Phloem-Expressed CLAVATA3/ESR-like Genes in Potato
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Plant Growth Conditions
2.2. Genetic Constructs
2.3. Analysis of Coding and Promoter Sequences of the StCLE19 Gene
2.4. RNA-Seq and Data Analysis
2.5. Histochemical Assays
3. Results
3.1. Screening for CLE Genes Expressed in the Phloem
3.2. Effects of Overexpression of StCLE12 and StCLE19 Genes on Root, Shoot, and Tuber Growth
3.3. Transcriptome Analysis of Transgenic Potato Stems with StCLE12 and StCLE19 Overexpression
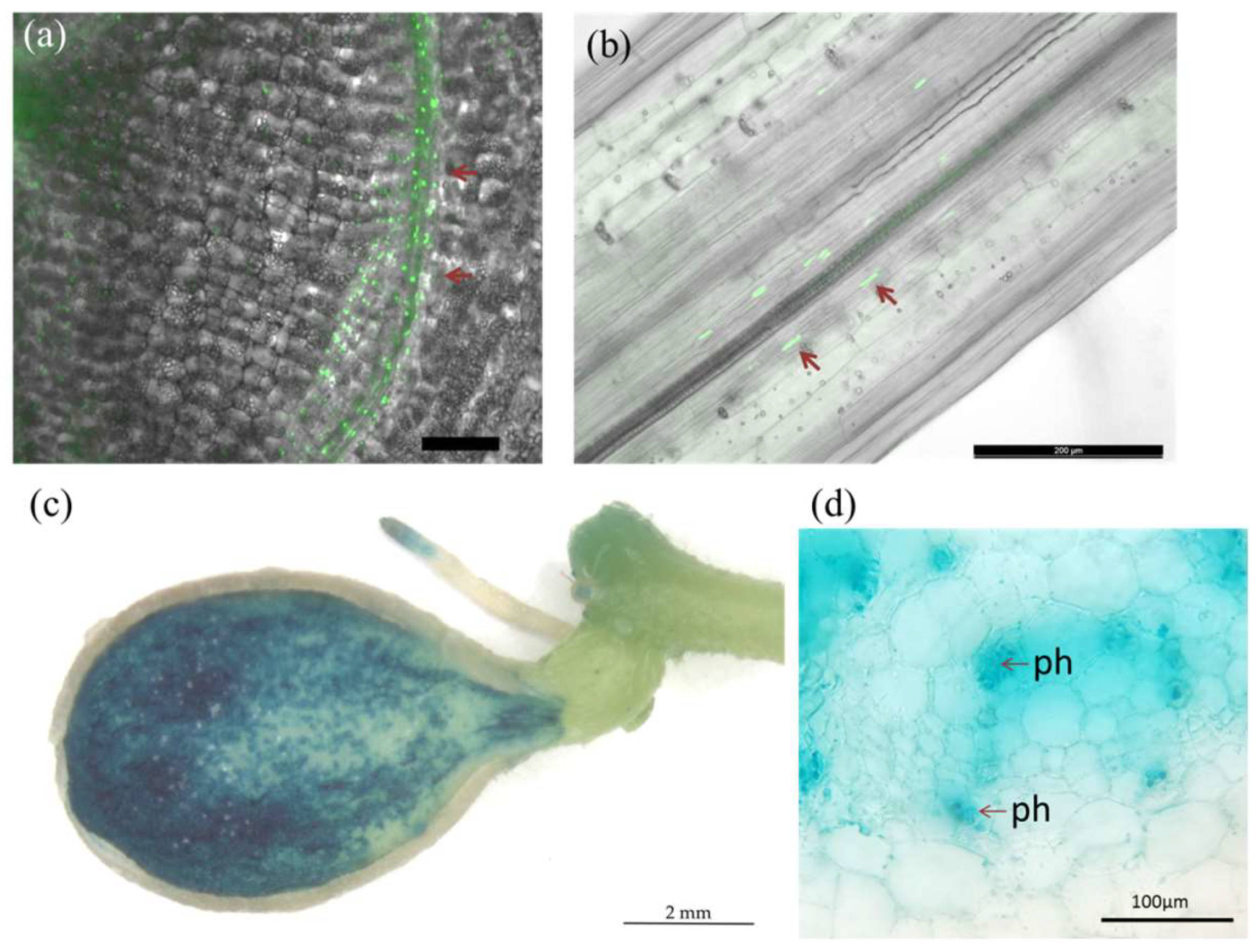
3.4. Promoter Activity of the StCLE8 and StCLE19 Genes in Potato
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fletcher, J.C. Recent Advances in Arabidopsis CLE Peptide Signaling. Trends Plant Sci. 2020, 25, 1005–1016. [Google Scholar] [CrossRef] [PubMed]
- Carbonnel, S.; Cornelis, S.; Hazak, O. The CLE33 Peptide Represses Phloem Differentiation via Autocrine and Paracrine Signaling in Arabidopsis. Commun. Biol. 2023, 6, 588. [Google Scholar] [CrossRef] [PubMed]
- Ren, S.C.; Song, X.F.; Chen, W.Q.; Lu, R.; Lucas, W.J.; Liu, C.M. CLE25 Peptide Regulates Phloem Initiation in Arabidopsis through a CLERK-CLV2 Receptor Complex. J. Integr. Plant Biol. 2019, 61, 1043–1061. [Google Scholar] [CrossRef] [PubMed]
- Depuydt, S.; Rodriguez-Villalon, A.; Santuari, L.; Wyser-Rmili, C.; Ragni, L.; Hardtke, C.S. Suppression of Arabidopsis Protophloem Differentiation and Root Meristem Growth by CLE45 Requires the Receptor-like Kinase BAM3. Proc. Natl. Acad. Sci. USA 2013, 110, 7074–7079. [Google Scholar] [CrossRef]
- Hazak, O.; Brandt, B.; Cattaneo, P.; Santiago, J.; Rodriguez-Villalon, A.; Hothorn, M.; Hardtke, C.S. Perception of Root-Active CLE Peptides Requires CORYNE Function in the Phloem Vasculature. EMBO Rep. 2017, 18, 1367–1381. [Google Scholar] [CrossRef] [PubMed]
- Czyzewicz, N.; Shi, C.L.; Vu, L.D.; Van De Cotte, B.; Hodgman, C.; Butenko, M.A.; De Smet, I. Modulation of Arabidopsis and Monocot Root Architecture by CLAVATA3/EMBRYO SURROUNDING REGION 26 Peptide. J. Exp. Bot. 2015, 66, 5229–5243. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, Y.; Shinohara, H.; Kondo, Y.; Inoue, A.; Nakanomyo, I.; Ogawa, M.; Sawa, S.; Ohashi-Ito, K.; Matsubayashi, Y.; Fukuda, H. Non-Cell-Autonomous Control of Vascular Stem Cell Fate by a CLE Peptide/Receptor System. Proc. Natl. Acad. Sci. USA 2008, 105, 15208. [Google Scholar] [CrossRef] [PubMed]
- Gancheva, M.S.; Lutova, L.A. Nitrogen-Activated CLV3/ESR-Related 4 (CLE4) Regulates Shoot, Root, and Stolon Growth in Potato. Plants 2023, 12, 3468. [Google Scholar] [CrossRef]
- Curtis, M.D.; Grossniklaus, U. A Gateway Cloning Vector Set for High-Throughput Functional Analysis of Genes in Planta. Plant Physiol. 2003, 133, 462–469. [Google Scholar] [CrossRef]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAYTM Vectors for Agrobacterium-Mediated Plant Transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A Flexible Trimmer for Illumina Sequence Data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A Fast Spliced Aligner with Low Memory Requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie Enables Improved Reconstruction of a Transcriptome from RNA-Seq Reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Bray, N.L.; Pimentel, H.; Melsted, P.; Pachter, L. Near-Optimal Probabilistic RNA-Seq Quantification. Nat. Biotechnol. 2016, 34, 525–527. [Google Scholar] [CrossRef] [PubMed]
- Schneider, C.A.; Rasband, W.S.; Eliceiri, K.W. NIH Image to ImageJ: 25 Years of Image Analysis. Nat. Methods 2012, 9, 671–675. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Lashbrook, C.C.; Cho, S.K.; Butler, N.M.; Sharma, P.; Muppirala, U.; Severin, A.J.; Hannapel, D.J. Transcriptional Analysis of Phloem-Associated Cells of Potato. BMC Genom. 2015, 16, 665. [Google Scholar] [CrossRef]
- Okonechnikov, K.; Golosova, O.; Fursov, M.; UGENE Team. Unipro UGENE: A unified bioinformatics toolkit. Bioinformatics 2012, 28, 1166–1167. [Google Scholar] [CrossRef]
- Liu, Y.; Yang, S.; Song, Y.; Men, S.; Wang, J. Gain-of-Function Analysis of Poplar CLE Genes in Arabidopsis by Exogenous Application and over-Expression Assays. J. Exp. Bot. 2016, 67, 2309–2324. [Google Scholar] [CrossRef]
- Hastwell, A.H.; Gresshoff, P.M.; Ferguson, B.J. Genome-Wide Annotation and Characterization of CLAVATA/ESR (CLE) Peptide Hormones of Soybean (Glycine max) and Common Bean (Phaseolus vulgaris), and Their Orthologues of Arabidopsis thaliana. J. Exp. Bot. 2015, 66, 5271–5287. [Google Scholar] [CrossRef] [PubMed]
- Strabala, T.J.; Phillips, L.; West, M.; Stanbra, L. Bioinformatic and Phylogenetic Analysis of the CLAVATA3/EMBRYO-SURROUNDING REGION (CLE) and the CLE-LIKE Signal Peptide Genes in the Pinophyta. BMC Plant Biol. 2014, 14, 47. [Google Scholar] [CrossRef]
- Takahashi, F.; Suzuki, T.; Osakabe, Y.; Betsuyaku, S.; Kondo, Y.; Dohmae, N.; Fukuda, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K. A Small Peptide Modulates Stomatal Control via Abscisic Acid in Long-Distance Signaling. Nature 2018, 556, 235–238. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.K.; Prat, S.; Hannapel, D.J. Efficient Production of Transgenic Potato (S. tuberosum L. ssp. andigena) Plants via Agrobacterium tumefaciens-Mediated Transformation. Plant Sci. 2006, 170, 732–738. [Google Scholar] [CrossRef]
- Smit, M.E.; McGregor, S.R.; Sun, H.; Gough, C.; Bågman, A.M.; Soyars, C.L.; Kroon, J.T.; Gaudinier, A.; Williams, C.J.; Yang, X.; et al. A PXY-Mediated Transcriptional Network Integrates Signaling Mechanisms to Control Vascular Development in Arabidopsis. Plant Cell 2020, 32, 319–335. [Google Scholar] [CrossRef] [PubMed]
- Knoblauch, M.; Froelich, D.R.; Pickard, W.F.; Peters, W.S. SEORious Business: Structural Proteins in Sieve Tubes and Their Involvement in Sieve Element Occlusion. J. Exp. Bot. 2014, 65, 1879–1893. [Google Scholar] [CrossRef] [PubMed]
- Ma, W.; Li, J.; Qu, B.; He, X.; Zhao, X.; Li, B.; Fu, X.; Tong, Y. Auxin Biosynthetic Gene TAR2 Is Involved in Low Nitrogen-Mediated Reprogramming of Root Architecture in Arabidopsis. Plant J. 2014, 78, 70–79. [Google Scholar] [CrossRef]
- Luo, J.; Zhou, J.J.; Zhang, J.Z. Aux/IAA Gene Family in Plants: Molecular Structure, Regulation, and Function. Int. J. Mol. Sci. 2018, 19, 259. [Google Scholar] [CrossRef]
- Swarup, R.; Bhosale, R. Developmental Roles of AUX1/LAX Auxin Influx Carriers in Plants. Front. Plant Sci. 2019, 10, 1306. [Google Scholar] [CrossRef]
- Ingram, P.; Dettmer, J.; Helariutta, Y.; Malamy, J.E. Arabidopsis Lateral Root Development 3 Is Essential for Early Phloem Development and Function, and Hence for Normal Root System Development. Plant J. 2011, 68, 455–467. [Google Scholar] [CrossRef]
- Goh, T.; Toyokura, K.; Wells, D.M.; Swarup, K.; Yamamoto, M.; Mimura, T.; Weijers, D.; Fukaki, H.; Laplaze, L.; Bennett, M.J.; et al. Quiescent Center Initiation in the Arabidopsis Lateral Root Primordia Is Dependent on the SCARECROW Transcription Factor. Development 2016, 143, 3363–3371. [Google Scholar] [CrossRef] [PubMed]
- Ren, H.; Gray, W.M. SAUR Proteins as Effectors of Hormonal and Environmental Signals in Plant Growth. Mol. Plant 2015, 8, 1153–1164. [Google Scholar] [CrossRef] [PubMed]
- Qiu, D.; Xu, S.; Wang, Y.; Zhou, M.; Hong, L. Primary Cell Wall Modifying Proteins Regulate Wall Mechanics to Steer Plant Morphogenesis. Front. Plant Sci. 2021, 12, 751372. [Google Scholar] [CrossRef]
- Ito, S.; Suzuki, Y.; Miyamoto, K.; Ueda, J.; Yamaguchi, I. AtFLA11, a Fasciclin-like Arabinogalactan-Protein, Specifically Localized in Screlenchyma Cells. Biosci. Biotechnol. Biochem. 2005, 69, 1963–1969. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Kucukoglu, M.; Zhang, L.; Chen, P.; Decker, D.; Nilsson, O.; Jones, B.; Sandberg, G.; Zheng, B. The Arabidopsis LRR-RLK, PXC1, Is a Regulator of Secondary Wall Formation Correlated with the TDIF-PXY/TDR-WOX4 Signaling Pathway. BMC Plant Biol. 2013, 13, 94. [Google Scholar] [CrossRef] [PubMed]
- Xu, K.; Jourquin, J.; Njo, M.F.; Nguyen, L.; Beeckman, T.; Fernandez, A.I. The Phloem Intercalated with Xylem-Correlated 3 Receptor-like Kinase Constitutively Interacts with Brassinosteroid Insensitive 1-Associated Receptor Kinase 1 and Is Involved in Vascular Development in Arabidopsis. Front. Plant Sci. 2022, 12, 706633. [Google Scholar] [CrossRef] [PubMed]
- Gancheva, M.; Dodueva, I.; Lebedeva, M.; Lutova, L. Clavata3/Embryo Surrounding Region (Cle) Gene Family in Potato (Solanum tuberosum L.): Identification and Expression Analysis. Agronomy 2021, 11, 984. [Google Scholar] [CrossRef]
- Gancheva, M.S.; Losev, M.R.; Gurina, A.A.; Poliushkevich, L.O.; Dodueva, I.E.; Lutova, L.A. Polymorphism of CLE Gene Sequences in Potato. Vavilovskii Zhurnal Genet. Sel. 2021, 25, 746–753. [Google Scholar] [CrossRef]
- Whitford, R.; Fernandez, A.; De Groodt, R.; Ortega, E.; Hilson, P. Plant CLE Peptides from Two Distinct Functional Classes Synergistically Induce Division of Vascular Cells. Proc. Natl. Acad. Sci. USA 2008, 105, 18625–18630. [Google Scholar] [CrossRef]
- Etchells, J.P.; Turner, S.R. The PXY-CLE41 Receptor Ligand Pair Defines a Multifunctional Pathway That Controls the Rate and Orientation of Vascular Cell Division. Development 2010, 137, 767–774. [Google Scholar] [CrossRef]
- Zhang, Y.; Tan, S.; Gao, Y.; Kan, C.; Wang, H.L.; Yang, Q.; Xia, X.; Ishida, T.; Sawa, S.; Guo, H.; et al. CLE42 Delays Leaf Senescence by Antagonizing Ethylene Pathway in Arabidopsis. New Phytol. 2022, 235, 550–562. [Google Scholar] [CrossRef] [PubMed]
- Gancheva, M.S.; Dodueva, I.E.; Lebedeva, M.A.; Tvorogova, V.E.; Tkachenko, A.A.; Lutova, L.A. Identification, Expression, and Functional Analysis of CLE Genes in Radish (Raphanus sativus L.) Storage Root. BMC Plant Biol. 2016, 16, 7. [Google Scholar] [CrossRef] [PubMed]
- Gancheva, M.S.; Dodueva, I.E.; Lutova, L.A. Role of CLE41 Peptide in the Development of Root Storage Parenchyma in Species of the Genus Raphanus L. Russ. J. Plant Physiol. 2018, 65, 498–511. [Google Scholar] [CrossRef]
- Kuznetsova, K.; Dodueva, I.; Gancheva, M.; Lutova, L. Transcriptomic Analysis of Radish (Raphanus sativus L.) Roots with CLE41 Overexpression. Plants 2022, 11, 2163. [Google Scholar] [CrossRef]
- Yue, J.; Yang, H.; Yang, S.; Wang, J. TDIF Regulates Auxin Accumulation and Modulates Auxin Sensitivity to Enhance Both Adventitious Root and Lateral Root Formation in Poplar Trees. Tree Physiol. 2020, 40, 1534–1547. [Google Scholar] [CrossRef] [PubMed]
- Hirakawa, Y.; Uchida, N.; Yamaguchi, Y.L.; Tabata, R.; Ishida, S.; Ishizaki, K.; Nishihama, R.; Kohchi, T.; Sawa, S.; Bowman, J.L. Control of Proliferation in the Haploid Meristem by CLE Peptide Signaling in Marchantia polymorpha. PLoS Genet. 2019, 15, e1007997. [Google Scholar] [CrossRef] [PubMed]
- Arae, T.; Nakakoji, M.; Noguchi, M.; Kamon, E.; Sano, R.; Demura, T.; Ohtani, M. Plant Secondary Cell Wall Proteome Analysis with an Inducible System for Xylem Vessel Cell Differentiation. Dev. Growth Differ. 2022, 64, 5–15. [Google Scholar] [CrossRef]
- Ito, Y.; Nakanomyo, I.; Motose, H.; Iwamoto, K.; Sawa, S.; Dohmae, N.; Fukuda, H. Dodeca-CLE as Peptides as Suppressors of Plant Stem Cell Differentiation. Science 2006, 313, 842–845. [Google Scholar] [CrossRef]
- Hirakawa, Y.; Kondo, Y.; Fukuda, H. TDIF Peptide Signaling Regulates Vascular Stem Cell Proliferation via the WOX4 Homeobox Gene in Arabidopsis. Plant Cell 2010, 22, 2618–2629. [Google Scholar] [CrossRef]
- Yang, S.; Bai, J.; Wang, J. TDIF Peptides Regulate Root Growth by Affecting Auxin Homeostasis and PINs Expression in Arabidopsis thaliana. Planta 2020, 251, 109. [Google Scholar] [CrossRef]
- Strabala, T.J.; O’Donnell, P.J.; Smit, A.M.; Ampomah-Dwamena, C.; Martin, E.J.; Netzler, N.; Nieuwenhuizen, N.J.; Quinn, B.D.; Foote, H.C.C.; Hudson, K.R. Gain-of-Function Phenotypes of Many CLAVATA3/ESR Genes, Including Four New Family Members, Correlate with Tandem Variations in the Conserved CLAVATA3/ESR Domain. Plant Physiol. 2006, 140, 1331–1344. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gancheva, M.S.; Losev, M.R.; Dodueva, I.E.; Lutova, L.A. Phloem-Expressed CLAVATA3/ESR-like Genes in Potato. Horticulturae 2023, 9, 1265. https://doi.org/10.3390/horticulturae9121265
Gancheva MS, Losev MR, Dodueva IE, Lutova LA. Phloem-Expressed CLAVATA3/ESR-like Genes in Potato. Horticulturae. 2023; 9(12):1265. https://doi.org/10.3390/horticulturae9121265
Chicago/Turabian StyleGancheva, Maria S., Maxim R. Losev, Irina E. Dodueva, and Lyudmila A. Lutova. 2023. "Phloem-Expressed CLAVATA3/ESR-like Genes in Potato" Horticulturae 9, no. 12: 1265. https://doi.org/10.3390/horticulturae9121265
APA StyleGancheva, M. S., Losev, M. R., Dodueva, I. E., & Lutova, L. A. (2023). Phloem-Expressed CLAVATA3/ESR-like Genes in Potato. Horticulturae, 9(12), 1265. https://doi.org/10.3390/horticulturae9121265





