Abstract
The vast diversity of traits exhibited by horticultural crops largely depends upon variation in gene expression regulation. The uppermost layer of gene expression regulation is chromatin compaction. In plants, the LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) is a member of the Polycomb Repressive Complex 1 (PRC1) that controls the spreading of the H3K27me3 mark throughout the genome to regulate gene expression. Much of the epigenetic control exerted by LHP1 has been deeply explored on the model species Arabidopsis thaliana. Recent advances in melon, tomato, and soybean highlight the relevance of LHP1 in controlling the development and physiology of a plethora of traits in crops. However, whether LHP1 exerts its diverse roles through similar mechanisms and through modulating the same target genes has been overlooked. In this review, we gather a wealth of knowledge about the LHP1 mode of action, which involves a tight connection with histone marks and long noncoding RNAs to modulate gene expression. Strikingly, we found that LHP1 may be linked to H3K27me3 regulation across the plant lineage, yet, through epigenetic regulation of a distinct set of target genes. This is supported by subtle differences in subcellular LHP1 localization between species found here. In addition, we summarize the variety of developmental outputs modulated by LHP1 across land plants pinpointing its importance for plant breeding. Hence, LHP1 has probably been co-opted in different lineages to modulate diverse traits contributing to crop diversification.
1. Introduction
Plant developmental diversity is the result of differential gene expression. The uppermost layer of gene expression regulation is governed by epigenetic mechanisms. Epigenetic gene repression is mainly controlled by the coordinated action of the Polycomb Repressive Complexes 1 and 2 (PRC1 and 2) through the deposition of the Trimethylation of Histone H3 Lysine 27 (H3K27me3) repressive mark. The LIKE HETEROCHROMATIN PROTEIN 1 (LHP1) is a pivotal component of the epigenetic machinery, being the first identified member of PRC1 in plants [1]. The main role exerted by LHP1 is to repress gene expression throughout maintaining the H3K27me3 mark over the 3′ end of target gene bodies in euchromatic regions. Moreover, through this regulatory path, LHP1 modulates the global genome topology in the model species Arabidopsis thaliana [2]. Consequently, LHP1 is crucial for several developmental processes in Arabidopsis, such as flowering time, flower morphology, plant height, biomass, and plant response to environmental cues [3,4,5]. Despite the progress in knowing the biological role and mode of action of LHP1 for Arabidopsis, an understanding of LHP1’s role in other plant species is in its infancy. For instance, what aspects of the molecular function of LHP1 are conserved or have diverged within the plant kingdom are misunderstood. In this review, we gathered all the knowledge about LHP1 roles in model and crop plants, and we identified the main open questions about conserved and divergent aspects related to its mode of action. Here we underpin that LHP1 acts as an epigenetic remodeler across the plant kingdom by controlling conserved and distinct molecular pathways. On the other hand, considering the diversity of biological roles depicted by LHP1 in different species, we postulate that this PRC1 component has been co-opted in different plant groups to exert specific functions.
2. LHP1 Regulates Gene Expression through Histone Modification and Chromatin Conformation
2.1. Origin, Structure, and Localization of LHP1
LHP1 was initially known as TERMINAL FLOWER 2 (TFL2) or TU8. The early flowering of tfl-2 and the altered leaf glucosinolate phenotype of TU8 were considered to determine their initial names, respectively [3,6]. Later, this protein was renamed LHP1 due to its similarity to the Drosophila melanogaster Heterochromatin Protein 1 (HP1) [7,8]. Phylogenetic analyses revealed that the LHP1 gene is present in all land plants (Embryophyte). Surprisingly, although chromodomain proteins exist in algae, an LHP1 homolog is absent in this clade. Therefore, a secondary loss scenario was suggested to account for the lack of LHP1 in this particular clade of green organisms [9,10]. Notably, LHP1 has been shown to be able to rescue the fission yeast mutant swi6−, the HP1 ortholog in Schizosaccharomyces pombe [7,8]. The homology between HP1 and LHP1 is postulated mainly due to the presence of two conserved protein domains, the chromodomain (CD) and the chromo shadow domain (CSD), being close to the N-terminus and the C-terminus, respectively. While the CD confers the ability of histone binding, the CDS is responsible for protein dimerization [7,11]. In animals, HP1 is tightly associated with the H3K9me3 epigenetic mark across heterochromatic genome regions [12]. Particularly, the CD domain is responsible for HP1 function as a histone reader [12]. Given the structural similarities between HP1 and LHP1, it was initially thought that the latest also functions as an epigenetic remodeler through the binding of the Histone H3 Lysine 9 (H3K9) [13,14]. Turck and collaborators [1] analyzed the affinity of LHP1 with histones. They observed that LHP1 binds Di- and Trimethylation of Histone H3 Lysine 9 (H3K9me2 and H3K9me3) as its animal homolog. Contrary to expectations, they also unveiled that LHP1 is mainly associated with the H3K27me3 epigenetic mark across euchromatic genome regions [1,15,16]. Several reports confirmed that the CD domain is the main responsible for histone binding [11,17,18,19]. Very recently, the crystal structure of the LHP1 CD domain was obtained and characterized. Surprisingly, in vitro affinity tests showed that the CD of AtLHP1 has no particular preference between Di and Tri-methylated forms of H3K9 or H3K27 peptides [17]. Although there is certainty about the dependence on the CD domain for histone binding, why LHP1 is associated in vivo mainly with H3K27me3 instead of H3K9me3 remains unclear. On the other hand, the CDS domain depicts a distinctive function in animal HP1. It was proposed that the CDS domain plays a determinant role in protein-protein interaction. This further promotes protein dimerization required for HP1 activity as an epigenetic remodeler [20]. Likewise, the CDS in plants is also required for LHP1 dimerization and protein interaction [3,7,14,19,21,22].
In addition, LHP1 has an intrinsically disordered region between the CD and the CDS-denominated Hinge [15]. This region has been thoroughly characterized as the one responsible for RNA binding. The capacity of LHP1 to bind RNAs has been substantially proven both in vitro and in vivo [19,23,24]. Notably, replacing several Arginine and Lysine residues in the Hinge region reduces or even abolishes LHP1’s capacity to bind RNAs. Therefore, these positively charged residues are necessary for RNA binding. Interestingly, the subnuclear localization of LHP1 was also affected when RNA binding was disrupted, highlighting the relevance of this interaction for LHP1 function. Nuclear speckles formation is visibly impaired when the Hinge region is perturbed, most likely affecting the function of LHP1 as an epigenetic remodeler [15,18,19] (Figure 1).
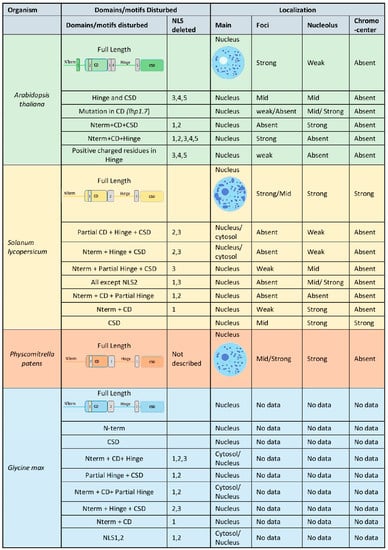
Figure 1.
Summary of LHP1 subcellular localization in model plants and crops. Arabidopsis thaliana: [3,4,7,11,15,25]; Solanum lycopersicum: [18]; Physcomitrella patens: [21,26]; Glycine max: [27].
Remarkably, LHP1 also contains two plant-specific motifs, motif I and II, buried within the Hinge region [9]. It was shown that motif I is necessary for LHP1 interaction with the ALPHA-THALASSEMIA MENTAL RETARDATION SYNDROME, X-LINKED (ATRX) complex, involved in H3.3 deposition similar to its ortholog in animals. ATRX interacts with HP1 in animals through the so-called “LxVxL” motif [25,26,27]. Although plant ATRX lacks the “LxVxL” motif, the interaction between LHP1 and ATRX is conserved [28]. Interestingly, it was revealed that this interaction in plants occurs through the plant-specific LAN (LHP1-interacting ATRX N-terminus) motifs in ATRX and the motif I of LHP1 [28], pinpointing a convergent evolutionary path.
Considering the origin of LHP1, which suggests a function as an epigenetic remodeler for being homolog to HP1, the analysis of the subcellular protein localization always attracts the attention of plant biologists. In Arabidopsis, LHP1 is strictly localized in the nuclei, likely due to the presence of several nuclear localization signals (NLS) in the LHP1 peptide [7] (Figure 1). In addition, LHP1 displays a distinctive foci patterning, which has been mainly associated with the CD/CSD domains and is almost absent from the nucleolus [7,15,19]. Nevertheless, LHP1 has nucleolar localizing signals (NoLS) within the hinge region. Only by removing the N-terminal part of the protein (including the CD domain), LHP1 invades the nucleolus implying that other functional domains prevent the localization in this compartment despite having a NoLS [11,15]. Strikingly, LHP1 was also found in the nuclei and nuclear foci of the moss Physcomitrella patens, meaning that this subcellular pattern is ancient. However, the abundant presence of LHP1 in the nucleolus of P. patens also reveals a distinctive pattern [29]. Moreover, an inspection of LHP1 localization in tomato (Solanum lycopersicum) revealed more similarities with the moss P. patens than with Arabidopsis, where nucleolus occupancy was also observed [18,29]. The LHP1 protein of soybean (Glycine max) also showed nuclear localization in Arabidopsis protoplast. Unfortunately, the subnuclear localization was not distinguished in this heterologous system [30]. Remarkably, the protein regions required for the different nuclear compartmentalization were dissected in tomato and soybean. Oppositely to the observations in Arabidopsis, the CDS domain of tomato is dispensable for nuclear localization and speckle formation, while disturbing the hinge region affected both patterns. In line with this, the CD can only generate a weak nuclear localization without nucleolar or speckles occupancy [18]. In soybean, LHP1 has three predicted NLS, one (NLS1) within the CD domain and two (NLS2 and 3) within the hinge region. Analyses of truncated versions of soybean LHP1 revealed the properties of the different protein domains and predicted peptide signals. In agreement with previous findings, the CDS was also found to be dispensable for nuclear localization in soybean. Notably, disruption of NLS1 and NLS2 resulted in protein accumulation in the whole cell [30], further validating their importance for LHP1 localization and, thereof, functioning. In agreement with these observations, in all the plant species studied to date, the LHP1 NLS located within the hinge region and close to the CD is highly conserved as a bipartite NLS (NLS class 3 and 4). Moreover, it has been shown that this specific NLS is redundantly bound by importin [alpha]-1 (IMPα-1), IMPα-2, and IMPα-3 in Arabidopsis. As a result, LHP1 is not correctly imported into the nuclei in plants lacking these importinα proteins [31]. Altogether, data obtained in different species show that while the NLSs located within the hinge region are required for nuclear and nucleolar localization, the CD and the CDS are mainly linked to foci patterning in the nucleoplasm (Figure 1). It is noteworthy to mention that Arabidopsis is the only species so far where LHP1 does not show a clear presence in the nucleolus.
2.2. LHP1 Is a Member of the Polycomb Repressive Complex 1 in Plants
Epigenetic repression of the FLOWERING LOCUS C (FLC) locus during vernalization was initially associated with histone deacetylation and H3K27me2 and H3K9me2 methylation [14,32]. Later, based on the flowering time of lhp1 plants, it was suggested that LHP1 participates in the vernalization process throughout maintaining the epigenetic repression of FLC [14]. Nevertheless, at that moment, the epigenetic mechanism by which LHP1 modulates flowering time throughout FLC was still enigmatic. Later, Turck and collaborators [1] proved that LHP1 binds the histone marks H3 Di- or Tri-methylated at Lysine 9 (H3K9me2 or H3K9me3), also recognized by HP1 in animals. Notably, they also revealed the association between LHP1 and the H3K27me3 repressive mark for the first time. Moreover, they found that LHP1 is almost exclusively associated with loci decorated with H3K27me3 (Figure 2). However, given that there is no re-distribution of H3K27me3 in lhp1 plants, they concluded that LHP1 does not act as an H3K27me3 writer. In animals, global epigenetic gene repression is modulated by the coordinated action of PRC1 and 2 throughout the deposition of the H3K27me3 mark. In plants, the PRC2 histone methyltransferase units CURLY LEAF (CLF) and SWINGER (SWN) deposit H3K27me3, being homolog components of the animal PRC2 EZH2 [33]. Interestingly, it was demonstrated that LHP1 interacts with MULTICOPY SUPPRESSOR OF IRA1 (MSI1), EMBRYONIC FLOWER 1 (EMF1), and indirectly with other PRC2 components such as SWN [34], and has very similar spatial and temporal localization over the FLC gene body. These combined writing (PRC2) and reading (LHP1) activities for H3K27me3 deposition may contribute to the distribution and maintain this repressive mark over the FLC gene body [35]. LHP1 was the first identified member of the PRC1 complex in plants [1]. However, despite the revealed association between LHP1 and H3K27me3, its PRC1 role was yet elusive. More recently, the role of LHP1 in H3K27me3 mark modulation was further unveiled through the combination of multiple biochemical approaches [2] (Figure 2) (See below).
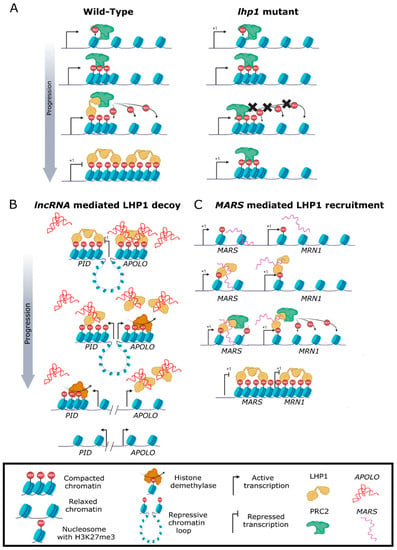
Figure 2.
LHP1 modifies the chromatin profile in plants. (A) LHP1 integrates the Polycomb Repressive Complex 1 to control the stability of the repressive histone mark H3K27me3. (B) LHP1 functions as an RNA Binding Protein (RBP) through its Hinge domain. An excess of the lncRNA APOLO can decoy LHP1 from target loci. (C) LHP1 binding over the MRN1 locus requires the activity of the lncRNA MARS to modulate marneral biosynthesis.
2.3. LHP1 Interacts with lncRNAs to Modulate the Transcriptional Machinery
The study of LHP1 was initially focused on its participation in global gene repression as a member of the PRC1. In the last few years, a growing interest in its novel function as an RNA-binding protein has emerged [5,19,23,24,36,37]. Interestingly, certain members of the CD-containing family from the animal kingdom also recognize RNA in vivo [38,39]. To date, three long noncoding RNAs (lncRNAs) have been identified to interact physically with LHP1. An antisense portion of the exon 1/intron 1 of FLC corresponding to the lncRNA COOLAIR was used to test the interaction in vitro, validating the relevance of the hinge domain for RNA interaction [19]. In addition, RNA immunoprecipitation (RIP) experiments performed in Arabidopsis served to demonstrate the interaction between LHP1 and the lncRNAs AUXIN-REGULATED PROMOTER LOOP (APOLO) and MARneral Silencing (MARS) in vivo [23,24,36] (Figure 2B,C). It was recently shown that APOLO could recognize its targets in trans through the establishment of RNA:DNA hybrids (R-loop) in the target locus. A subset of APOLO bona fide targets was classified according to its co-localization with LHP1-bound regions. In addition, there is a significant and positive correlation with H3K27me3 levels over those direct targets highlighting the relevance of APOLO-LHP1 interaction [24]. For instance, one of the common targets is the auxin biosynthesis gene YUCCA2 (YUC2), required for plant thermomorphogenic response. Most notably, excess APOLO can act as a decoy for LHP1 and remove it from the YUC2 locus at warm temperatures de-repressing its transcription [5]. It was also shown that APOLO interacts with hemimethylated DNA binding protein VARIANT IN METHYLATION 1 (VIM1) and that LHP1-VIM1 direct interaction also occurs. Interestingly, VIM1 human homolog UHRF1 was shown to co-localize with HP1 [40], and they both interact with lncRNAs [40,41], hinting at similar ribonucleoprotein complexes involved in epigenetics across kingdoms [5]. Furthermore, it was reported that LHP1 is able to bind the human lncRNA UHRF1 Protein Associated Transcript (UPAT) when expressed in plant cells [5], highlighting the ability of LHP1 to recognize sequence-unrelated lncRNAs. Interestingly, a more general regulation of YUCCA genes by LHP1 was also uncovered in Arabidopsis, hinting at a key role of LHP1 in auxin homeostasis [42]. On the other hand, the interaction between LHP1 and MARS unveiled a distinct set of co-regulated genes. Yet, in response to abscisic acid (ABA), MARS can act as a decoy for LHP1 and remove it from the marneral cluster gene responsible for marneral biosynthesis, MARNERAL SYNTHASE (MRN1). Conversely, under normal conditions, LHP1 binding over MRN1 is dependent on MARS activity [36]. Therefore, depending on the lncRNA interacting partner, LHP1 directly regulates the expression of distinct sets of targets. Contrary to the knowledge about the conserved association between LHP1 and histone modifications in plants, there is no report of lncRNAs LHP1-interactors besides the ones identified in Arabidopsis. Considering that the majority of lncRNAs are species-specific, this might provide a highly dynamic regulatory mechanism across different plant species that deserves further attention.
2.4. PRC1 Activity of LHP1 Modulates Genome Topology to Ultimately Alter Gene Transcription
Another breakthrough discovery related to the molecular role of LHP1 as an epigenetic remodeler was conducted by Veluchamy and collaborators [2]. Throughout combining genome-wide transcriptomic (RNA-seq), chromatin binding (ChIP-seq), and chromatin tridimensional (Hi-C) analyses, this work unveiled that LHP1 modulates the spreading of H3K27me3 towards the 3′ end of gene bodies at a genome-wide scale in Arabidopsis. Outstandingly, altered accumulation of the H3K27me3 repressive mark exhibited by lhp1 plants is correlated with differential gene looping. The 76.8% of chromosomal specific-LHP1 associated interactions are affected in the mutant lhp1 background displaying reduced H3K27me3 levels. Thus, the resultant reorganization of the genome conformation utterly alters the transcription of euchromatic genes. For instance, the formation of the chromatin loop encompassing PINOID (PID) and APOLO loci is disrupted, causing altered transcription of PID [2]. PID encodes a kinase involved in auxin efflux, further linking LHP1 and auxin signaling. Moreover, the lncRNAs APOLO and MARS can decoy LHP1 from the chromatin in cis and/or in trans, de-repressing gene expression of specific targets by modifying chromatin accessibility [5,24,36,37].
Besides the PRC1 gene repression exerted, LHP1 can also activate the expression of a significant set of genes in a direct manner [2]. Nevertheless, the mechanism behind gene activation is far less understood.
3. LHP1 Modulates a Plethora of Traits in Model Plants and Crops
3.1. In Model Plants
The biological relevance of LHP1/TFL2 was first unveiled in the A. thaliana knockout mutant tfl2-1 that exhibited an accelerated flowering time [3]. Inspecting several tfl2 mutants, it was observed that these plants had approximately four fewer leaves compared to the wild type at bolting [8]. Moreover, altered plant height and the development of other organs, such as leaves and shoots, reflect a pleiotropic effect of LHP1 abolition [7]. Many efforts helped to decipher the molecular basis lying behind the accelerated flowering time in the lhp1 mutants. LHP1 modulates several flowering time genes. Among them, LHP1 represses the expression of the FLOWERING LOCUS T (FT), a key player in promoting the transition to flowering. Interestingly, the lack of TFL2/LHP1 completely rescues the late flowering phenotype of ft-1 plants [8,43]. Later, the layer of epigenetic control exerted by LHP1 in the flowering pathway was revealed. Remarkably, Turck and collaborators [1] uncovered the epigenetic mechanism by which LHP1 associates with H3K27me3 at the FT locus to repress its expression in a canonical PRC1-dependent manner.
Until recently, given the importance of flowering time as an adaptive trait, other phenotypes associated with LHP1 biological roles were somehow unattended. Notably, Ramirez-Prado and collaborators [4] revealed that LHP1 plays important roles in plant immunity and abiotic stress response in Arabidopsis. For instance, LHP1 reduces ABA sensitivity by directly repressing the expression of the ABA-responsive genes NAC Transcription Factor ANAC019, ANAC055, and VEGETATIVE STORAGE PROTEIN 1 (VSP1) [4]. The plant response to the stress produced by salinity or drought is mainly regulated by ABA. Accordingly, plants lacking LHP1 have an increased tolerance to water deprivation. On the other hand, VSP1 is also required for plant response to biotic stress. Insect attack on Arabidopsis plants is counteracted by VSP1 [44,45]. In agreement, the augmented resistance observed of lhp1 mutant plants to the green peach aphid Myzus persicae is linked to increased levels of VPS1. In contrast, lhp1 plants exhibit increased susceptibility to the hemibiotrophic pathogen Pseudomonas syringae pv. tomato DC3000, probably linked to reduced salicylic acid (SA) content in these plants. Notably, BSMT1 is required for SA content. Therefore, the reduced content of SA is due to the direct repression exerted over the SA biosynthesis gene ISOCHORISMATE SYNTHASE 1 (ICS) and the upregulation of the inactivating enzyme SALICYLATE/BENZOATE CARBOXYL METHYLTRANSFERASE (BSMT1) by LHP1 [4]. It was recently uncovered that root hair development depends upon LHP1 function [37]. The master regulator of root hair elongation, ROOT HAIR DEFECTIVE 6 (RHD6), is targeted by LHP1 and the lncRNA APOLO at 22 °C. Notoriously, in low-temperature treatments (10 °C), APOLO expression is induced, precluding LHP1 binding over the RHD6 promoter, thereof impacting its transcriptional output. In agreement, the response to cold is impaired in lhp1 plants producing roots that are less elongated and with shorter hairs [37]. Therefore, soil exploration through root hair elongation requires LHP1 action in Arabidopsis. In addition, it was shown that the complex integrated by LHP1, VIM1, and APOLO is required for normal plant response to warm temperatures implicated in hypocotyl development [5]. This complex is responsible for the epigenetic control of YUCCA2 (YUC2) expression, involved in auxin biosynthesis during thermo-morphogenesis [46,47,48]. Under warm temperatures, the down-regulation of APOLO releases epigenetic control over YUC2. Accordingly, plants lacking LHP1 cannot elongate the hypocotyl at warm temperatures (29 °C) properly (Figure 3).
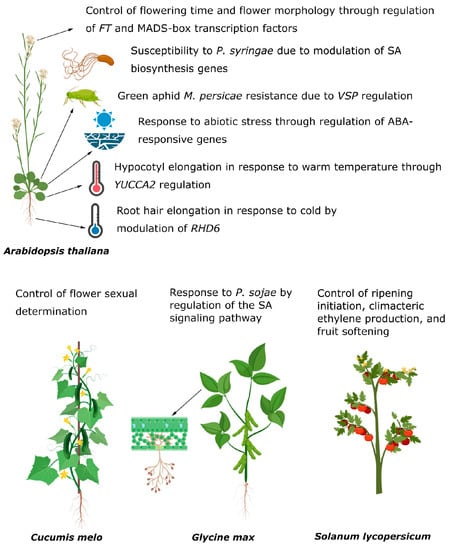
Figure 3.
Plant traits modulated by LHP1 in different species [3,4,5,14,30,32,37,44,45,46,47,48,49,50,51]. Illustrations were prepared with Biorender ©.
In addition to the model vascular plant A. thaliana, the function of LHP1 was also explored in the model moss Physcomitrella patens. Interestingly, PpLHP1 is also an important epigenetic remodeler despite the fact that it diverged from Arabidopsis more than 450 my ago [21,29,52]. Interestingly, the loss of LHP1 also produces pleiotropic defects in P. patens highlighting its biological relevance [29]. For instance, the colonies formed in the loss-of-function mutant exhibit a smaller size compared with WT plants, implying that LHP1 is required for normal growth. In addition, primary chloronema length and secondary chloronema number are also impaired in the Pplhp1 mutant. Moreover, normal chloroplast development is also affected by the absence of LHP1 across the gamethophore leaf cells. Outstandingly, PpLHP1 displays a subnuclear localization in foci, nucleolus, and interaction with other PRC1 members, revealing a conserved ancestral participation in the epigenetic machinery [29] (Figure 1). On the other hand, BiFC and Y2H assays showed that PpLHP1 could interact with DNA methylation 1 (DDM1), a component of DNA methylation machinery [52]. Considering that LHP1 and DDM1 interaction was only reported for moss, this might reveal an interesting and unexplored mode of epigenetic control specific to this group of plants. LHP1 interacts with the DNA methyltransferase CHROMOMETHYLASE3 (CMT3) in Arabidopsis [13]. In addition, the complex integrated by LHP1, VIM1, and APOLO can recruit the methylation machinery over YUC2 [5]. VIM1 was shown to directly interact with the DNA methyltransferase MET1 [53], whereas in humans, MET1 homolog DNMT1 directly interacts with HP1 and VIM1 homolog UHRF1 [54,55], further revealing a common interplay between HP1/LHP1 and the DNA methylation machinery between animals and plants. Altogether, these reports suggest that LHP1 also exerts an indirect role in regulating DNA methylation through multiple mechanisms.
3.2. In Crops
In the last few years, there has been an increasing interest in unraveling the function of LHP1 in crops. In this trend, the role of tomato (Solanum lycopersicum) LHP1 was recently explored. Interestingly, the tomato has two copies of LHP1 denominated SlLHP1a and SlLHP1b [49]. While SlLHP1a is stably expressed across all tissues, SlLHP1b expression is restricted to fruit during ripening. Functional experiments to disentangle the function of SlLHP1b showed that this protein is involved in ripening initiation, climacteric ethylene production, and fruit softening. While down-regulation of SlLHP1b promotes those events in tomato fruits, overexpressing tomato plants triggers the opposite effect [49]. SlLHP1b directly regulates the loci of several fruit-ripening genes (1-AMINOCYCLOPROPANE-1-CARBOXYLATE SYNTHASE 2 and 4 [ACS2, ACS4], POLYGALACTURONASE-2A [PG2a] and MADS-box transcription factor RIN) that are decorated with the H3K27me3 repressive mark, exhibiting the expected behavior as a member of the PRC1 complex. Moreover, enrichment levels of H3K27me3 are increased when SiLHP1b is overexpressed with a concomitant delay of the aforementioned events. In addition, similarly to AtLHP1, SlLHP1b interacts with the PRC2 member MSI1, further supporting its epigenetic control exerted over fruit-ripening genes. It is worth noting that although SiLHP1a is ubiquitously expressed in all the tissues analyzed, it remains unknown if both LHP1 proteins in tomatoes have a similar role or not [49]. Considering the relevance of the above-mentioned traits for tomato breeding, SlLHP1b might stand as a key player in the optimization of fruit ripening of this important vegetable crop.
LHP1 function was also recently dissected in melon (Cucumis melo), unveiling an unexpected role in sex determination (Figure 3). This economically important fruit crop is known for its monoecious reproductive system, and therefore, it is also considered a model for sex determination studies. Melon exhibits two LHP1 members, named CmLHP1A and CmLHP1B, that modulate sex determination. Single mutants of CmLHP1A or CmLHP1B do not display appreciable phenotypes suggesting that both proteins act redundantly [51]. Interestingly, cmlhp1a/cmlhp1b double mutant plants have an increased male:female ratio. To explain if the differential sex determination in cmlhp1/cmlhp1b plants is related to transcriptional changes associated with PRC1 activity, global profiling of H3K27me3 was performed in wild type and cmlhp1a/cmlhp1b plants. Strikingly, this showed a globally altered accumulation profile of H3K27me3 correlated with a PRC1-dependent activity. Hence, CmLHP1A and CmLHP1B cooperatively regulate the levels of H3K27me3 across the melon genome. Notoriously, the authors also reported a similar behavior for LHP1 and H3K27me3 association as the one found in Arabidopsis. Down-regulated and up-regulated loci exhibited H3K27me3 hypermethylation and H3K27me3 hypomethylation in cmlhp1/cmlhp1b plants, respectively [51].
Soybean (Glycine max) is one of the main extensive crops for protein production. The productivity of soybean is, however, severely affected by several pathogens that cause significant economic losses. One of the most lethal pathogens for soybean is Phytophthora sojae which causes stem and root rot [56]. Notably, up and down-regulation of GmLHP1 causes a reduction in soybean response to P. sojae by repressing the SA signaling pathway [30] (Figure 3). GmLHP1 represses the expression of the GmWRKY40 gene required for SA-signaling by two mechanisms. Through direct regulation, the promoter of GmWRKY40 is recognized and bound by GmLHP1, resulting in its transcriptional repression. On the other hand, GmWRKY40 expression is induced by SA. Hence, considering that GmLHP1 impairs SA accumulation in soybean, an alternative indirect mechanism by which GmWRKY40 is repressed was also suggested [30]. Nevertheless, it is relevant to point out that although a direct mechanism for GmWRKY40 repression by GmLHP1 was proposed, yet, it is unknown if this repression involves PRC1 activity. Strikingly, the work by Zhang and collaborators [30] also contributed to the understanding of an unknown dimension of LHP1 function modulation, which involves the regulation of protein action mediated by post-translational modifications. They provided solid evidence about the mechanism of GmLHP1 degradation by ubiquitination throughout the action of the GmBTB/POZ complex. Moreover, GmBTB/POZ increases the defense response of soybean against P. sojae [50]. Hence, this novel post-translational mechanism of repression of GmLHP1 action is required for soybean immunity. Notably, LHP1 acts as a negative regulator of SA in soybean and Arabidopsis hinting at a conserved role. Strikingly, it was reported that GmLHP1 also plays a role during stress response in soybean through a non-PCR1/2 canonical mechanism. The PLANT HOMEODOMAIN FINGER 6 (GmPHD6) interacts with the H3K4me0/1/2 marks and binds the G-rich elements in the promoters of the target loci. In turn, LHP1 is recruited by GmPHD6 and activates the expression of stress-associated genes [57] (Figure 3).
Altogether, considering the plant stress responses modulated by LHP1 in different species (Figure 3), this epigenetic remodeler may be used to mitigate biotic and abiotic stress in crops.
4. Future Directions to Explore LHP1 Function and Suitability for Crop Trait Optimization
Structural analysis of LHP1 revealed that it has a similar affinity for H3K9me3 and H3K27me3. Nevertheless, there is a vast amount of evidence showing LHP1 preference for H3K27me3 across plant genomes. Hence, the lying bases behind LHP1 association with H3K27me3 are still elusive. Note that structural analyses were performed by crystalizing the CD domain alone. Therefore, it is probable that other protein domains are also required to dictate LHP1 histone mark preference finally. Moreover, the CDS and the Hinge domains were observed to affect LHP1 localization and/or function when disturbed. In addition, considering that LHP1 can modulate the tridimensional chromatin structure, it is also likely that, in turn, the genomic and epigenomic context determines LHP1 affinity.
The conserved presence of the CD and the CSD between animals HP1 and plants LHP1 is considered the main evidence to support a common evolutionary origin. However, several crucial observations challenge this hypothesis. First, the absence of LHP1 in the genome of the basal clade of all green organisms, the algae, masks the phylogenetic origin of this protein. In a secondary loss scenario, as postulated [9,10], this event should have occurred earlier in the diversification of algae (given the lack of LHP1 in all extant sequenced algae) but not in the branch of the phylogenetic tree from which embryophyte has emerged. At least, we consider that this hypothesis needs to be revised in light of a complete picture of the genome evolution of earlier photosynthetic organisms. Second, despite the high similarities between HP1 and LHP1 chromodomains, they bind two different histone marks in different chromatin contexts. This difference entails much of the contrasting mode of action, also defying the homology claim. Therefore, the CD has acquired its preference for H3K27me3 early in plant history since the moss LHP1 already binds this epigenetic mark. Hence, other evolutionary scenarios may need to be considered and explored in the future. For example, (i) the possible existence of a horizontal gene transfer event in early land plants from a yet unknown host; (ii) the de novo origin of LHP1 in early land plants.
Gene and genome duplication have a profound effect on plant evolution, especially in flowering plants from which most of the horticultural crops have been domesticated. In this sense, duplicated copies of LHP1 might have acquired novel unknown functions, which need attention. In tomato, only one of the two LHP1 duplicated copies has been functionally characterized so far, while the role of the other copy remains unknown. In melon, the duplicated copies exhibit a redundant function related to sub-functionalization. Although duplicated copies of LHP1 were identified in a variety of plant species [9], a neo-functionalized duplicated copy remains hidden. Therefore, another interesting edge of exploring deeper from an evolutionary point of view is the function of paralog copies of LHP1.
An important piece of missing information is the molecular and biological role of LHP1 function in one of the most important groups of flowering plants, the Monocots. This clade is one of the most diverse extant plant groups and contains numerous economically important horticultural crops with a wide variety of uses in the food industry, ornaments, and medicinal products, among others. Considering the importance and diversity of biological roles of LHP1 in Dicots, it can be speculated that its characterization in Monocots is promising.
The only report explaining how post-translational regulation can affect LHP1 activity is based on a crop species. As mentioned earlier, LHP1 is ubiquitinated by BTB/POZ for degradation in soybean [30]. Interestingly, from all the sets of possible post-translational modifications that may affect protein activity, it is unknown what other mechanisms are involved in the control of LHP1. For instance, they might be of importance also for the biotechnological use of LHP1 for plant trait optimization.
In the last few years, given the importance and abundance of noncoding transcripts in plants, extensive research has been undertaken to identify and characterize them. Interestingly, while LHP1 is conserved in all plant species from Bryophyte, lncRNAs can be highly species-specific. Moreover, the disordered hinge region responsible for RNA binding is not as conserved as other functional domains. Thus, given the exposed functional significance of the interaction between LHP1 and RNAs, elucidating potential lncRNAs partners conserved and/or species-specific emerges as a promising field.
5. Conclusions
Here we summarized the knowledge about the molecular function of the epigenetic remodeler LHP1 in model and crop plants, deciphering conserved and divergent aspects of its mode of action. Strikingly, LHP1 controls several developmental and physiological traits in different plant species highlighting its importance for plant breeding. Moreover, biotic and abiotic stress responses are modulated by this component of the epigenetic machinery. Modern plant crop systems are characterized by the use of biotechnological tools. Both our knowledge of LHP1’s role and modern biotechnological tools can be combined to fine-tune LHP1 action to optimize desired traits in horticultural crops. In this review, we spotted the future directions to explore deeper LHP1 function and its use as a potential target for crop improvement.
Author Contributions
The manuscript was written by L.E.L., with contributions of N.M., L.F., and F.D.A. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Agencia Nacional de Promoción de la Investigación, el Desarrollo Tecnológico y la Innovación (ANPCyT) (PICT-2019-00034, PICT-2019-04137, PICT-2020-00218). N.M. is a fellow of ANPCyT.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
We thank Agencia Nacional de Promoción de la Investigación, el Desarrollo Tecnológico y la Innovación (ANPCyT).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Turck, F.; Roudier, F.; Farrona, S.; Martin-Magniette, M.L.; Guillaume, E.; Buisine, N.; Gagnot, S.; Martienssen, R.A.; Coupland, G.; Colot, V.; et al. Arabidopsis TFL2/LHP1 Specifically Associates with Genes Marked by Trimethylation of Histone H3 Lysine 27. PLoS Genet. 2007, 3, 855–866. [Google Scholar] [CrossRef] [PubMed]
- Veluchamy, A.; Jégu, T.; Ariel, F.; Latrasse, D.; Mariappan, K.G.; Kim, S.K.; Crespi, M.; Hirt, H.; Bergounioux, C.; Raynaud, C.; et al. LHP1 Regulates H3K27me3 Spreading and Shapes the Three-Dimensional Conformation of the Arabidopsis Genome. PLoS ONE 2016, 11, e0158936. [Google Scholar] [CrossRef] [PubMed]
- Larsson, A.S.; Landberg, K.; Meeks-Wagner, D.R. The TERMINAL FLOWER2 (TFL2) Gene Controls the Reproductive Transition and Meristem Identity in Arabidopsis Thaliana. Genetics 1998, 149, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Prado, J.S.; Latrasse, D.; Rodriguez-Granados, N.Y.; Huang, Y.; Antipolis, N.S.; Agrobiotech, I.S.; Antipolis, S.; Manza-Mianza, D.; Brik-Chaouche, R.; Jaouannet, M.; et al. The Polycomb Protein LHP1 Regulates Arabidopsis Thaliana Stress Responses through the Repression of the MYC2-Dependent Branch of Immunity. Plant J. 2019, 100, 1118–1131. [Google Scholar] [CrossRef] [PubMed]
- Fonouni-Farde, C.; Christ, A.; Blein, T.; Legascue, M.F.; Ferrero, L.; Moison, M.; Lucero, L.; Ramírez-Prado, J.S.; Latrasse, D.; Gonzalez, D.; et al. The Arabidopsis APOLO and Human UPAT Sequence-Unrelated Long Noncoding RNAs Can Modulate DNA and Histone Methylation Machineries in Plants. Genome Biol. 2022, 23, 181. [Google Scholar] [CrossRef]
- Kim, J.H.; Durrett, T.P.; Last, R.L.; Jander, G. Characterization of the Arabidopsis TU8 Glucosinolate Mutation, an Allele of TERMINAL FLOWER2. Plant Mol. Biol. 2004, 54, 671–682. [Google Scholar] [CrossRef]
- Gaudin, V.; Libault, M.; Pouteau, S.; Juul, T.; Zhao, G.; Lefebvre, D.; Grandjean, O. Mutations in LIKE HETEROCHROMATIN PROTEIN 1 Affect Flowering Time and Plant Architecture in Arabidopsis. Development 2001, 128, 4847–4858. [Google Scholar] [CrossRef]
- Kotake, T.; Takada, S.; Nakahigashi, K.; Ohto, M.; Goto, K. Arabidopsis Terminal Flower 2 Gene Encodes a Heterochromatin Protein 1 Homolog and Represses Both FLOWERING LOCUS T to Regulate Flowering Time and Several Floral Homeotic Genes. Plant Cell Physiol. 2003, 44, 555–564. [Google Scholar] [CrossRef]
- Berke, L.; Snel, B. The Plant Polycomb Repressive Complex 1 (PRC1) Existed in the Ancestor of Seed Plants and Has a Complex Duplication History. BMC Evol. Biol. 2015, 15, 44. [Google Scholar] [CrossRef]
- Huang, Y.; Jiang, L.; Liu, B.Y.; Tan, C.F.; Chen, D.H.; Shen, W.H.; Ruan, Y. Evolution and Conservation of Polycomb Repressive Complex 1 Core Components and Putative Associated Factors in the Green Lineage. BMC Genom. 2019, 20, 533. [Google Scholar] [CrossRef]
- Exner, V.; Aichinger, E.; Shu, H.; Wildhaber, T.; Alfarano, P.; Caflisch, A.; Gruissem, W.; Köhler, C.; Hennig, L.; Ko, C.; et al. The Chromodomain of LIKE HETEROCHROMATIN PROTEIN 1 Is Essential for H3K27me3 Binding and Function during Arabidopsis Development. PLoS ONE 2009, 4, e5335. [Google Scholar] [CrossRef] [PubMed]
- Kaustov, L.; Ouyang, H.; Amaya, M.; Lemak, A.; Nady, N.; Duan, S.; Wasney, G.A.; Li, Z.; Vedadi, M.; Schapira, M.; et al. Recognition and Specificity Determinants of the Human Cbx Chromodomains. J. Biol. Chem. 2011, 286, 521–529. [Google Scholar] [CrossRef] [PubMed]
- Jackson, J.P.; Lindroth, A.M.; Cao, X.; Jacobsen, S.E. Control of CpNpG DNA Methylation by the KRYPTONITE Histone H3 Methyltransferase. Nature 2002, 416, 556–560. [Google Scholar] [CrossRef]
- Mylne, J.S.; Barrett, L.; Tessadori, F.; Mesnage, S.; Johnson, L.; Bernatavichute, Y.V.; Jacobsen, S.E.; Fransz, P.; Dean, C. LHP1, the Arabidopsis Homologue of HETEROCHROMATIN PROTEIN1, Is Required for Epigenetic Silencing of FLC. Proc. Natl. Acad. Sci. USA 2006, 103, 5012–5017. [Google Scholar] [CrossRef]
- Libault, M.; Tessadori, F.; Germann, S.; Snijder, B.; Fransz, P.; Gaudin, V. The Arabidopsis LHP1 Protein Is a Component of Euchromatin. Planta 2005, 222, 910–925. [Google Scholar] [CrossRef] [PubMed]
- Nakahigashi, K.; Jasencakova, Z.; Schubert, I.; Goto, K. The Arabidopsis HETEROCHROMATIN PROTEIN1 Homolog (TERMINAL FLOWER2) Silences Genes Within the Euchromatic Region but Not Genes Positioned in Heterochromatin. Plant Cell Physiol. 2005, 46, 1747–1756. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, X.; Zhou, M.; Yang, Y.; Li, F.; Yan, X.; Zhang, M.; Wei, Z.; Qin, S.; Min, J. Structural Basis for the Recognition of Methylated Histone H3 by the Arabidopsis LHP1 Chromodomain. J. Biol. Chem. 2022, 298, 101623. [Google Scholar] [CrossRef]
- Zemach, A.; Li, Y.; Ben-Meir, H.; Oliva, M.; Mosquna, A.; Kiss, V.; Avivi, Y.; Ohad, N.; Grafi, G. Different Domains Control the Localization and Mobility of LIKE HETEROCHROMATIN PROTEIN1 in Arabidopsis Nuclei. Plant Cell 2006, 18, 133–145. [Google Scholar] [CrossRef]
- Berry, S.; Rosa, S.; Howard, M.; Bühler, M.; Dean, C. Disruption of an RNA-Binding Hinge Region Abolishes LHP1-Mediated Epigenetic Repression. Genes Dev. 2017, 31, 2115–2120. [Google Scholar] [CrossRef]
- Cowieson, N.P.; Partridge, J.F.; Allshire, R.C.; McLaughlin, P.J. Dimerisation of a Chromo Shadow Domain and Distinctions from the Chromodomain as Revealed by Structural Analysis. Curr. Biol. 2000, 10, 517–525. [Google Scholar] [CrossRef]
- Dangwal, M.; Kapoor, S.; Kapoor, M. The PpCMT Chromomethylase Affects Cell Growth and Interacts with the Homolog of LIKE HETEROCHROMATIN PROTEIN 1 in the Moss Physcomitrella Patens. Plant J. 2014, 77, 589–603. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Gu, X.; Yuan, W.; Schmitz, R.J.; He, Y. Photoperiodic Control of the Floral Transition through a Distinct Polycomb Repressive Complex. Dev. Cell 2014, 28, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.; Jegu, T.; Latrasse, D.; Romero-Barrios, N.; Christ, A.; Benhamed, M.; Crespi, M. Noncoding Transcription by Alternative Rna Polymerases Dynamically Regulates an Auxin-Driven Chromatin Loop. Mol. Cell 2014, 55, 383–396. [Google Scholar] [CrossRef] [PubMed]
- Ariel, F.; Lucero, L.; Christ, A.; Mammarella, M.F.; Jegu, T.; Veluchamy, A.; Mariappan, K.; Latrasse, D.; Blein, T.; Liu, C.; et al. R-Loop Mediated Trans Action of the APOLO Long Noncoding RNA. Mol. Cell 2020, 77, 1055–1065.e4. [Google Scholar] [CrossRef] [PubMed]
- Lechner, M.S.; Schultz, D.C.; Negorev, D.; Maul, G.G.; Rauscher, F.J. The Mammalian Heterochromatin Protein 1 Binds Diverse Nuclear Proteins through a Common Motif That Targets the Chromoshadow Domain. Biochem. Biophys. Res. Commun. 2005, 331, 929–937. [Google Scholar] [CrossRef]
- Ryu, H.W.; Lee, D.H.; Florens, L.; Swanson, S.K.; Washburn, M.P.; Kwon, S.H. Analysis of the Heterochromatin Protein 1 (HP1) Interactome in Drosophila. J. Proteom. 2014, 102, 137–147. [Google Scholar] [CrossRef]
- Eissenberg, J.C.; Elgin, S.C.R. HP1a: A Structural Chromosomal Protein Regulating Transcription. Trends Genet. 2014, 30, 103–110. [Google Scholar] [CrossRef]
- Wang, H.; Jiang, D.; Axelsson, E.; Lorković, Z.J.; Montgomery, S.; Holec, S.; Pieters, B.J.G.E.; Al Temimi, A.H.K.; Mecinović, J.; Berger, F. LHP1 Interacts with ATRX through Plant-Specific Domains at Specific Loci Targeted by PRC2. Mol. Plant 2018, 11, 1038–1052. [Google Scholar] [CrossRef]
- Parihar, V.; Arya, D.; Walia, A.; Tyagi, V.; Dangwal, M.; Verma, V.; Khurana, R.; Boora, N.; Kapoor, S.; Kapoor, M. Functional Characterization of LIKE HETEROCHROMATIN PROTEIN 1 in the Moss Physcomitrella Patens: Its Conserved Protein Interactions in Land Plants. Plant J. 2019, 97, 221–239. [Google Scholar] [CrossRef]
- Zhang, C.; Cheng, Q.; Wang, H.; Gao, H.; Fang, X.; Chen, X.; Zhao, M.; Wei, W.; Song, B.; Liu, S.; et al. GmBTB/POZ Promotes the Ubiquitination and Degradation of LHP1 to Regulate the Response of Soybean to Phytophthora Sojae. Commun. Biol. 2021, 4, 372. [Google Scholar] [CrossRef]
- Chen, C.; Kim, D.; Yun, H.R.; Lee, Y.M.; Yogendra, B.; Bo, Z.; Kim, H.E.; Min, J.H.; Lee, Y.; Rim, Y.G.; et al. Nuclear Import of the LIKE HETEROCHROMATIN PROTEIN1 Is Redundantly Mediated by Importin$α$-1, Importin$α$-2, and Importin$α$-3. Plant J. 2020, 53, tpj.14796. [Google Scholar] [CrossRef] [PubMed]
- Sung, S.; He, Y.; Eshoo, T.W.; Tamada, Y.; Johnson, L.; Nakahigashi, K.; Goto, K.; Jacobsen, S.E.; Amasino, R.M. Epigenetic Maintenance of the Vernalized State in Arabidopsis Thaliana Requires LIKE HETEROCHROMATIN PROTEIN 1. Nat. Genet. 2006, 38, 706–710. [Google Scholar] [CrossRef] [PubMed]
- Shu, J.; Chen, C.; Thapa, R.K.; Bian, S.; Nguyen, V.; Yu, K.; Yuan, Z.C.; Liu, J.; Kohalmi, S.E.; Li, C.; et al. Genome-Wide Occupancy of Histone H3K27 Methyltransferases CURLY LEAF and SWINGER in Arabidopsis Seedlings. Plant Direct 2019, 3, e00100. [Google Scholar] [CrossRef] [PubMed]
- Derkacheva, M.; Steinbach, Y.; Wildhaber, T.; Mozgová, I.; Mahrez, W.; Nanni, P.; Bischof, S.; Gruissem, W.; Hennig, L. Arabidopsis MSI1 Connects LHP1 to PRC2 Complexes. EMBO J. 2013, 32, 2073–2085. [Google Scholar] [CrossRef]
- Yang, H.; Berry, S.; Olsson, T.S.G.; Hartley, M.; Howard, M.; Dean, C. Distinct Phases of Polycomb Silencing to Hold Epigenetic Memory of Cold in Arabidopsis. Science 2017, 357, 1142–1145. [Google Scholar] [CrossRef] [PubMed]
- Roulé, T.; Christ, A.; Hussain, N.; Huang, Y.; Hartmann, C.; Benhamed, M.; Gutierrez-Marcos, J.; Ariel, F.; Crespi, M.; Blein, T. The LncRNA MARS Modulates the Epigenetic Reprogramming of the Marneral Cluster in Response to ABA. Mol. Plant 2022, 15, 840–856. [Google Scholar] [CrossRef]
- Moison, M.; Pacheco, J.M.; Lucero, L.; Fonouni-Farde, C.; Rodríguez-Melo, J.; Mansilla, N.; Christ, A.; Bazin, J.; Benhamed, M.; Ibañez, F.; et al. The LncRNA APOLO Interacts with the Transcription Factor WRKY42 to Trigger Root Hair Cell Expansion in Response to Cold. Mol. Plant 2021, 14, 145576. [Google Scholar] [CrossRef]
- Piacentini, L.; Fanti, L.; Negri, R.; Del Vescovo, V.; Fatica, A.; Altieri, F.; Pimpinelli, S. Heterochromatin Protein 1 (HP1a) Positively Regulates Euchromatic Gene Expression through RNA Transcript Association and Interaction with HnRNPs in Drosophila. PLoS Genet. 2009, 5, e1000670. [Google Scholar] [CrossRef]
- Yap, K.L.; Li, S.; Muñoz-Cabello, A.M.; Raguz, S.; Zeng, L.; Mujtaba, S.; Gil, J.; Walsh, M.J.; Zhou, M.M. Molecular Interplay of the Noncoding RNA ANRIL and Methylated Histone H3 Lysine 27 by Polycomb CBX7 in Transcriptional Silencing of INK4a. Mol. Cell 2010, 38, 662–674. [Google Scholar] [CrossRef]
- Rose, N.R.; Klose, R.J. Understanding the Relationship between DNA Methylation and Histone Lysine Methylation. Biochim. Biophys. Acta-Gene Regul. Mech. 2014, 1839, 1362–1372. [Google Scholar] [CrossRef]
- Taniue, K.; Kurimoto, A.; Sugimasa, H.; Nasu, E.; Takeda, Y.; Iwasaki, K.; Nagashima, T.; Okada-Hatakeyama, M.; Oyama, M.; Kozuka-Hata, H.; et al. Long Noncoding RNA UPAT Promotes Colon Tumorigenesis by Inhibiting Degradation of UHRF1. Proc. Natl. Acad. Sci. USA 2016, 113, 1273–1278. [Google Scholar] [CrossRef] [PubMed]
- Rizzardi, K.; Landberg, K.; Nilsson, L.; Ljung, K.; Sundås-Larsson, A. TFL2/LHP1 Is Involved in Auxin Biosynthesis through Positive Regulation of YUCCA Genes. Plant J. 2011, 65, 897–906. [Google Scholar] [CrossRef] [PubMed]
- Takada, S.; Goto, K. Terminal Flower2, an Arabidopsis Homolog of Heterochromatin Protein1, Counteracts the Activation of Flowering Locus T by Constans in the Vascular Tissues of Leaves to Regulate Flowering Time. Plant Cell 2003, 15, 2856–2865. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Ahn, J.E.; Datta, S.; Salzman, R.A.; Moon, J.; Huyghues-Despointes, B.; Pittendrigh, B.; Murdock, L.L.; Koiwa, H.; Zhu-Salzman, K. Arabidopsis Vegetative Storage Protein Is an Anti-Insect Acid Phosphatase. Plant Physiol. 2005, 139, 1545–1556. [Google Scholar] [CrossRef] [PubMed]
- Jaouannet, M.; Morris, J.A.; Hedley, P.E.; Bos, J.I.B. Characterization of Arabidopsis Transcriptional Responses to Different Aphid Species Reveals Genes That Contribute to Host Susceptibility and Non-Host Resistance. PLoS Pathog. 2015, 11, e1004918. [Google Scholar] [CrossRef] [PubMed]
- Mashiguchi, K.; Tanaka, K.; Sakai, T.; Sugawara, S.; Kawaide, H.; Natsume, M.; Hanada, A.; Yaeno, T.; Shirasu, K.; Yao, H.; et al. The Main Auxin Biosynthesis Pathway in Arabidopsis. Proc. Natl. Acad. Sci. USA 2011, 108, 18512–18517. [Google Scholar] [CrossRef]
- Cheng, Y.; Dai, X.; Zhao, Y. Auxin Biosynthesis by the YUCCA Flavin Monooxygenases Controls the Formation of Floral Organs and Vascular Tissues in Arabidopsis. Genes Dev. 2006, 20, 1790–1799. [Google Scholar] [CrossRef]
- Sakata, T.; Oshino, T.; Miura, S.; Tomabechi, M.; Tsunaga, Y.; Higashitani, N.; Miyazawa, Y.; Takahashi, H.; Watanabe, M.; Higashitani, A. Auxins Reverse Plant Male Sterility Caused by High Temperatures. Proc. Natl. Acad. Sci. USA 2010, 107, 8569–8574. [Google Scholar] [CrossRef]
- Liang, Q.; Deng, H.; Li, Y.; Liu, Z.; Shu, P.; Fu, R.; Zhang, Y.; Pirrello, J.; Zhang, Y.; Grierson, D.; et al. Like Heterochromatin Protein 1b Represses Fruit Ripening via Regulating the H3K27me3 Levels in Ripening-Related Genes in Tomato. New Phytol. 2020, 227, 485–497. [Google Scholar] [CrossRef]
- Zhang, C.; Gao, H.; Li, R.; Han, D.; Wang, L.; Wu, J.; Xu, P.; Zhang, S. GmBTB/POZ, a Novel BTB/POZ Domain-Containing Nuclear Protein, Positively Regulates the Response of Soybean to Phytophthora Sojae Infection. Mol. Plant Pathol. 2019, 20, 78–91. [Google Scholar] [CrossRef]
- Rodriguez-Granados, N.Y.; Ramirez-Prado, J.S.; Brik-Chaouche, R.; An, J.; Manza-Mianza, D.; Sircar, S.; Troadec, C.; Hanique, M.; Soulard, C.; Costa, R.; et al. CmLHP1 Proteins Play a Key Role in Plant Development and Sex Determination in Melon (Cucumis melo). Plant J. 2022, 109, 1213–1228. [Google Scholar] [CrossRef] [PubMed]
- Parihar, V.; Dangwal, M.; Arya, D.; Kapoor, S.; Kapoor, M. Decrease in DNA Methylation 1 Interacts with Chromomethylase and like Heterochromatin Protein 1 in Physcomitrella Patens. FEBS Lett. 2019, 593, 2686–2697. [Google Scholar] [CrossRef] [PubMed]
- Feng, S.; Cokus, S.J.; Zhang, X.; Chen, P.Y.; Bostick, M.; Goll, M.G.; Hetzel, J.; Jain, J.; Strauss, S.H.; Halpern, M.E.; et al. Conservation and Divergence of Methylation Patterning in Plants and Animals. Proc. Natl. Acad. Sci. USA 2010, 107, 8689–8694. [Google Scholar] [CrossRef]
- Smallwood, A.; Estève, P.O.; Pradhan, S.; Carey, M. Functional Cooperation between HP1 and DNMT1 Mediates Gene Silencing. Genes Dev. 2007, 21, 1169–1178. [Google Scholar] [CrossRef] [PubMed]
- Nady, N.; Lemak, A.; Walker, J.R.; Avvakumov, G.V.; Kareta, M.S.; Achour, M.; Xue, S.; Duan, S.; Allali-Hassani, A.; Zuo, X.; et al. Recognition of Multivalent Histone States Associated with Heterochromatin by UHRF1 Protein. J. Biol. Chem. 2011, 286, 24300–24311. [Google Scholar] [CrossRef]
- Zhang, S.; Xu, P.; Wu, J.; Xue, A.G.; Zhang, J.; Li, W.; Chen, C.; Chen, W.; Lv, H. Races of Phytophthora Sojae and Their Virulences on Soybean Cultivars in Heilongjiang, China. Plant Dis. 2010, 94, 87–91. [Google Scholar] [CrossRef]
- Wei, W.; Tao, J.J.; Chen, H.W.; Li, Q.T.; Zhang, W.K.; Ma, B.; Lin, Q.; Zhang, J.S.; Chen, S.Y. A Histone Code Reader and a Transcriptional Activator Interact to Regulate Genes for Salt Tolerance. Plant Physiol. 2017, 175, 1304–1320. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).