Genetically Modified Legume Plants as a Basis for Studying the Signal Regulation of Symbiosis with Nodule Bacteria
Abstract
:1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Conditions for Their Cultivation
2.2. Plant Growth Conditions
2.3. Molecular Cloning
2.3.1. Cloning of the MtSPHK1 Gene Fragment Encoding Domain for Phosphatidic Acid Binding
2.3.2. Cloning of the MAPK6 Gene
2.4. Transformation of Pea Pisum sativum Plants
2.5. Transformation of Medicago truncatula Plants
2.6. Isolation of Total RNA
2.7. Quantitative Reverse Transcription-Polymerase Chain Reaction (RT-qPCR) Analysis
2.8. Statistical Data Analysis
3. Results
3.1. The Influence of MtSPHK1 Gene Fragment Encoding Domain for Phosphatidic Acid Binding in Symbiosis
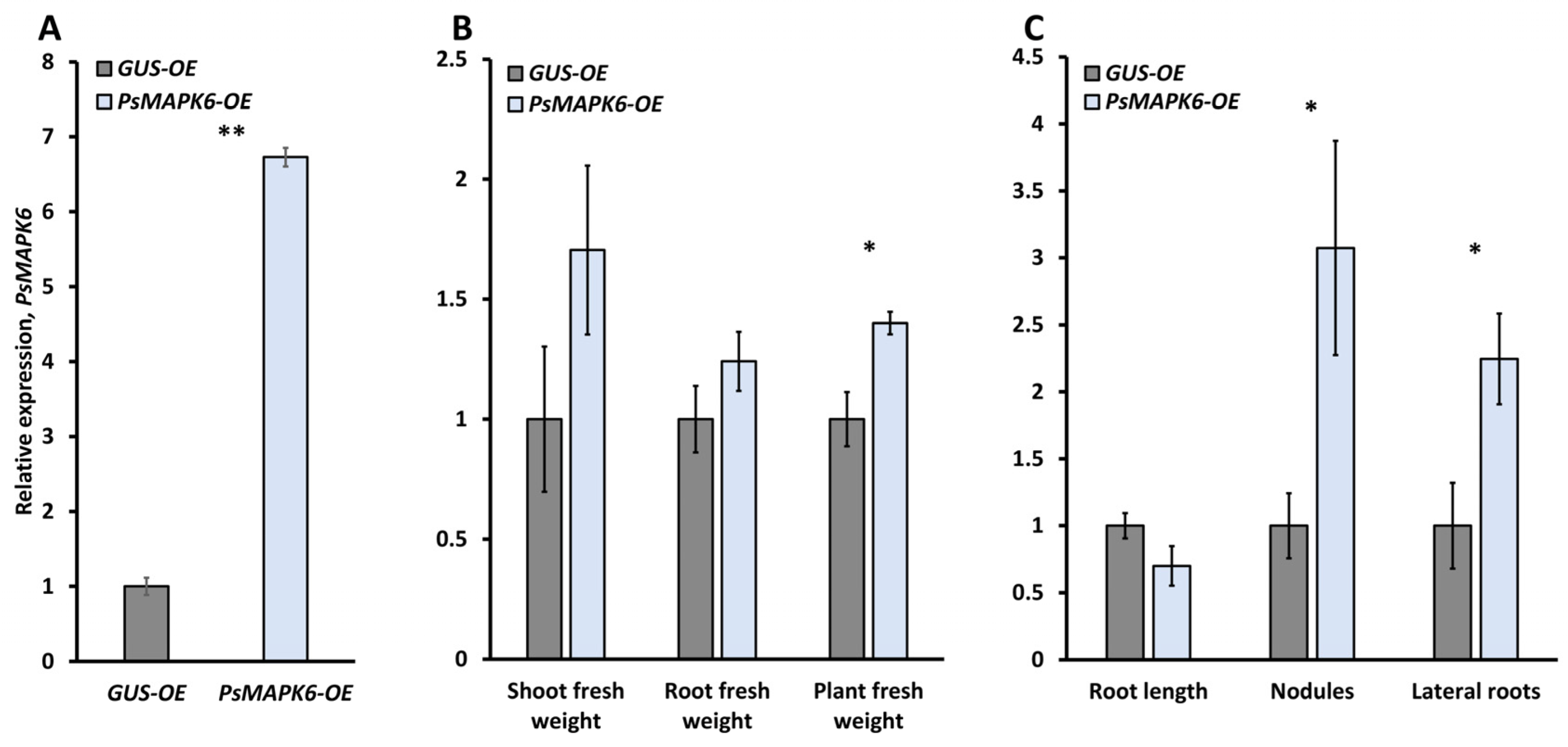
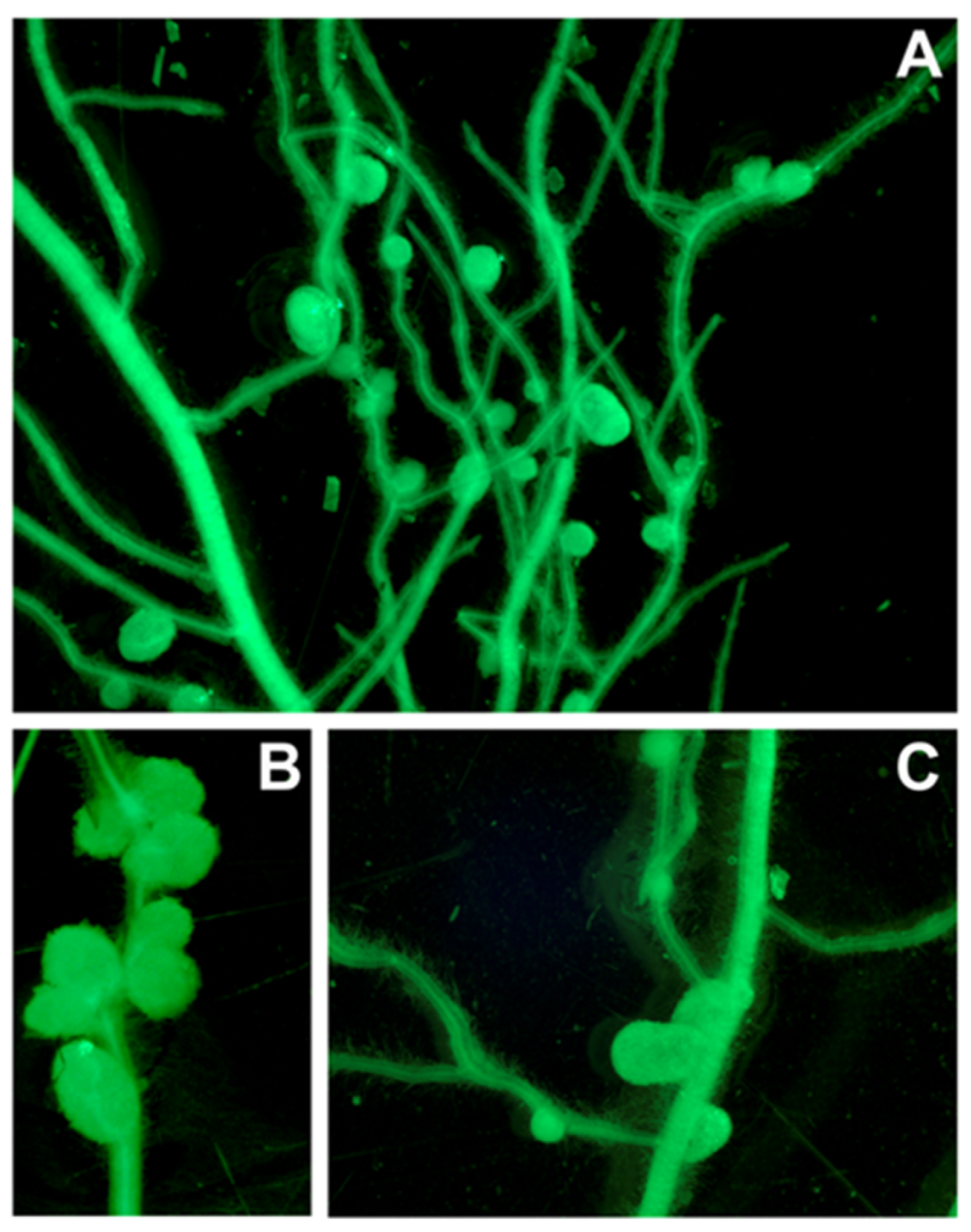
3.2. The Effect of MAPK6 Overexpression on the Development of Symbiosis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ben Amor, B.; Shaw, S.L.; Oldroyd, G.E.D.; Maillet, F.; Penmetsa, R.V.; Cook, D.; Long, S.R.; Dénarié, J.; Gough, C. The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J. 2003, 34, 495–506. [Google Scholar] [CrossRef] [PubMed]
- Madsen, E.B.; Madsen, L.H.; Radutoiu, S.; Olbryt, M.; Rakwalska, M.; Szczyglowski, K.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; et al. A receptor kinase gene of the LysM type is involved in legumeperception of rhizobial signals. Nature 2003, 425, 637–640. [Google Scholar] [CrossRef] [PubMed]
- Zhukov, V.; Radutoiu, S.; Madsen, L.H.; Rychagova, T.; Ovchinnikova, E.; Borisov, A.; Tikhonovich, I.; Stougaard, J. The pea Sym37 receptor kinase gene controls infection-thread initiation and nodule development. Mol. Plant-Microbe Interact. 2008, 21, 1600–1608. [Google Scholar] [CrossRef] [PubMed]
- Radutoiu, S.; Madsen, L.H.; Madsen, E.B.; Felle, H.H.; Umehara, Y.; Grønlund, M.; Sato, S.; Nakamura, Y.; Tabata, S.; Sandal, N.; et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 2003, 425, 585–592. [Google Scholar] [CrossRef] [PubMed]
- Smit, P.; Limpens, E.; Geurts, R.; Fedorova, E.; Dolgikh, E.; Gough, C.; Bisseling, T. Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol. 2007, 145, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Kirienko, A.N.; Porozov, Y.B.; Malkov, N.V.; Akhtemova, G.A.; Le Signor, C.; Thompson, R.; Saffray, C.; Dalmais, M.; Bendahmane, A.; Tikhonovich, I.A.; et al. Role of a receptor-like kinase K1 in pea Rhizobium symbiosis development. Planta 2018, 248, 1101–1120. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lan, L.; Jin, Y.; Yu, N.; Wang, D.; Wang, E. Mechanisms underlying legume–rhizobium symbioses. J. Integr. Plant Biol. 2022, 64, 244–267. [Google Scholar] [CrossRef]
- Oldroyd, G.E.D.; Downie, J.A. Coordinating nodule morphogenesis with rhizobial infection in legumes. Annu. Rev. Plant Biol. 2008, 59, 519–546. [Google Scholar] [CrossRef]
- Roy, S.; Liu, W.; Nandety, R.S.; Crook, A.; Mysore, K.S.; Pislariu, C.I.; Frugoli, J.; Dickstein, R.; Udvardi, M.K. Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 2020, 32, 15–41. [Google Scholar] [CrossRef]
- Endre, G.; Kereszt, A.; Kevei, Z.; Mihacea, S.; Kaló, P.; Kiss, G.B. A receptor kinase gene regulating symbiotic nodule development. Nature 2002, 417, 962–966. [Google Scholar] [CrossRef]
- Stracke, S.; Kistner, C.; Yoshida, S.; Mulder, L.; Sato, S.; Kaneko, T.; Tabata, S.; Sandal, N.; Stougaard, J.; Szczyglowski, K.; et al. A plant receptor-like kinase required for both bacterial and fungal symbiosis. Nature 2002, 417, 959–962. [Google Scholar] [CrossRef] [PubMed]
- Ané, J.-M.; Lévy, J.; Thoquet, P.; Kulikova, O.; de Billy, F.; Penmetsa, V.; Kim, D.-J.; Debellé, F.; Rosenberg, C.; Cook, D.R.; et al. Genetic and cytogenetic mapping of DMI1, DMI2, and DMI3 genes of Medicago truncatula involved in Nod factor transduction, nodulation, and mycorrhization. Mol. Plant-Microbe Interact. 2002, 15, 1108–1118. [Google Scholar] [CrossRef]
- Imaizumi-Anraku, H.; Takeda, N.; Charpentier, M.; Perry, J.; Miwa, H.; Umehara, Y.; Kouchi, H.; Murakami, Y.; Mulder, L.; Vickers, K.; et al. Plastid proteins crucial for symbiotic fungal and bacterial entry into plant roots. Nature 2005, 433, 527–531. [Google Scholar] [CrossRef]
- Charpentier, M.; Bredemeier, R.; Wanner, G.; Takeda, N.; Schleiff, E.; Parniske, M. Lotus japonicus CASTOR and POLLUX are ion channels essential for perinuclear calcium spiking in legume root endosymbiosis. Plant Cell 2009, 20, 3467–3479. [Google Scholar] [CrossRef] [PubMed]
- Kanamori, N.; Madsen, L.H.; Radutoiu, S.; Frantescu, M.; Quistgaard, E.M.H.; Miwa, H.; Downie, J.A.; James, E.K.; Felle, H.H.; Haaning, L.L.; et al. A nucleoporin is required for induction of Ca2+ spiking in legume nodule development and essential for rhizobial and fungal symbiosis. Proc. Natl. Acad. Sci. USA 2006, 103, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Yoshikawa, M.; Yano, K.; Miwa, H.; Uchida, H.; Asamizu, E.; Sato, S.; Tabata, S.; Imaizumi-Anraku, H.; Umehara, Y.; et al. NUCLEOPORIN85 is required for calcium spiking, fungal and bacterial symbioses, and seed production in Lotus japonicus. Plant Cell 2007, 19, 610–624. [Google Scholar] [CrossRef] [PubMed]
- Groth, M.; Takeda, N.; Perry, J.; Uchida, H.; Dräxl, S.; Brachmann, A.; Sato, S.; Tabata, S.; Kawaguchi, M.; Wang, T.L.; et al. NENA, a Lotus japonicus homolog of Sec13, is required for rhizodermal infection by arbuscular mycorrhiza fungi and rhizobia but dispensable for cortical endosymbiotic development. Plant Cell 2010, 22, 2509–2526. [Google Scholar] [CrossRef]
- Capoen, W.; Sun, J.; Wysham, D.; Otegui, M.S.; Venkateshwaran, M.; Hirsch, S.; Miwa, H.; Downie, J.A.; Morris, R.J.; Ané, J.-M.; et al. Nuclear membranes control symbiotic calcium signaling of legumes. Proc. Natl. Acad. Sci. USA 2011, 108, 14348–14353. [Google Scholar] [CrossRef]
- Lévy, J.; Bres, C.; Geurts, R.; Chalhoub, B.; Kulikova, O.; Duc, G.; Journet, E.-P.; Ané, J.-M.; Lauber, E.; Bisseling, T.; et al. A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 2004, 303, 1361–1364. [Google Scholar] [CrossRef]
- Tirichine, L.; James, E.K.; Sandal, N.; Stougaard, J. Spontaneous root-nodule formation in the model legume Lotus japonicus: A novel class of mutants nodulates in the absence of rhizobia. Mol. Plant-Microbe Interact. 2006, 19, 373–382. [Google Scholar] [CrossRef]
- Yano, K.; Yoshida, S.; Müller, J.; Singh, S.; Banba, M.; Vickers, K.; Markmann, K.; White, C.; Schuller, B.; Sato, S.; et al. CYCLOPS, a mediator of symbiotic intracellular accommodation. Proc. Natl. Acad. Sci. USA 2008, 105, 20540–20545. [Google Scholar] [CrossRef] [PubMed]
- Messinese, E.; Mun, J.-H.; Yeun, L.H.; Jayaraman, D.; Rougé, P.; Barre, A.; Lougnon, G.; Schornack, S.; Bono, J.-J.; Cook, D.R.; et al. A novel nuclear protein interacts with the symbiotic DMI3 calcium- and calmodulin-dependent protein kinase of Medicago truncatula. Mol. Plant-Microbe Interact. 2007, 20, 912–921. [Google Scholar] [CrossRef] [PubMed]
- Cerri, M.R.; Frances, L.; Laloum, T.; Auriac, M.-C.; Niebel, A.; Oldroyd, G.E.D.; Barker, D.G.; Fournier, J.; de Carvalho-Niebel, F. Medicago truncatula ERN transcription factors: Regulatory interplay with NSP1/NSP2 GRAS factors and expression dynamics throughout rhizobial infection. Plant Physiol. 2012, 160, 2155–2172. [Google Scholar] [CrossRef] [PubMed]
- Schauser, L.; Roussis, A.; Stiller, J.; Stougaard, J. A plant regulator controlling development of symbiotic root nodules. Nature 1999, 402, 191–195. [Google Scholar] [CrossRef]
- Marsh, J.F.; Rakocevic, A.; Mitra, R.M.; Brocard, L.; Sun, J.; Eschstruth, A.; Long, S.R.; Schultze, M.; Ratet, P.; Oldroyd, G.E.D. Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol. 2007, 144, 324–335. [Google Scholar] [CrossRef]
- Soyano, T.; Kouchi, H.; Hirota, A.; Hayashi, M. NODULE INCEPTION directly targets NF-Y subunit genes to regulate essential processes of root nodule development in Lotus japonicus. PLoS Genet. 2013, 9, e1003352. [Google Scholar] [CrossRef]
- Michell, R.H. Inositol derivatives: Evolution and functions. Nat. Rev. Mol. Cell Biol. 2008, 9, 151–161. [Google Scholar] [CrossRef]
- Charron, D.; Pingret, J.-L.; Chabaud, M.; Journet, E.-P.; Barker, D.G. Pharmacological evidence that multiple phospholipid signaling pathways link Rhizobium nodulation factor perception in Medicago truncatula root hairs to intracellular responses, including Ca2+ spiking and specific ENOD gene expression. Plant Physiol. 2004, 136, 3582–3593. [Google Scholar] [CrossRef]
- Ichimura, K.; Shinozaki, K.; Tena, G.; Sheen, J.; Henry, Y.; Champion, A.; Kreis, M.; Zhang, S.; Hirt, H.; Wilson, C.; et al. Mitogen-activated protein kinase cascades in plants: A new nomenclature. Trends Plant Sci. 2002, 7, 301–308. [Google Scholar] [CrossRef]
- Chen, J.; Wang, L.; Yuan, M. Update on the roles of rice MAPK cascades. Int. J. Mol. Sci. 2021, 22, 1679. [Google Scholar] [CrossRef]
- Urano, D.; Jones, A.M. Heterotrimeric G Protein–Coupled Signaling in Plants. Annu. Rev. Plant Biol. 2014, 65, 365–384. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, S.R.; Pandey, S. Specific Subunits of Heterotrimeric G Proteins Play Important Roles during Nodulation in Soybean. Plant Physiol. 2013, 162, 522–533. [Google Scholar] [CrossRef] [PubMed]
- Roy Choudhury, S.; Pandey, S. SymRK-dependent phosphorylation of Gα protein and its role in signaling during soybean (Glycine max) nodulation. Plant J. 2022, 110, 277–291. [Google Scholar] [CrossRef] [PubMed]
- Bovin, A.D.; Pavlova, O.A.; Dolgikh, A.V.; Leppyanen, I.V.; Dolgikh, E.A. The Role of Heterotrimeric G-Protein Beta Subunits During Nodulation in Medicago truncatula Gaertn and Pisum sativum L. Front. Plant Sci. 2022, 12, 808573. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Gong, R.; Yuan, S.; Su, Y.; Lv, W.; Zhou, Y.; Zhang, Q.; Deng, X.; Tong, P.; Liang, S.; et al. Phospholipase D a 6 and phosphatidic acid regulate gibberellin signaling in rice. EMBO Rep. 2021, 22, e51871. [Google Scholar] [CrossRef] [PubMed]
- Tei, R.; Baskin, J.M. Spatiotemporal control of phosphatidic acid signaling with optogenetic, engineered phospholipase Ds. J. Cell Biol. 2020, 219, e201907013. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Yang, J.; Chen, X.; Zhao, D.; Zhou, X.; Zhang, Y.; Wang, X. Phospholipase D- and phosphatidic acid-mediated phospholipid metabolism and signaling modulate symbiotic interaction and nodulation in soybean (Glycine max). Plant J. 2021, 106, 142–158. [Google Scholar] [CrossRef]
- Den Hartog, M.; Musgrave, A.; Munnik, T. Nod factor-induced phosphatidic acid and diacylglycerol pyrophosphate formation: A role for phospholipase C and D in root hair deformation. Plant J. 2001, 25, 55–65. [Google Scholar] [CrossRef]
- den Hartog, M.; Verhoef, N.; Munnik, T. Nod factor and elicitors activate different phospholipid signaling pathways in suspension-cultured alfalfa cells. Plant Physiol. 2003, 132, 311–317. [Google Scholar] [CrossRef]
- Kwiatek, J.M.; Carman, G.M. Yeast phosphatidic acid phosphatase Pah1 hops and scoots along the membrane phospholipid bilayer. J. Lipid Res. 2020, 61, 1232–1243. [Google Scholar] [CrossRef]
- Pandit, S.; Dalal, V.; Mishra, G. Identification of novel phosphatidic acid binding domain on sphingosine kinase 1 of Arabidopsis thaliana. Plant Physiol. Biochem. 2018, 128, 178–184. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Zhu, H.; Ke, D.; Cai, K.; Wang, C.; Gou, H.; Hong, Z.; Zhang, Z. A MAP kinase kinase interacts with SymRK and regulates nodule organogenesis in Lotus japonicus. Plant Cell 2012, 24, 823–838. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Zhou, B.; Duan, L.; Zhu, H.; Zhang, Z. MtMAPKK4 is an essential gene for growth and reproduction of Medicago truncatula. Physiol. Plant. 2017, 159, 492–503. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Guan, X.; Zhang, H.; Wang, L.; Li, H.; Zhang, Q.; Chen, T.; Xu, Z.; Hong, Z.; Cao, Y.; et al. An MAP kinase interacts with LHK1 and regulates nodule organogenesis in Lotus japonicus. Sci. China Life Sci. 2019, 62, 1203–1217. [Google Scholar] [CrossRef] [PubMed]
- Hrbáčková, M.; Luptovčiak, I.; Hlaváčková, K.; Dvořák, P.; Tichá, M.; Šamajová, O.; Novák, D.; Bednarz, H.; Niehaus, K.; Ovečka, M.; et al. Overexpression of alfalfa SIMK promotes root hair growth, nodule clustering and shoot biomass production. Plant Biotechnol. J. 2021, 19, 767–784. [Google Scholar] [CrossRef]
- van Brussel, A.; Planqué, K.; Quispel, A. The wall of Rhizobium leguminosarum in bacteroid and free-living forms. Microbiology-sgm 1977, 101, 51–56. [Google Scholar] [CrossRef]
- Pecrix, Y.; Staton, S.E.; Sallet, E.; Lelandais-Brière, C.; Moreau, S.; Carrère, S.; Blein, T.; Jardinaud, M.-F.; Latrasse, D.; Zouine, M.; et al. Whole-genome landscape of Medicago truncatula symbiotic genes. Nat. Plants 2018, 4, 1017–1025. [Google Scholar] [CrossRef]
- Kreplak, J.; Madoui, M.A.; Cápal, P.; Novák, P.; Labadie, K.; Aubert, G.; Bayer, P.E.; Gali, K.K.; Syme, R.A.; Main, D.; et al. A reference genome for pea provides insight into legume genome evolution. Nat. Genet. 2019, 51, 1411–1422. [Google Scholar] [CrossRef]
- van Brussel, A.A.; Tak, T.; Wetselaar, A.; Pees, E.; Wijffelman, C. Small leguminosae as test plants for nodulation of Rhizobium leguminosarum and other rhizobia and agrobacteria harbouring a leguminosarum sym-plasmid. Plant Sci. Lett. 1982, 27, 317–325. [Google Scholar] [CrossRef]
- Leppyanen, I.V.; Kirienko, A.N.; Dolgikh, E.A. Agrobacterium rhizogenes—Mediated transformation of Pisum sativum L. roots as a tool for studying the mycorrhizal and root nodule symbioses. PeerJ 2019, 7, e6552. [Google Scholar] [CrossRef]
- Boisson-dernier, A.; Chabaud, M.; Rosenberg, C.; Barker, D.G.; De Biologie, L.; Garcia, F.; Bécard, G.; Umr, I.; Rosenberg, C.; Barker, D.G.; et al. Agrobacterium rhizogenes-based transformation of Medicago truncatula Protocol for A. rhizogenes transformation of M. truncatula. Am. Phytopathol. Soc. 2001, 14, 695–700. [Google Scholar]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Huo, Y.; Yang, N.; Wei, T. Phosphatidic acid: From biophysical properties to diverse functions. FEBS J. 2023, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kolesnikov, Y.; Kretynin, S.; Bukhonska, Y.; Pokotylo, I.; Ruelland, E.; Martinec, J.; Kravets, V. Phosphatidic acid in plant hormonal signaling: From target proteins to membrane conformations. Int. J. Mol. Sci. 2022, 23, 3227. [Google Scholar] [CrossRef] [PubMed]
- Zhukovsky, M.A.; Filograna, A.; Luini, A.; Corda, D.; Valente, C. Phosphatidic acid in membrane rearrangements. FEBS Lett. 2019, 593, 2428–2451. [Google Scholar] [CrossRef] [PubMed]
- Cardinale, F.; Jonak, C.; Ligterink, W.; Niehaus, K.; Boller, T.; Hirt, H. Differential activation of four specific MAPK pathways by distinct elicitors. J. Biol. Chem. 2000, 275, 36734–36740. [Google Scholar] [CrossRef]
- Wang, X.; Devaiah, S.; Zhang, W.; Welti, R. Signaling functions of phosphatidic acid. Prog. Lipid Res. 2006, 45, 250–278. [Google Scholar] [CrossRef]
- Hong, Y.; Zhao, J.; Guo, L.; Kim, S.-C.; Deng, X.; Wang, G.; Zhang, G.; Li, M.; Wang, X. Plant phospholipases D and C and their diverse functions in stress responses. Prog. Lipid Res. 2016, 62, 55–74. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bovin, A.D.; Dolgikh, A.V.; Dymo, A.M.; Kantsurova, E.S.; Pavlova, O.A.; Dolgikh, E.A. Genetically Modified Legume Plants as a Basis for Studying the Signal Regulation of Symbiosis with Nodule Bacteria. Horticulturae 2024, 10, 9. https://doi.org/10.3390/horticulturae10010009
Bovin AD, Dolgikh AV, Dymo AM, Kantsurova ES, Pavlova OA, Dolgikh EA. Genetically Modified Legume Plants as a Basis for Studying the Signal Regulation of Symbiosis with Nodule Bacteria. Horticulturae. 2024; 10(1):9. https://doi.org/10.3390/horticulturae10010009
Chicago/Turabian StyleBovin, Andrey D., Alexandra V. Dolgikh, Alina M. Dymo, Elizaveta S. Kantsurova, Olga A. Pavlova, and Elena A. Dolgikh. 2024. "Genetically Modified Legume Plants as a Basis for Studying the Signal Regulation of Symbiosis with Nodule Bacteria" Horticulturae 10, no. 1: 9. https://doi.org/10.3390/horticulturae10010009
APA StyleBovin, A. D., Dolgikh, A. V., Dymo, A. M., Kantsurova, E. S., Pavlova, O. A., & Dolgikh, E. A. (2024). Genetically Modified Legume Plants as a Basis for Studying the Signal Regulation of Symbiosis with Nodule Bacteria. Horticulturae, 10(1), 9. https://doi.org/10.3390/horticulturae10010009






