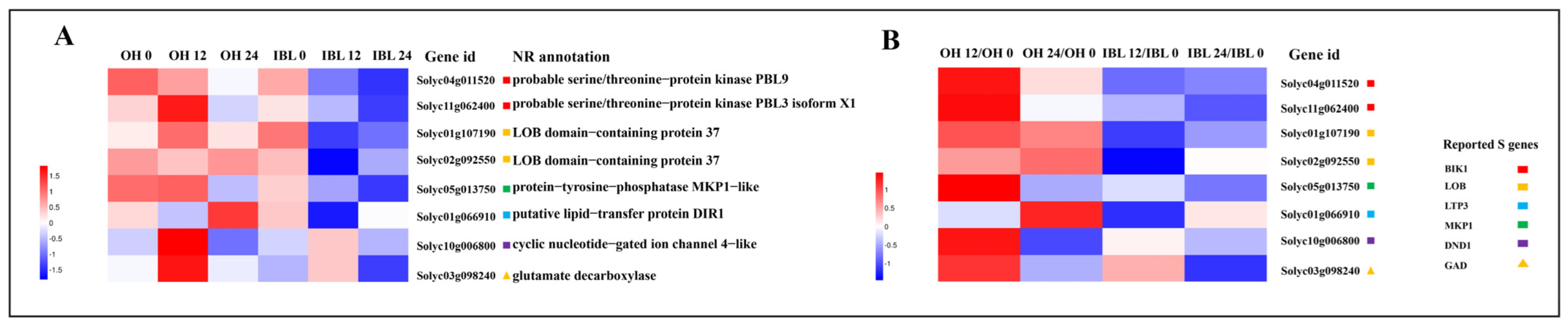
2.5. Candidate Susceptibility Genes
2.6. Validation of RNA-Seq by RT-qPCR
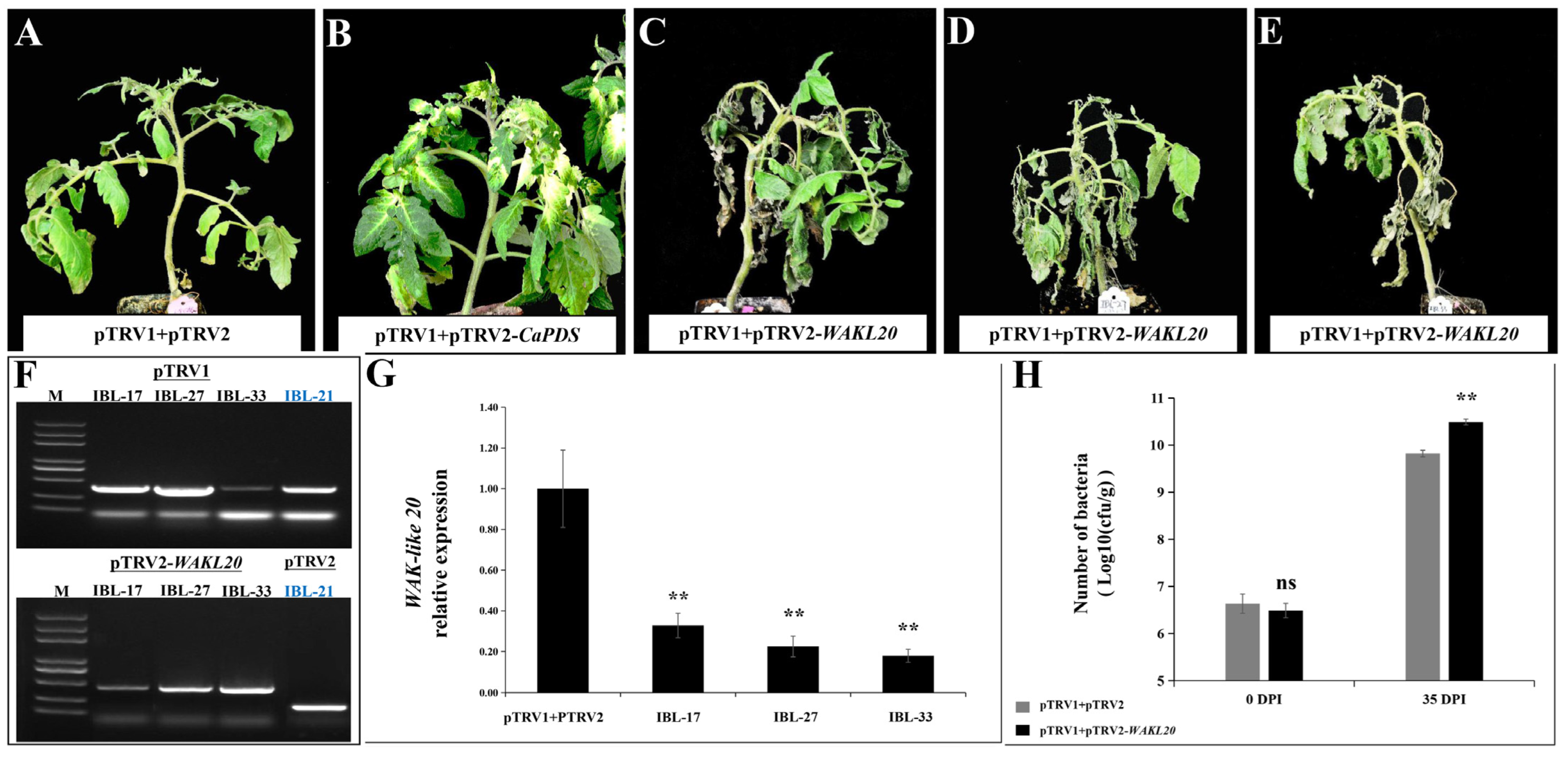
2.7. Silencing of WAKL20 Enhanced Susceptibility to Cm
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Pathogenic Cm Strain
4.2. Growth Conditions and Inoculation Treatments
4.3. RNA Extraction and Library Preparation for Illumina Sequencing
4.4. Transcriptome Data Processing
4.5. Gene Expression Validation with RT-qPCR Analysis
4.6. Virus-Induced Gene Silencing (VIGS)
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Li, X.; Tambong, J.; Yuan, K.X.; Chen, W.; Xu, H.; Lévesque, C.A.; De Boer, S.H. Re-classification of Clavibacter michiganensis subspecies on the basis of whole-genome and multi-locus sequence analyses. Int. J. Syst. Evol. Micr. 2018, 68, 234–240. [Google Scholar] [CrossRef] [PubMed]
- Peritore-Galve, F.C.; Tancos, M.A.; Smart, C.D. Bacterial canker of tomato: Revisiting a global and economically damaging seedborne pathogen. Plant Dis. 2021, 105, 1581–1595. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Deng, S.; Li, Z.; Yang, W. Advances in the characterization of the mechanism underlying bacterial canker development and tomato plant resistance. Horticulturae 2022, 8, 209. [Google Scholar] [CrossRef]
- Smith, E.F. A new tomato disease of economic importance. Science 1910, 31, 794–796. [Google Scholar]
- EPPO. Distribution of Clavibacter michiganesis subsp. michiganesis. 2016. Available online: https://gd.eppo.int/taxon/CORBMI/distribution (accessed on 11 October 2022).
- Wang, Y.; Zhang, Y.; Gao, Z.; Yang, W. Breeding for resistance to tomato bacterial diseases in China: Challenges and prospects. Hortic. Plant J. 2018, 4, 193–207. [Google Scholar] [CrossRef]
- Chalupowicz, L.; Zellermann, E.M.; Fluegel, M.; Dror, O.; Eichenlaub, R.; Gartemann, K.H.; Savidor, A.; Sessa, G.; Iraki, N.; Barash, I.; et al. Colonization and movement of GFP-labeled Clavibacter michiganensis subsp. michiganensis during tomato infection. Phytopathology 2012, 102, 23–31. [Google Scholar] [CrossRef]
- Sen, Y.; Feng, Z.; Vandenbroucke, H.; van der Wolf, J.; Visser, R.G.F.; van Heusden, A.W. Screening for new sources of resistance to Clavibacter michiganensis subsp. michiganensis (Cmm) in tomato. Euphytica 2013, 190, 309–317. [Google Scholar] [CrossRef]
- Carlton, W.M.; Braun, E.J.; Gleason, M.L. Ingress of Clavibacter michiganensis subsp. michiganensis into tomato leaves through hydathodes. Phytopathology 1998, 88, 525–529. [Google Scholar] [CrossRef]
- Xu, X.; Rajashekara, G.; Paul, P.A.; Miller, S.A. Colonization of tomato seedlings by bioluminescent Clavibacter michiganensis subsp. michiganensis under different humidity regimes. Phytopathology 2012, 102, 177–184. [Google Scholar] [CrossRef]
- Medina-Mora, C.M.; Hausbeck, M.K.; Fulbright, D.W. Bird‘s eye lesions of tomato fruit produced by aerosol and direct application of Clavibacter michiganensis subsp. michiganensis. Plant Dis. 2001, 85, 88–91. [Google Scholar] [CrossRef]
- Francis, D.M.; Kabelka, E.; Bell, J.; Franchino, B.; St, C.D. Resistance to bacterial canker in tomato (Lycopersicon hirsutum LA407) and its Progeny Derived from Crosses to L. esculentum. Plant Dis. 2001, 85, 1171–1176. [Google Scholar] [CrossRef] [PubMed]
- Yokotani, N.; Hasegawa, Y.; Sato, M.; Hirakawa, H.; Kouzai, Y.; Nishizawa, Y.; Yamamoto, E.; Naito, Y.; Isobe, S. Transcriptome analysis of Clavibacter michiganensis subsp. michiganensis-infected tomatoes: A role of salicylic acid in the host response. BMC Plant Biol. 2021, 21, 476. [Google Scholar] [CrossRef] [PubMed]
- Basim, H.; Basim, E.; Tombuloglu, H.; Unver, T. Comparative transcriptome analysis of resistant and cultivated tomato lines in response to Clavibacter michiganensis subsp. michiganensis. Genomics 2021, 113, 2455–2467. [Google Scholar] [CrossRef] [PubMed]
- Pereyra-Bistraín, L.I.; Ovando-Vázquez, C.; Rougon-Cardoso, A.; Alpuche-Solís, Á.G. Comparative RNA-Seq analysis reveals potentially resistance-related genes in response to bacterial canker of tomato. Genes 2021, 12, 1745. [Google Scholar] [CrossRef] [PubMed]
- Savidor, A.; Teper, D.; Gartemann, K.; Eichenlaub, R.; Chalupowicz, L.; Manulis-Sasson, S.; Barash, I.; Tews, H.; Mayer, K.; Giannone, R.J.; et al. The Clavibacter michiganensis subsp. michiganensis-tomato interactome reveals the perception of pathogen by the host and suggests mechanisms of infection. J. Proteome Res. 2012, 11, 736–750. [Google Scholar] [CrossRef] [PubMed]
- Kourelis, J.; van der Hoorn, R.A.L. Defended to the Nines: 25 Years of resistance gene cloning identifies nine mechanisms for R protein function. Plant Cell 2018, 30, 285–299. [Google Scholar] [CrossRef]
- Pavan, S.; Jacobsen, E.; Visser, R.G.F.; Bai, Y. Loss of susceptibility as a novel breeding strategy for durable and broad-spectrum resistance. Mol. Breed. 2010, 25, 1–12. [Google Scholar] [CrossRef]
- Koseoglou, E.; van der Wolf, J.M.; Visser, R.G.F.; Bai, Y. Susceptibility reversed: Modified plant susceptibility genes for resistance to bacteria. Trends Plant Sci. 2022, 27, 69–79. [Google Scholar] [CrossRef]
- Kabelka, E.; Franchino, B.; Francis, D.M. Two Loci from Lycopersicon hirsutum LA407 confer resistance to strains of Clavibacter michiganensis subsp. michiganensis. Phytopathology 2002, 92, 504–510. [Google Scholar] [CrossRef]
- Zhang, N.; Pombo, M.A.; Rosli, H.G.; Martin, G.B. Tomato wall-Associated kinase SlWak1 depends on Fls2/Fls3 to promote apoplastic immune responses to Pseudomonas syringae. Plant Physiol. 2020, 183, 1869–1882. [Google Scholar] [CrossRef]
- Li, Q.; Hu, A.; Qi, J.; Dou, W.; Qin, X.; Zou, X.; Xu, L.; Chen, S.; He, Y. CsWAKL08, a pathogen-induced wall-associated receptor-like kinase in sweet orange, confers resistance to citrus bacterial canker via ROS control and JA signaling. Hortic. Res. 2020, 7, 42. [Google Scholar] [CrossRef] [PubMed]
- Harkenrider, M.; Sharma, R.; De Vleesschauwer, D.; Tsao, L.; Zhang, X.; Chern, M.; Canlas, P.; Zuo, S.; Ronald, P.C. Overexpression of rice wall-associated kinase 25 (OsWAK25) alters resistance to bacterial and fungal pathogens. PLoS ONE 2016, 11, e147310. [Google Scholar] [CrossRef] [PubMed]
- Sen, Y.; van der Wolf, J.; Visser, R.G.F.; van Heusden, S. Bacterial canker of tomato: Current knowledge of detection, management, resistance, and interactions. Plant Dis. 2015, 99, 4–13. [Google Scholar] [CrossRef] [PubMed]
- Coaker, G.L.; Francis, D.M. Mapping, genetic effects, and epistatic interaction of two bacterial canker resistance QTLs from Lycopersicon hirsutum. Theor. Appl. Genet. 2004, 108, 1047–1055. [Google Scholar] [CrossRef]
- Heusden, A.W.V.; Koornneef, M.; Voorrips, R.E.; Bruggemann, W.; Pet, G.; Vrielink-van Ginkel, R.; Chen, X.; Lindhout, P. Three QTLs from Lycopersicon peruvianum confer a high level of resistance to Clavibacter michiganensis ssp. michiganensis. Theor. Appl. Genet. 1999, 99, 1068–1074. [Google Scholar] [CrossRef]
- Ramachandran, P.; Joshi, B.J.; Maupin-Furlow, J.A.; Uthandi, S. Bacterial effectors mimicking ubiquitin-proteasome pathway tweak plant immunity. Microbiol. Res. 2021, 250, 126810. [Google Scholar] [CrossRef]
- Furlan, G.; Klinkenberg, J.; Trujillo, M. Regulation of plant immune receptors by ubiquitination. Front. Plant Sci. 2012, 3, 238. [Google Scholar] [CrossRef]
- Guerra, D.D.; Callis, J. Ubiquitin on the move: The ubiquitin modification system plays diverse roles in the regulation of endoplasmic reticulum- and plasma membrane-localized proteins. Plant Physiol. 2012, 160, 56–64. [Google Scholar] [CrossRef]
- Li, W.; Zhong, S.; Li, G.; Li, Q.; Mao, B.; Deng, Y.; Zhang, H.; Zeng, L.; Song, F.; He, Z. Rice RING protein OsBBI1 with E3 ligase activity confers broad-spectrum resistance against Magnaporthe oryzae by modifying the cell wall defense. Cell Res. 2011, 21, 835–848. [Google Scholar] [CrossRef]
- Liu, L.; Jin, L.; Huang, X.; Geng, Y.; Li, F.; Qin, Q.; Wang, R.; Ji, S.; Zhao, S.; Xie, Q.I.; et al. OsRFPH2-10, a RING-H2 Finger E3 Ubiquitin Ligase, is involved in rice antiviral defense in the early stages of rice dwarf virus infection. Mol. Plant 2014, 7, 1057–1060. [Google Scholar]
- Ishikawa, K.; Yamaguchi, K.; Sakamoto, K.; Yoshimura, S.; Inoue, K.; Tsuge, S.; Kojima, C.; Kawasaki, T. Bacterial effector modulation of host E3 ligase activity suppresses PAMP-triggered immunity in rice. Nat. Commun. 2014, 5, 5430. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Meng, G.; Wang, M.; Qian, Z.; Zhang, Y.; Yang, W. Tomato SlPUB24 enhances resistance to Xanthomonas euvesicatoria pv. perforans race T3. Hortic. Res. 2021, 8, 30. [Google Scholar] [CrossRef]
- Schwessinger, B.; Ronald, P.C. Plant innate immunity: Perception of conserved microbial signatures. Annu. Rev. Plant Biol. 2012, 63, 451–482. [Google Scholar] [CrossRef] [PubMed]
- Sarowar, S.; Oh, H.W.; Cho, H.S.; Baek, K.; Seong, E.S.; Joung, Y.H.; Choi, G.J.; Lee, S.; Choi, D. Capsicum annuum CCR4-associated factor CaCAF1 is necessary for plant development and defence response. Plant J. 2007, 51, 792–802. [Google Scholar] [CrossRef] [PubMed]
- Shimo, H.M.; Terassi, C.; Lima Silva, C.C.; Zanella, J.D.L.; Mercaldi, G.F.; Rocco, S.A.; Benedetti, C.E. Role of the Citrus sinensis RNA deadenylase CsCAF1 in citrus canker resistance. Mol. Plant Pathol. 2019, 20, 1105–1118. [Google Scholar] [CrossRef]
- Chen, Q.; Tian, Z.; Jiang, R.; Zheng, X.; Xie, C.; Liu, J. StPOTHR1, a NDR1/HIN1-like gene in Solanum tuberosum, enhances resistance against Phytophthora infestans. Biochem. Biophys. Res. Commun. 2018, 496, 1155–1161. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, C.; Zhang, L.; Huang, J.; Dang, C.; Xie, C.; Wang, Z. TaRPP13-3, a CC-NBS-LRR-like gene located on chr 7D, promotes disease resistance to wheat powdery mildew in Brock. J. Phytopathol. 2020, 168, 688–699. [Google Scholar] [CrossRef]
- Bittner-Eddy, P.D.; Crute, I.R.; Holub, E.B.; Beynon, J.L. RPP13 is a simple locus in Arabidopsis thaliana for alleles that specify downy mildew resistance to different avirulence determinants in Peronospora parasitica. Plant J. 2000, 21, 177–188. [Google Scholar] [CrossRef]
- Liu, L.; Liu, C.; Wang, H.; Yu, S.; Guan, T.; Huang, Y.; Li, R. The abscisic acid receptor gene VvPYL4 positively regulates grapevine resistance to Plasmopara viticola. Plant Cell Tissue Organ Culture 2020, 142, 483–492. [Google Scholar] [CrossRef]
- Campos, M.D.; Félix, M.D.R.; Patanita, M.; Materatski, P.; Albuquerque, A.; Ribeiro, J.A.; Varanda, C. Defense strategies: The role of transcription factors in tomato–pathogen interaction. Biology 2022, 11, 235. [Google Scholar] [CrossRef]
- Soler, M.; Camargo, E.L.; Carocha, V.; Cassan-Wang, H.; San, C.H.; Savelli, B.; Hefer, C.A.; Paiva, J.A.; Myburg, A.A.; Grima-Pettenati, J. The Eucalyptus grandis R2R3-MYB transcription factor family: Evidence for woody growth-related evolution and function. New Phytol 2015, 206, 1364–1377. [Google Scholar] [CrossRef] [PubMed]
- Qiu, Z.; Yan, S.; Xia, B.; Jiang, J.; Yu, B.; Lei, J.; Chen, C.; Chen, L.; Yang, Y.; Wang, Y.; et al. The eggplant transcription factor MYB44 enhances resistance to bacterial wilt by activating the expression of spermidine synthase. J. Exp. Bot. 2019, 70, 5343–5354. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Dong, L.; Gao, T.; Liu, T.; Li, N.; Wang, L.; Chang, X.; Wu, J.; Xu, P.; Zhang, S. The bHLH transcription factor GmPIB1 facilitates resistance to Phytophthora sojae in Glycine max. J. Exp. Bot. 2018, 69, 2527–2541. [Google Scholar] [CrossRef]
- Onohata, T.; Gomi, K. Overexpression of jasmonate-responsive OsbHLH034 in rice results in the induction of bacterial blight resistance via an increase in lignin biosynthesis. Plant Cell Rep. 2020, 39, 1175–1184. [Google Scholar] [CrossRef] [PubMed]
- Guo, W.; Chen, B.; Guo, Y.; Chen, X.; Li, Q.; Yang, H.; Li, X.; Zhou, J.; Wang, G. Expression of pumpkin CmbHLH87 gene improves powdery mildew resistance in Tobacco. Front. Plant Sci. 2020, 11, 163. [Google Scholar] [CrossRef]
- Li, Q.; Jia, R.; Dou, W.; Qi, J.; Qin, X.; Fu, Y.; He, Y.; Chen, S. CsBZIP40, a BZIP transcription factor in sweet orange, plays a positive regulatory role in citrus bacterial canker response and tolerance. PLoS ONE 2019, 14, e223498. [Google Scholar] [CrossRef]
- Lim, C.W.; Baek, W.; Lim, S.; Han, S.; Lee, S.C. Expression and functional roles of the pepper pathogen–induced bZIP transcription factor CabZIP2 in enhanced disease resistance to bacterial pathogen infection. Mol. Plant Microbe Interact. 2015, 28, 825–833. [Google Scholar] [CrossRef]
- Lee, S.C.; Choi, H.W.; Hwang, I.S.; Choi, D.S.; Hwang, B.K. Functional roles of the pepper pathogen-induced bZIP transcription factor, CAbZIP1, in enhanced resistance to pathogen infection and environmental stresses. Planta 2006, 224, 1209–1225. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.; Lai, Z.; Fan, B.; Chen, Z. Arabidopsis WRKY38 and WRKY62 transcription factors interact with histone deacetylase 19 in basal defense. Plant Cell 2008, 20, 2357–2371. [Google Scholar] [CrossRef] [PubMed]
- Xing, D.; Lai, Z.; Zheng, Z.; Vinod, K.M.; Fan, B.; Chen, Z. Stress- and pathogen-induced Arabidopsis WRKY48 is a transcriptional activator that represses plant basal defense. Mol. Plant 2008, 1, 459–470. [Google Scholar] [CrossRef]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef] [PubMed]
- Oh, S.K.; Baek, K.H.; Park, J.M.; Yi, S.Y.; Yu, S.H.; Kamoun, S.; Choi, D. Capsicum annuum WRKY protein CaWRKY1 is a negative regulator of pathogen defense. New Phytol. 2008, 177, 977–989. [Google Scholar] [CrossRef] [PubMed]
- Ifnan Khan, M.; Zhang, Y.; Liu, Z.; Hu, J.; Liu, C.; Yang, S.; Hussain, A.; Furqan Ashraf, M.; Noman, A.; Shen, L.; et al. CaWRKY40b in pepper acts as a negative regulator in response to Ralstonia solanacearum by directly modulating defense genes including CaWRKY40. Int. J. Mol. Sci. 2018, 19, 1403. [Google Scholar] [CrossRef]
- Gu, C.; Guo, Z.; Hao, P.; Wang, G.; Jin, Z.; Zhang, S. Multiple regulatory roles of AP2/ERF transcription factor in angiosperm. Bot. Stud. 2017, 58, 6. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Tian, Y.; Xu, J.; Fu, X.; Gao, J.; Wang, B.; Han, H.; Wang, L.; Peng, R.; Yao, Q. A tomato ERF transcription factor, SlERF84, confers enhanced tolerance to drought and salt stress but negatively regulates immunity against Pseudomonas syringae pv. tomato DC3000. Plant Physiol. Biochem. 2018, 132, 683–695. [Google Scholar] [CrossRef]
- Balaji, V.; Mayrose, M.; Sherf, O.; Jacob-Hirsch, J.; Eichenlaub, R.; Iraki, N.; Manulis-Sasson, S.; Rechavi, G.; Barash, I.; Sessa, G. Tomato transcriptional changes in response to Clavibacter michiganensis subsp. michiganensis reveal a role for ethylene in disease development. Plant Physiol. 2008, 146, 1797–1809. [Google Scholar] [CrossRef]
- Wang, S.; Sun, J.; Fan, F.; Tan, Z.; Zou, Y.; Lu, D. A Xanthomonas oryzae pv. oryzae effector, XopR, associates with receptor-like cytoplasmic kinases and suppresses PAMP-triggered stomatal closure. Sci. China Life Sci. 2016, 59, 897–905. [Google Scholar] [CrossRef]
- Thatcher, L.F.; Powell, J.J.; Aitken, E.A.B.; Kazan, K.; Manners, J.M. The Lateral organ boundaries domain transcription factor LBD20 functions in fusarium wilt susceptibility and jasmonate signaling in Arabidopsis. Plant Physiol. 2012, 160, 407–418. [Google Scholar] [CrossRef]
- Hu, Y.; Zhang, J.; Jia, H.; Sosso, D.; Li, T.; Frommer, W.B.; Yang, B.; White, F.F.; Wang, N.; Jones, J.B. Lateral organ boundaries 1 is a disease susceptibility gene for citrus bacterial canker disease. Proc. Natl. Acad. Sci. USA 2014, 111, 521–529. [Google Scholar] [CrossRef]
- Jia, H.; Zhang, Y.; Orbović, V.; Xu, J.; White, F.F.; Jones, J.B.; Wang, N. Genome editing of the disease susceptibility gene CsLOB1 in citrus confers resistance to citrus canker. Plant Biotechnol. J. 2017, 15, 817–823. [Google Scholar] [CrossRef]
- Kanneganti, V.; Gupta, A.K. Wall associated kinases from plants—An overview. Physiol. Mol. Biol. Plants 2008, 14, 109–118. [Google Scholar] [CrossRef]
- Coaker, G.L.; Willard, B.; Kinter, M.; Stockinger, E.J.; Francis, D.M. Proteomic analysis of resistance mediated by Rcm 2.0 and Rcm 5.1, two Loci controlling resistance to bacterial canker of tomato. Mol. Plant Microbe Interact. 2004, 17, 1019–1028. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.; Mendell, J.T.; Salzberg, S.L. String Tie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, 480–484. [Google Scholar] [CrossRef]
- Zheng, Y.; Jiao, C.; Sun, H.; Rosli, H.G.; Pombo, M.A.; Zhang, P.; Banf, M.; Dai, X.; Martin, G.B.; Giovannoni, J.J.; et al. iTAK: A program for genome-wide prediction and classification of plant transcription factors, transcriptional regulators, and protein kinases. Mol. Plant 2016, 9, 1667–1670. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Dinesh-Kumar, S.P. Virus-induced gene silencing in tomato. Plant J. 2002, 31, 777–786. [Google Scholar] [CrossRef]
- Guo, J.; Liu, C.; Wang, P.; Cheng, Q.; Sun, L.; Yang, W.; Shen, H. The Aborted Microspores (AMS)-Like gene is required for anther and microspore development in pepper (Capsicum annuum L.). Int. J. Mol. Sci. 2018, 19, 1341. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Sacks, E.J.; Lewis Ivey, M.L.; Miller, S.A.; Francis, D.M. Resistance in Lycopersicon esculentum intraspecific crosses to race T1 strains of Xanthomonas campestris pv. vesicatoria causing bacterial spot of tomato. Phytopathology 2005, 95, 519–527. [Google Scholar] [CrossRef] [PubMed]
- Pathuri, I.P.; Reitberger, I.E.; Huckelhoven, R.; Proels, R.K. Alcohol dehydrogenase 1 of barley modulates susceptibility to the parasitic fungus Blumeria graminis f.sp. hordei. J. Exp. Bot. 2011, 62, 3449–3457. [Google Scholar] [CrossRef] [PubMed]
- Ma, C.; Liu, Y.; Bai, B.; Han, Z.; Tang, J.; Zhang, H.; Yaghmaiean, H.; Zhang, Y.; Chai, J. Structural basis for BIR1-mediated negative regulation of plant immunity. Cell Res. 2017, 27, 1521–1524. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Bai, Y.; Wei, Y.; Shi, H. CAMTA3 negatively regulates disease resistance through modulating immune response and extensive transcriptional reprogramming in cassava. Tree Physiol. 2020, 40, 1520–1533. [Google Scholar] [CrossRef] [PubMed]
- Thomazella, D.P.D.T.; Seong, K.; Mackelprang, R.; Dahlbeck, D.; Geng, Y.; Gill, U.S.; Qi, T.; Pham, J.; Giuseppe, P.; Lee, C.Y.; et al. Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. USA 2021, 118, e2026152118. [Google Scholar] [CrossRef]
- Sun, K.; Wolters, A.A.; Loonen, A.E.H.M.; Huibers, R.P.; van der Vlugt, R.; Goverse, A.; Jacobsen, E.; Visser, R.G.F.; Bai, Y. Down-regulation of Arabidopsis DND1 orthologs in potato and tomato leads to broad-spectrum resistance to late blight and powdery mildew. Transgenic Res. 2016, 25, 123–138. [Google Scholar] [CrossRef]
- Zhang, Y.; Bai, Y.; Wu, G.; Zou, S.; Chen, Y.; Gao, C.; Tang, D. Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 2017, 91, 714–724. [Google Scholar] [CrossRef]
- Chen, H.; Xue, L.; Chintamanani, S.; Germain, H.; Lin, H.; Cui, H.; Cai, R.; Zuo, J.; Tang, X.; Li, X.; et al. ETHYLENE INSENSITIVE3 and ETHYLENE INSENSITIVE3-LIKE1 repress SALICYLIC ACID INDUCTION DEFICIENT2 expression to negatively regulate plant innate immunity in Arabidopsis. Plant Cell 2009, 21, 2527–2540. [Google Scholar] [CrossRef]
- Anderson, J.C.; Wan, Y.; Kim, Y.; Pasa-Tolic, L.; Metz, T.O.; Peck, S.C. Decreased abundance of type III secretion system-inducing signals in Arabidopsis mkp1 enhances resistance against Pseudomonas syringae. Proc. Natl. Acad. Sci. USA 2014, 111, 6846–6851. [Google Scholar] [CrossRef]
- La Camera, S.; Geoffroy, P.; Samaha, H.; Ndiaye, A.; Rahim, G.; Legrand, M.; Heitz, T. A pathogen-inducible patatin-like lipid acyl hydrolase facilitates fungal and bacterial host colonization in Arabidopsis. Plant J. 2005, 44, 810–825. [Google Scholar] [CrossRef] [PubMed]
- Vogel, J.P.; Raab, T.K.; Somerville, C.R.; Somerville, S.C. Mutations in PMR5 result in powdery mildew resistance and altered cell wall composition. Plant J. 2004, 40, 968–978. [Google Scholar] [CrossRef] [PubMed]
- Huibers, R.P.; Loonen, A.E.; Gao, D.; Van den Ackerveken, G.; Visser, R.G.; Bai, Y. Powdery mildew resistance in tomato by impairment of SlPMR4 and SlDMR1. PLoS ONE 2013, 8, e67467. [Google Scholar] [CrossRef]
- Curtis, R.H.C.; Pankaj; Powers, S.J.; Napier, J.; Matthes, M.C. The Arabidopsis F-box/Kelch-repeat protein At2g44130 Is upregulated in giant cells and promotes nematode susceptibility. Mol. Plant Microbe Interact. 2013, 26, 36–43. [Google Scholar] [CrossRef] [PubMed]
- Peng, Z.; Hu, Y.; Zhang, J.; Huguet-Tapia, J.C.; Block, A.K.; Park, S.; Sapkota, S.; Liu, Z.; Liu, S.; White, F.F. Xanthomonas translucens commandeers the host rate-limiting step in ABA biosynthesis for disease susceptibility. Proc. Natl. Acad. Sci. USA 2019, 116, 20938–20946. [Google Scholar] [CrossRef]
- Liu, L.; Wang, Y.; Cui, F.; Fang, A.; Wang, S.; Wang, J.; Wei, C.; Li, S.; Sun, W. The type III effector AvrXccB in Xanthomonas campestris pv. campestris targets putative methyltransferases and suppresses innate immunity in Arabidopsis. Mol. Plant Pathol. 2017, 18, 768–782. [Google Scholar] [CrossRef]
- Campa, M.; Piazza, S.; Righetti, L.; Oh, C.; Conterno, L.; Borejsza-Wysocka, E.; Nagamangala, K.C.; Beer, S.V.; Aldwinckle, H.S.; Malnoy, M. HIPM is a susceptibility gene of Malus spp.: Reduced expression reduces susceptibility to Erwinia amylovora. Mol. Plant Microbe Interact. 2019, 32, 167–175. [Google Scholar] [CrossRef]
- Cohn, M.; Bart, R.S.; Shybut, M.; Dahlbeck, D.; Gomez, M.; Morbitzer, R.; Hou, B.; Frommer, W.B.; Lahaye, T.; Staskawicz, B.J. Xanthomonas axonopodis Virulence is promoted by a transcription activator-like effector-mediated induction of a SWEET sugar transporter in Cassava. Mol. Plant Microbe Interact. 2014, 27, 1186–1198. [Google Scholar] [CrossRef]
- Li, P.; Zhang, L.; Mo, X.; Ji, H.; Bian, H.; Hu, Y.; Majid, T.; Long, J.; Pang, H.; Tao, Y.; et al. Rice aquaporin PIP1;3 and harpin Hpa1 of bacterial blight pathogen cooperate in a type III effector translocation. J. Exp. Bot. 2019, 70, 3057–3073. [Google Scholar] [CrossRef]
- Hui, S.; Shi, Y.; Tian, J.; Wang, L.; Li, Y.; Wang, S.; Yuan, M. TALE-carrying bacterial pathogens trap host nuclear import receptors for facilitation of infection of rice. Mol. Plant Pathol. 2019, 20, 519–532. [Google Scholar] [CrossRef]
- Zhang, D.; Tian, C.; Yin, K.; Wang, W.; Qiu, J. Postinvasive bacterial resistance conferred by open stomata in Rice. Mol. Plant Microbe Interact. 2019, 32, 255–266. [Google Scholar] [CrossRef] [PubMed]
- Gao, S.; Guo, W.; Feng, W.; Liu, L.; Song, X.; Chen, J.; Hou, W.; Zhu, H.; Tang, S.; Hu, J. LTP3 contributes to disease susceptibility in Arabidopsis by enhancing abscisic acid (ABA) biosynthesis. Mol. Plant Pathol. 2016, 17, 412–426. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Zhang, Z.; Lei, Y.; Hu, G.; Liu, J.; Hao, M.; Chen, A.; Peng, Q.; Wu, J. Cotton WATs Modulate SA biosynthesis and local lignin deposition participating in plant resistance against Verticillium dahliae. Front. Plant Sci. 2019, 10, 526. [Google Scholar] [CrossRef]
- Kay, S.; Hahn, S.; Marois, E.; Hause, G.; Bonas, U. A bacterial effector acts as a plant transcription factor and induces a cell size regulator. Science 2007, 318, 648–651. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Shi, Y.; Zou, L.; Huang, J.; Shen, L.; Wang, Y.; Guan, D.; He, S. Pepper CaMLO6 negatively regulates ralstonia solanacearum resistance and positively regulates high temperature and high humidity responses. Plant Cell Physiol. 2020, 61, 1223–1238. [Google Scholar] [CrossRef]
- Xian, L.; Yu, G.; Wei, Y.; Rufian, J.S.; Li, Y.; Zhuang, H.; Xue, H.; Morcillo, R.J.L.; Macho, A.P. A Bacterial effector protein hijacks plant metabolism to support pathogen nutrition. Cell Host Microbe 2020, 28, 548–557. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deng, S.; Li, Z.; Liu, X.; Yang, W.; Wang, Y. Comparative Transcriptome Analysis Reveals Potential Genes Conferring Resistance or Susceptibility to Bacterial Canker in Tomato. Horticulturae 2023, 9, 242. https://doi.org/10.3390/horticulturae9020242
Deng S, Li Z, Liu X, Yang W, Wang Y. Comparative Transcriptome Analysis Reveals Potential Genes Conferring Resistance or Susceptibility to Bacterial Canker in Tomato. Horticulturae. 2023; 9(2):242. https://doi.org/10.3390/horticulturae9020242
Chicago/Turabian StyleDeng, Shuozhen, Ziyan Li, Xinyu Liu, Wencai Yang, and Yuqing Wang. 2023. "Comparative Transcriptome Analysis Reveals Potential Genes Conferring Resistance or Susceptibility to Bacterial Canker in Tomato" Horticulturae 9, no. 2: 242. https://doi.org/10.3390/horticulturae9020242
APA StyleDeng, S., Li, Z., Liu, X., Yang, W., & Wang, Y. (2023). Comparative Transcriptome Analysis Reveals Potential Genes Conferring Resistance or Susceptibility to Bacterial Canker in Tomato. Horticulturae, 9(2), 242. https://doi.org/10.3390/horticulturae9020242




