The Search of a Molecular “Swiss Knife” for Chloroplast Genomic Editing
Abstract
:1. Introduction
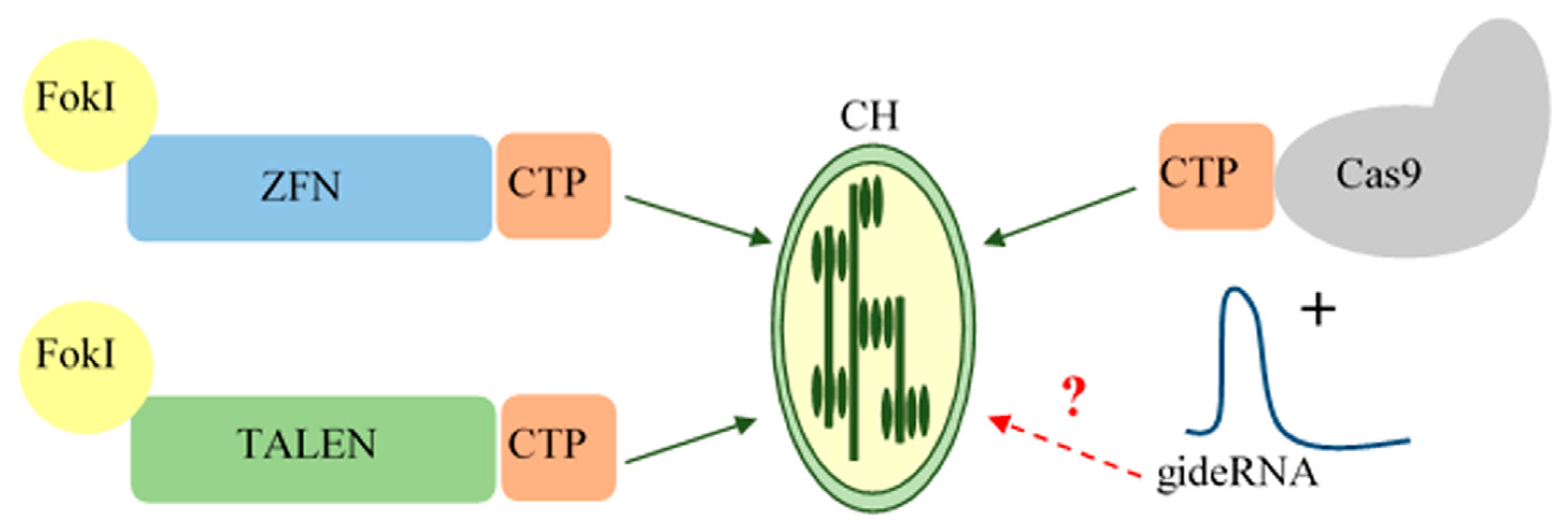
2. Genome Editing Systems Applicable to Chloroplasts
3. Delivery of Genome-Editing Tools into Chloroplasts
3.1. Delivery of DNA Plasmids into Chloroplasts Using Bioballistics
3.2. Nuclear Transformation: Vector-Mediated Transfer and Direct Delivery
3.3. gRNA Delivery into Chloroplasts
3.4. Delivery of DNA Plasmids into Chloroplasts Using Nanoparticles
3.5. Delivery of DNA into Chloroplasts Using Signal Peptides
4. Practical Application of Chloroplast Genomic Editing Platforms
4.1. Base-Editing Technologies
| Target Gene | Genome Editor | Species | Transformation Method | References |
|---|---|---|---|---|
| psaA (chlorophyll a of photosystem I) | ptTALED (plastid-targeted TALE deaminase); CRISPR/Cas9 | rice, lettuce, arabidopsis, chlamydomonas | Agrobacterium, mRNA, biolistic | [25,39,55] |
| psbA (photosynthetic protein, D1) | ptTALED | lettuce, rapeseed, arabidopsis | Agrobacterium, mRNA, polyethylene glycol (PEG)-mediated | [23,25,26] |
| psbB (photosynthetic protein, CP-47) | ptTALED | lettuce, rapeseed | Agrobacterium | [23] |
| rrn16 (16SrRNA) | ptTALED | lettuce, rapeseed, arabidopsis | Agrobacterium, PEG-mediated | [22] |
| rbcL (large, catalytic subunit of RuBisCo) | ptTALED | lettuce, arabidopsis | Agrobacterium | [22] |
| rpoC1 (β subunit of RNA polymerase) | ptTALED | arabidopsis | Agrobacterium | [26] |
4.2. Development of CRISPR/Cas9 System to Increase the Efficiency of Plastid Transformation
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rashid, B.; Tariq, M.; Khalid, A.; Shams, F.; Ali, Q.; Ashraf, F.; Ghaffar, I.; Khan, M.I.; Rehman, R.; Husnain, T. Crop improvement: New approaches and modern techniques. Plant Gene Trait. 2017, 8, 18–30. [Google Scholar] [CrossRef]
- Fiaz, S.; Ahmar, S.; Saeed, S.; Riaz, A.; Mora-Poblete, F.; Jung, K.H. Evolution and application of genome editing techniques for achieving food and nutritional security. Int. J. Mol. Sci. 2021, 22, 5585. [Google Scholar] [CrossRef]
- Fichtner, F.; Urrea Castellanos, R.; Ülker, B. Precision genetic modifications: A new era in molecular biology and crop improvement. Planta 2014, 239, 921–939. [Google Scholar] [CrossRef]
- Govindan, G.; Ramalingam, S. Programmable Site-Specific Nucleases for targeted genome engineering in higher Eukaryotes. J. Cell Physiol. 2016, 231, 2380–2392. [Google Scholar] [CrossRef] [PubMed]
- Das, D.; Singha, D.L.; Paswan, R.R.; Chowdhury, N.; Sharma, M.; Reddy, P.S.; Chikkaputtaiah, C. Recent advancements in CRISPR/Cas technology for accelerated crop improvement. Planta 2022, 255, 109. [Google Scholar] [CrossRef]
- Liu, H.; Chen, W.; Li, Y.; Sun, L.; Chai, Y.; Chen, H.; Nie, H.; Huang, C. CRISPR/Cas9 technology and its utility for crop improvement. Int. J. Mol. Sci. 2022, 23, 10442. [Google Scholar] [CrossRef]
- Basu, U.; Riaz Ahmed, S.; Bhat, B.A.; Anwar, Z.; Ali, A.; Ijaz, A.; Gulzar, A.; Bibi, A.; Tyagi, A.; Nebapure, S.M.; et al. A CRISPR way for accelerating cereal crop improvement: Progress and challenges. Front. Genet. 2023, 13, 866976. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chang, L.; Zhang, J. Advancing organelle genome transformation and editing for crop improvement. Plant Commun. 2021, 4, 100141. [Google Scholar] [CrossRef]
- An, Y.; Wang, Y.; Wang, X.; Xiao, J. Development of chloroplast transformation and gene expression regulation technology in land plants. Front. Plant Sci. 2022, 13, 1037038. [Google Scholar] [CrossRef]
- Yu, Y.; Yu, P.C.; Chang, W.J.; Yu, K.; Lin, C.S. Plastid transformation: How does it work? Can it be applied to crops? What can it offer? Int. J. Mol. Sci. 2020, 21, 4854. [Google Scholar] [CrossRef]
- Bock, R. Engineering plastid genomes: Methods, tools, and applications in basic research and biotechnology. Annu. Rev. Plant Biol. 2015, 66, 211–241. [Google Scholar] [CrossRef] [PubMed]
- Rozov, S.M.; Sidorchuk, Y.V.; Deineko, E.V. Transplastomic plants: Problems of production and their solution. Russ. J. Plant Physiol. 2022, 69, 20. [Google Scholar] [CrossRef]
- Son, S.; Park, S.R. Challenges facing CRISPR/Cas9-based genome editing in plants. Front. Plant Sci. 2022, 13, 902413. [Google Scholar] [CrossRef] [PubMed]
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast genomes: Diversity, evolution, and applications in genetic engineering. Genome Biol. 2016, 23, 134. [Google Scholar] [CrossRef] [PubMed]
- Zhu, G.; Zhu, H. Modified gene editing systems: Diverse bioengineering tools and crop improvement. Front. Plant Sci. 2022, 13, 847169. [Google Scholar] [CrossRef]
- Bracher, A.; Whitney, S.M.; Hartl, F.U.; Hayer-Hartl, M. Biogenesis and metabolic maintenance of rubisco. Annu. Rev. Plant Biol. 2017, 68, 29–60. [Google Scholar] [CrossRef]
- Khalil, A.M. The genome editing revolution: Review. J. Genet. Eng. Biotechnol. 2020, 18, 68. [Google Scholar] [CrossRef] [PubMed]
- Tariq, A.; Mushtaq, M.; Yaqoob, H.; Bhat, B.A.; Zargar, S.M.; Raza, A.; Ali, S.; Charagh, S.; Mubarik, M.S.; Zaman, Q.U.; et al. Putting CRISPR-Cas system in action: A golden window for efficient and precise genome editing for crop improvement. GM Crops Food 2023, 14, 1–27. [Google Scholar] [CrossRef]
- Sakuma, T.; Woltjen, K. Nuclease-mediated genome editing: At the front-line of functional genomics technology. Dev. Growth Differ. 2014, 56, 2–13. [Google Scholar] [CrossRef]
- Bhardwaj, A.; Nain, V. TALENs-an indispensable tool in the era of CRISPR: A mini review. J. Genet. Eng. Biotechnol. 2021, 21, 125. [Google Scholar] [CrossRef]
- Cermak, T.; Doyle, E.L.; Christian, M.; Wang, L.; Zhang, Y.; Schmidt, C.; Baller, J.A.; Somia, N.V.; Bogdanove, A.J.; Voytas, D.F. Efcient design and assembly of custom TALEN and other TAL efector-based constructs for DNA targeting. Nucleic Acids Res. 2011, 39, e82. [Google Scholar] [CrossRef]
- Ahmad, N.; Nielsen, B.L.; Mansoor, S. Editing the plastid genome of recalcitrant plant species. Trends Genet. 2021, 37, 955–957. [Google Scholar] [CrossRef]
- Kang, B.C.; Bae, S.J.; Lee, S.; Lee, J.S.; Kim, A.; Lee, H.; Baek, G.; Seo, H.; Kim, J.; Kim, J.S. Chloroplast and mitochondrial DNA editing in plants. Nat. Plants 2021, 7, 899–905. [Google Scholar] [CrossRef]
- Mok, B.Y.; de Moraes, M.H.; Zeng, J.; Bosch, D.E.; Kotrys, A.V.; Raguram, A.; Hsu, F.; Radey, M.C.; Peterson, S.B.; Mootha, V.K.; et al. A bacterial cytidine deaminase toxin enables CRISPR-free mitochondrial base editing. Nature 2020, 583, 631–637. [Google Scholar] [CrossRef]
- Mok, Y.G.; Hong, S.; Bae, S.J.; Cho, S.I.; Kim, J.S. Targeted A-to-G base editing of chloroplast DNA in plants. Nat. Plants 2022, 8, 1378–1384. [Google Scholar] [CrossRef] [PubMed]
- Nakazato, I.; Okuno, M.; Yamamoto, H.; Tamura, Y.; Itoh, T.; Shikanai, T.; Takanashi, H.; Tsutsumi, N.; Arimura, S.I. Targeted base editing in the plastid genome of Arabidopsis thaliana. Nat. Plants 2021, 7, 906–913. [Google Scholar] [CrossRef]
- Maliga, P. Engineering the plastid and mitochondrial genomes of flowering plants. Nat. Plants 2022, 8, 996–1006. [Google Scholar] [CrossRef] [PubMed]
- Nakazato, I.; Okuno, M.; Itoh, T.; Tsutsumi, N.; Arimura, S.I. Characterization and development of a plastid genome base editor, ptpTALECD. Plant J. 2023, 115, 1151–1162. [Google Scholar] [CrossRef]
- Hille, F.; Richter, H.; Wong, S.P.; Bratovic, M.; Ressel, S.; Charpentier, E. The Biology of CRISPR-Cas: Backward and Forward. Cell 2018, 172, 1239–1259. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex genome engineering using CRISPR/Cas systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Jiang, F.; Taylor, D.W.; Sternberg, S.H.; Kaya, E.; Ma, E.; Anders, C.; Hauer, M.; Zhou, K.; Lin, S.; et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science 2014, 343, 1247997. [Google Scholar] [CrossRef]
- Kim, H.; Kim, J.S. A guide to genome engineering with programmable nucleases. Nat. Rev. Genet. 2014, 15, 321–334. [Google Scholar] [CrossRef]
- Razzaq, A.; Saleem, F.; Kanwal, M.; Mustafa, G.; Yousaf, S.; Imran Arshad, H.M.; Hameed, M.K.; Khan, M.S.; Joyia, F.A. Modern trends in plant genome editing: An inclusive review of the CRISPR/Cas9 toolbox. Int. J. Mol. Sci. 2019, 20, 4045. [Google Scholar] [CrossRef]
- Jasin, M.; Haber, J.E. The democratization of gene editing: Insights from site-specific cleavage and double-strand break repair. DNA Repair 2016, 44, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Luan, X.; Liu, Y.; Wang, L.; Wang, J.; Yang, S.; Liu, S.; Zhang, J.; Liu, H.; Yao, D. Strategies and methods for improving the efficiency of CRISPR/Cas9 gene editing in plant molecular breeding. Plants 2023, 12, 1478. [Google Scholar] [CrossRef] [PubMed]
- Ozyigit, I.I.; Yucebilgili Kurtoglu, K. Particle bombardment technology and its applications in plants. Mol. Biol. Rep. 2020, 47, 9831–9847. [Google Scholar] [CrossRef] [PubMed]
- Kumar, A.U.; Ling, A.P.K. Gene introduction approaches in chloroplast transformation and its applications. J. Genet. Eng. Biotechnol. 2021, 19, 148. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Nannas, N.J.; Fu, F.F.; Shi, J.; Aspinwall, B.; Parrott, W.A.; Dawe, R.K. Genome-scale sequence disruption following biolistic transformation in rice and maize. Plant Cell 2019, 31, 368–383. [Google Scholar] [CrossRef] [PubMed]
- Yoo, B.C.; Yadav, N.S.; Orozco, E.M., Jr.; Sakai, H. Cas9/gRNA-mediated genome editing of yeast mitochondria and Chlamydomonas chloroplasts. PeerJ 2020, 8, e8362. [Google Scholar] [CrossRef]
- Tang, N.; Xia, Y.; Zhan, Y.; Dan, J.; Yu, M.; Bu, X.; Cao, M. Improvement of chloroplast transformation using CRISPR/Cas9. J. Biobased Mater. Bioenergy 2020, 14, 401. [Google Scholar] [CrossRef]
- Glass, Z.; Lee, M.; Li, Y.; Xu, Q. Engineering the delivery system for CRISPR-based genome editing. Trends Biotechnol. 2017, 36, 173–185. [Google Scholar] [CrossRef]
- Tian, L.; Chou, H.L.; Fukuda, M.; Kumamaru, T.; Okita, T.W. mRNA Localization in Plant Cells. Plant Physiol. 2020, 182, 97–109. [Google Scholar] [CrossRef]
- Gómez, G.; Pallás, V. Noncoding RNA mediated traffic of foreign mRNA into chloroplasts reveals a novel signaling mechanism in plants. PLoS ONE 2010, 19, e12269. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.F.; Huang, Y.P.; Chen, L.H.; Hsu, Y.H.; Tsai, C.H. Chloroplast phosphoglycerate kinase is involved in the targeting of Bamboo mosaic virus to chloroplasts in Nicotiana benthamiana plants. Plant Physiol. 2013, 3, 1598–1608. [Google Scholar] [CrossRef] [PubMed]
- Newkirk, G.M.; de Allende, P.; Jinkerson, R.E.; Giraldo, J.P. Nanotechnology approaches for chloroplast biotechnology advancements. Front. Plant Sci. 2021, 12, 691295. [Google Scholar] [CrossRef] [PubMed]
- Santana, I.; Hu, P.; Jeon, S.J.; Castillo, C.; Tu, H.; Giraldo, J.P. Peptide-mediated targeting of nanoparticles with chemical cargoes to chloroplasts in Arabidopsis plants. Bio-Protocol 2021, 20, e4060. [Google Scholar] [CrossRef] [PubMed]
- Kwak, S.Y.; Lew, T.T.S.; Sweeney, C.J.; Koman, V.B.; Wong, M.H.; Bohmert-Tatarev, K.; Snell, K.D.; Seo, J.S.; Chua, N.H.; Strano, M.S. Chloroplast-selective gene delivery and expression in planta using chitosan-complexed single-walled carbon nanotube carriers. Nat. Nanotechnol. 2019, 14, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Odahara, M.; Miyamoto, T.; Numata, K. Fusion peptide-based biomacromolecule delivery system for plant cells. ACS Biomater. Sci. Eng. 2021, 7, 2246–2254. [Google Scholar] [CrossRef]
- Liu, B.R.; Chen, C.W.; Huang, Y.W.; Lee, H.J. Cell-penetrating peptides for use in development of transgenic plants. Molecules 2023, 28, 3367. [Google Scholar] [CrossRef]
- Yoshizumi, T.; Oikawa, K.; Chuah, J.A.; Kodama, Y.; Numata, K. Selective gene delivery for integrating exogenous DNA into plastid and mitochondrial genomes using peptide-DNA complexes. Biomacromolecules 2018, 19, 1582–1591. [Google Scholar] [CrossRef]
- Thagun, C.; Chuah, J.A.; Numata, K. Targeted gene delivery into various plastids mediated by clustered cell-penetrating and chloroplast-targeting eptides. Adv. Sci. 2019, 6, 1902064. [Google Scholar] [CrossRef] [PubMed]
- Oikawa, K.; Tateishi, A.; Odahara, M.; Kodama, Y.; Numata, K. Imaging of the entry pathway of a cell-penetrating peptide-DNA complex from the extracellular space to chloroplast nucleoids across multiple membranes in Arabidopsis leaves. Front. Plant Sci. 2021, 12, 759871. [Google Scholar] [CrossRef] [PubMed]
- Odahara, M.; Horii, Y.; Itami, J.; Watanabe, K.; Numata, K. Functional peptide-mediated plastid transformation in tobacco, rice, and kenaf. Front. Plant Sci. 2022, 13, 989310. [Google Scholar] [CrossRef]
- Lee, H.; Lee, S.; Baek, G.; Kim, A.; Kang, B.-C.; Seo, H.; Kim, J.-S. Mitochondrial DNA editing in mice with DddA-TALE fusion deaminases. Nat. Commun. 2021, 12, 1190. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Char, S.N.; Liu, B.; Liu, H.; Li, X.; Yang, B. High-efficiency plastome base editing in rice with TAL cytosine deaminase. Mol. Plant. 2021, 6, 1412–1414. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dorogova, N.V.; Sidorchuk, Y.V. The Search of a Molecular “Swiss Knife” for Chloroplast Genomic Editing. Horticulturae 2023, 9, 1338. https://doi.org/10.3390/horticulturae9121338
Dorogova NV, Sidorchuk YV. The Search of a Molecular “Swiss Knife” for Chloroplast Genomic Editing. Horticulturae. 2023; 9(12):1338. https://doi.org/10.3390/horticulturae9121338
Chicago/Turabian StyleDorogova, Natalya V., and Yuriy V. Sidorchuk. 2023. "The Search of a Molecular “Swiss Knife” for Chloroplast Genomic Editing" Horticulturae 9, no. 12: 1338. https://doi.org/10.3390/horticulturae9121338
APA StyleDorogova, N. V., & Sidorchuk, Y. V. (2023). The Search of a Molecular “Swiss Knife” for Chloroplast Genomic Editing. Horticulturae, 9(12), 1338. https://doi.org/10.3390/horticulturae9121338





