Transcriptome Analysis Identifies Genes Associated with Chlorogenic Acid Biosynthesis during Apple Fruit Development
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Measurement of Chlorogenic Acid Content
2.3. Determination of HCT and C3H Enzymatic Activities
2.4. Transcriptome Sequencing
2.5. RNA Extraction and cDNA Synthesis
2.6. Oligonucleotide Primers and Quantitative Real-Time PCR Analysis
2.7. Statistical Analysis
3. Results
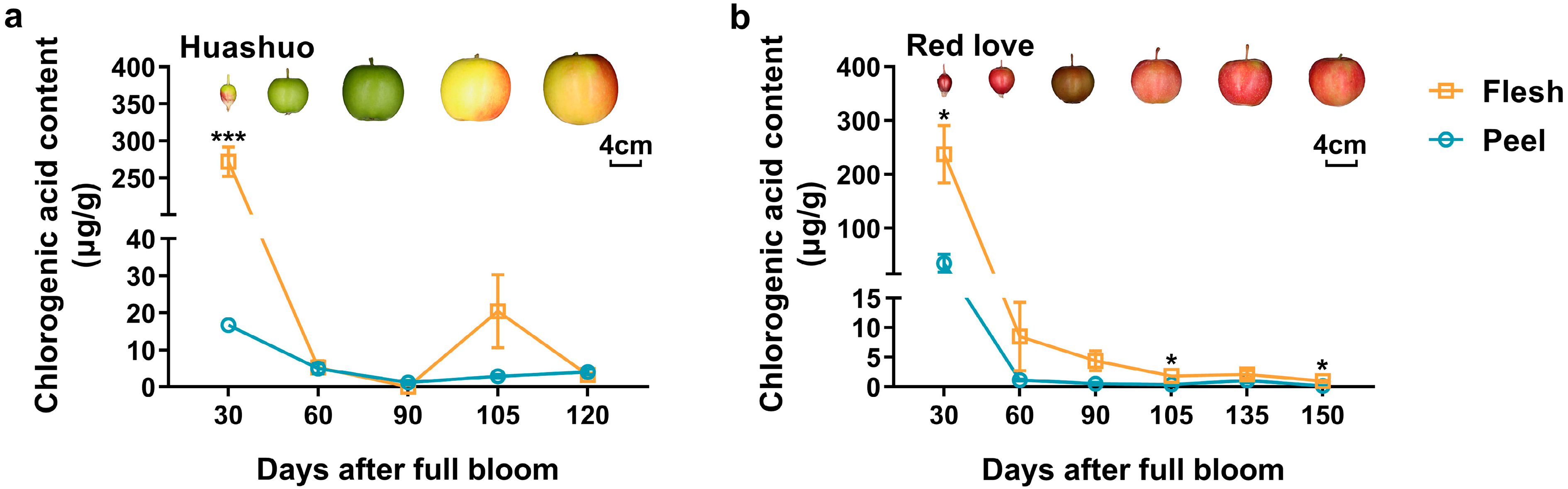
3.1. Changes of Chlorogenic Acid Content during Fruit Development
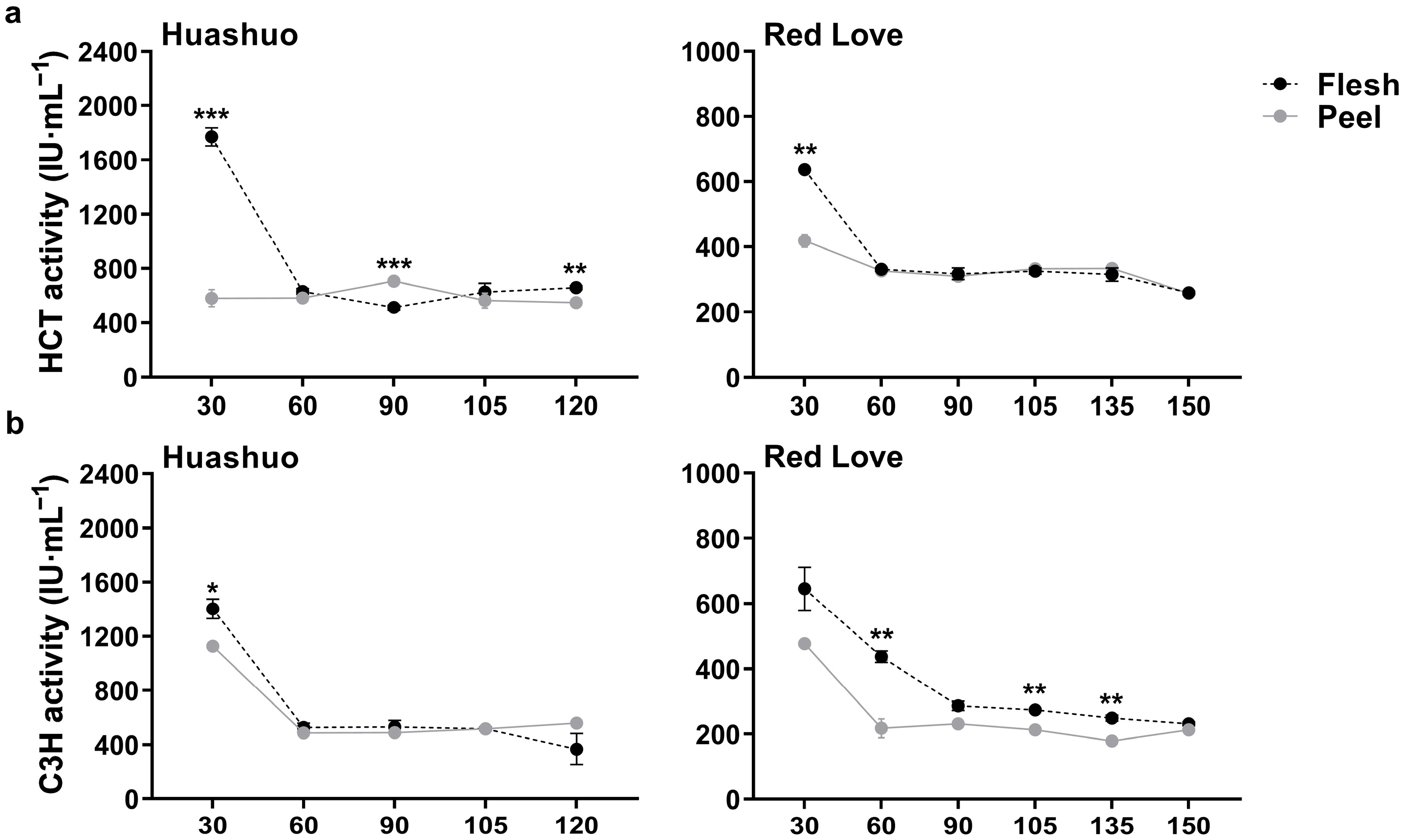
3.2. Changes of HCT and C3H Enzymatic Activities during Apple Fruit Development
3.3. Transcriptomic Analysis of Differentially Expressed Genes
3.4. Analysis of Differentially Expressed Genes Associated with CGA Biosynthesis during Apple Fruit Development
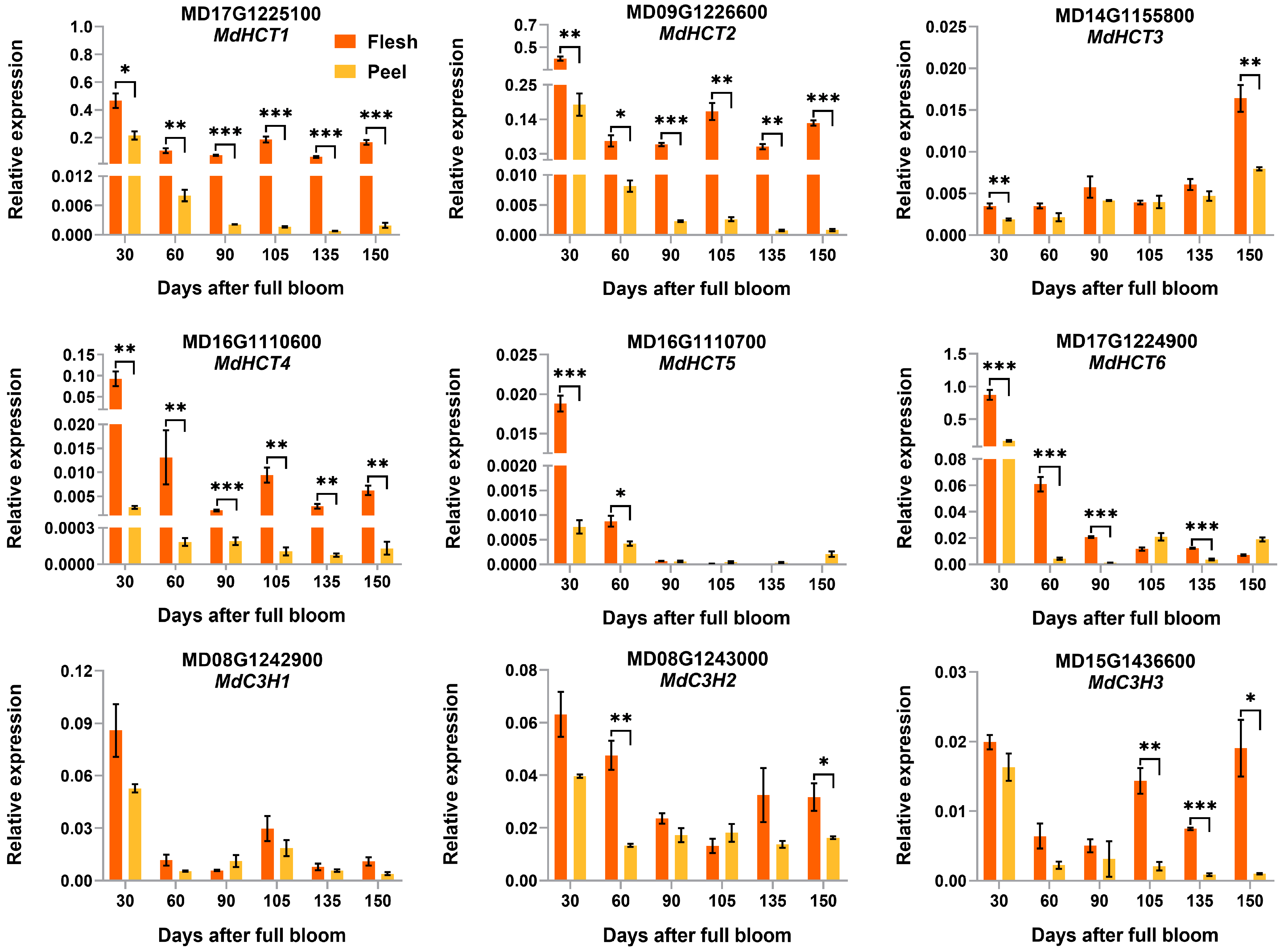
3.5. Validation of the Differentially Expressed CGA Biosynthesis-Associated Genes by qRT-PCR Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Liao, L.; Zhang, W.; Zhang, B.; Cai, Y.; Gao, L.; Ogutu, C.; Sun, J.; Zheng, B.; Wang, L.; Li, L.; et al. Evaluation of chlorogenic acid accumulation in cultivated and wild apples. J. Food Compos. Anal. 2021, 104, 104156. [Google Scholar] [CrossRef]
- Liaudanskas, M.; Viškelis, P.; Kviklys, D.; Raudonis, R.; Janulis, V. A comparative study of phenolic content in apple fruits. Int. J. Food Prop. 2015, 18, 945–953. [Google Scholar] [CrossRef]
- Shu, C.; Zhang, W.; Zhao, H.; Cao, J.; Jiang, W. Chlorogenic acid treatment alleviates the adverse physiological responses of vibration injury in apple fruit through the regulation of energy metabolism. Postharvest Biol. Technol. 2020, 159, 110997. [Google Scholar] [CrossRef]
- Chen, X.; Cai, W.; Xia, J.; Yu, H.; Wang, Q.; Pang, F.; Zhao, M. Metabolomic and transcriptomic analyses reveal that blue light promotes chlorogenic acid synthesis in strawberry. J. Agric. Food Chem. 2020, 68, 12485–12492. [Google Scholar] [CrossRef] [PubMed]
- Su, X.; Zhang, J.; Wang, H.; Xu, J.; He, J.; Liu, L.; Zhang, T.; Chen, R.; Kang, J. Phenolic acid profiling, antioxidant, and anti-inflammatory activities, and mirna regulation in the polyphenols of 16 blueberry samples from China. Molecules 2017, 22, 312. [Google Scholar] [CrossRef]
- Wen, H.; Wang, W.; Jiang, X.; Wu, M.; Bai, H.; Wu, C.; Shen, L. Transcriptome analysis to identify candidate genes related to chlorogenic acid biosynthesis during development of Korla fragrant pear in Xinjiang. Food Sci. Hum. Wellness 2022, 11, 854–864. [Google Scholar] [CrossRef]
- Santana-Gálvez, J.; Cisneros-Zevallos, L.; Jacobo-Velázquez, D.A. Chlorogenic acid: Recent advances on its dual role as a food additive and a nutraceutical against metabolic syndrome. Molecules 2017, 22, 358. [Google Scholar] [CrossRef]
- Jiao, W.; Shu, C.; Li, X.; Cao, J.; Fan, X.; Jiang, W. Preparation of a chitosan-chlorogenic acid conjugate and its application as edible coating in postharvest preservation of peach fruit. Postharvest Biol. Technol. 2019, 154, 129–136. [Google Scholar] [CrossRef]
- Silva, N.; Mazzafera, P.; Cesarino, I. Should I stay or should I go: Are chlorogenic acids mobilized towards lignin biosynthesis? Phytochemistry 2019, 166, 112063. [Google Scholar] [CrossRef]
- Hoffmann, L.; Besseau, S.; Geoffroy, P.; Ritzenthaler, C.; Meyer, D.; Lapierre, C.; Pollet, B.; Legrand, M. Silencing of hydroxycinnamoyl-coenzyme a shikimate/quinate hydroxycinnamoyltransferase affects phenylpropanoid biosynthesis. Plant Cell 2004, 16, 1446–1465. [Google Scholar] [CrossRef]
- Pina, A.; Errea, P. Differential induction of phenylalanine ammonia-lyase gene expression in response to in vitro callus unions of Prunus spp. J. Plant Physiol. 2008, 165, 705–714. [Google Scholar] [CrossRef] [PubMed]
- Villegas, R.J.A.; Kojima, M. Purification and characterization of hydroxycinnamoyl d-glucose. Quinate hydroxycinnamoyl transferase in the root of the sweet potato, Ipomoea batatas lam. J. Biol. Chem. 1986, 261, 8729–8733. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Qin, S.; Li, C.; Wu, Q.; Jiang, C.; Yang, J.; Guo, X.; Ou, C. Differential gene expressions and phytohormone changes altered Lonicera japonica quality after plant introduction. Pharmacogn. Mag. 2019, 15, 18–23. [Google Scholar] [CrossRef]
- Strack, D.; Gross, W. Properties and activity changes of chlorogenic acid:glucaric acid caffeoyltransferase from tomato (Lycopersicon escufenium). Plant Physiol. 1990, 92, 41–47. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Wang, Y.; Yu, Y.; Ma, P.; Jia, Z.; Guo, X.; Xie, Y.; Bian, X. Overexpression of IbPAL1 promotes chlorogenic acid biosynthesis in sweetpotato. Crop J. 2021, 9, 204–215. [Google Scholar] [CrossRef]
- Zhao, L.; Wang, D.; Liu, J.; Yu, X.; Wang, R.; Wei, Y.; Wen, C.; Ouyang, Z. Transcriptomic analysis of key genes involved in chlorogenic acid biosynthetic pathway and characterization of MaHCT from Morus alba L. Protein Expr. Purif. 2019, 156, 25–35. [Google Scholar] [CrossRef]
- Yang, Y.; Cui, S.; Zhang, Y.; Wang, X.; Li, D.; Wang, R. PbHCT4 regulates growth through affecting chlorogenic acid (cga) content in pear. Sci. Hortic. 2022, 303, 111225. [Google Scholar] [CrossRef]
- Niggeweg, R.; Michael, A.J.; Martin, C. Engineering plants with increased levels of the antioxidant chlorogenic acid. Nat. Biotechnol. 2004, 22, 746–754. [Google Scholar] [CrossRef]
- Heo, J.; Adhikari, K.; Choi, K.S.; Lee, J. Analysis of caffeine, chlorogenic acid, trigonelline, and volatile compounds in cold brew coffee using high-performance liquid chromatography and solid-phase microextraction—Gas chromatography-mass spectrometry. Foods 2020, 9, 1746. [Google Scholar] [CrossRef]
- Peng, Q.; Zhu, Y.; Liu, Z.; Du, C.; Li, K.; Xie, D. An integrated approach to demonstrating the ANR pathway of proanthocyanidin biosynthesis in plants. Planta 2012, 236, 901–918. [Google Scholar] [CrossRef]
- Wang, M.; Li, T.; Wu, Y.; Song, S.; Bai, T.; Jiao, J.; Song, C.; Zheng, X. Genome-wide identification of microRNAs involved in the regulation of fruit ripening in apple (Malus domestica). Sci. Hortic. 2021, 289, 110416. [Google Scholar] [CrossRef]
- Hu, G.; Dong, Y.; Zhang, Z.; Fan, X.; Ren, F. Elimination of apple necrosis mosaic virus from potted apple plants by thermotherapy combined with shoot-tip grafting. Sci. Hortic. 2019, 252, 310–315. [Google Scholar] [CrossRef]
- Zheng, X.; Wu, Y.; Wang, H.; Song, S.; Bai, T.; Jiao, J.; Song, C.; Pang, H.; Wang, M. Genome-wide investigation of the zinc finger-homeodomain family genes reveals potential roles in apple fruit ripening. Front. Genet. 2022, 12, 783482. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Su, Z.; Jia, H.; Sun, M.; Cai, Z.; Shen, Z.; Zhao, B.; Li, J.; Ma, R.; Yu, M.; Yan, J. Integrative analysis of the metabolome and transcriptome reveals the molecular mechanism of chlorogenic acid synthesis in peach fruit. Front. Nutr. 2022, 9, 1–16. [Google Scholar] [CrossRef]
- Ju, Z.; Yuan, Y.; Liou, C.; Xin, S. Relationships among phenylalanine ammonia-iyase activity, simple phenol concentrations and anthocyanin accumulation in apple. Sci. Hortic. 1995, 61, 215–226. [Google Scholar] [CrossRef]
- Wojdyło, A.; Oszmiański, J. Antioxidant activity modulated by polyphenol contents in apple and leaves during fruit development and ripening. Antioxidants 2020, 9, 567. [Google Scholar] [CrossRef]
- Han, G.; Bai, G.; Wu, Y.; Zhou, Y.; Yao, W.; Li, L. Comparative transcriptome analysis to identify candidate genes related to chlorogenic acid and flavonoids biosynthesis in iridaceae. Forests 2022, 13, 1632. [Google Scholar] [CrossRef]
- Yin, S.; Cui, H.; Zhang, L.; Yan, J.; Qian, L.; Ruan, S. Transcriptome and metabolome integrated analysis of two ecotypes of Tetrastigma hemsleyanum reveals candidate genes involved in chlorogenic acid accumulation. Plants 2021, 10, 1288. [Google Scholar] [CrossRef]
- Zhao, L.; Shan, C.; Shan, T.; Xu, J.; Zhang, S.; Tao, Y.; Wu, J. Probing the transcriptome of Boehmeria nivea reveals candidate genes associated with the biosynthesis of chlorogenic acid. Gene 2022, 833, 146579. [Google Scholar] [CrossRef]
- Tang, N.; Cao, Z.; Yang, C.; Ran, D.; Wu, P.; Gao, H.; He, N.; Liu, G.; Chen, Z. A R2R3-MYB transcriptional activator LmMYB15 regulates chlorogenic acid biosynthesis and phenylpropanoid metabolism in Lonicera macranthoides. Plant Sci. 2021, 308, 110924. [Google Scholar] [CrossRef] [PubMed]
- Schoch, G.; Goepfert, S.; Morant, M.; Hehn, A.; Meyer, D.; Ullmann, P.; Werck-Reichhart, D. CYP98A3 from Arabidopsis thaliana is a 3′-hydroxylase of phenolic esters, a missing link in the phenylpropanoid pathway. J. Biol. Chem. 2001, 276, 36566–36574. [Google Scholar] [CrossRef] [PubMed]
- St-Pierre, B.; Laflamme, P.; Alarco, A.M.; De Luca, V. The terminal o-acetyltransferase involved in vindoline biosynthesis defines a new class of proteins responsible for coenzyme A-dependent acyl transfer. Plant J. 1998, 14, 703–713. [Google Scholar] [CrossRef] [PubMed]
- He, J.; Cheng, Y.; Guan, J.; Ge, W.; Zhao, Z. Changes of chlorogenic acid content and its synthesis-associated genes expression in Xuehua pear fruit during development. J. Integr. Agric. 2017, 16, 471–477. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, H.; Zheng, X.; Wu, Y.; Zhan, W.; Guo, Y.; Chen, M.; Bai, T.; Jiao, J.; Song, C.; Song, S.; et al. Transcriptome Analysis Identifies Genes Associated with Chlorogenic Acid Biosynthesis during Apple Fruit Development. Horticulturae 2023, 9, 217. https://doi.org/10.3390/horticulturae9020217
Wang H, Zheng X, Wu Y, Zhan W, Guo Y, Chen M, Bai T, Jiao J, Song C, Song S, et al. Transcriptome Analysis Identifies Genes Associated with Chlorogenic Acid Biosynthesis during Apple Fruit Development. Horticulturae. 2023; 9(2):217. https://doi.org/10.3390/horticulturae9020217
Chicago/Turabian StyleWang, Hao, Xianbo Zheng, Yao Wu, Wenduo Zhan, Yanfei Guo, Ming Chen, Tuanhui Bai, Jian Jiao, Chunhui Song, Shangwei Song, and et al. 2023. "Transcriptome Analysis Identifies Genes Associated with Chlorogenic Acid Biosynthesis during Apple Fruit Development" Horticulturae 9, no. 2: 217. https://doi.org/10.3390/horticulturae9020217
APA StyleWang, H., Zheng, X., Wu, Y., Zhan, W., Guo, Y., Chen, M., Bai, T., Jiao, J., Song, C., Song, S., & Wang, M. (2023). Transcriptome Analysis Identifies Genes Associated with Chlorogenic Acid Biosynthesis during Apple Fruit Development. Horticulturae, 9(2), 217. https://doi.org/10.3390/horticulturae9020217




