Genome-Wide Analysis of the SWEET Transporters and Their Potential Role in Response to Cold Stress in Rosa rugosa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification and Analysis of RrSWEET
2.2. Expression Analysis of the RrSWEETs
2.3. Cloning and Subcellular Localization Analysis of RrSWEETs
3. Results
3.1. Identification of RrSWEET Family Members
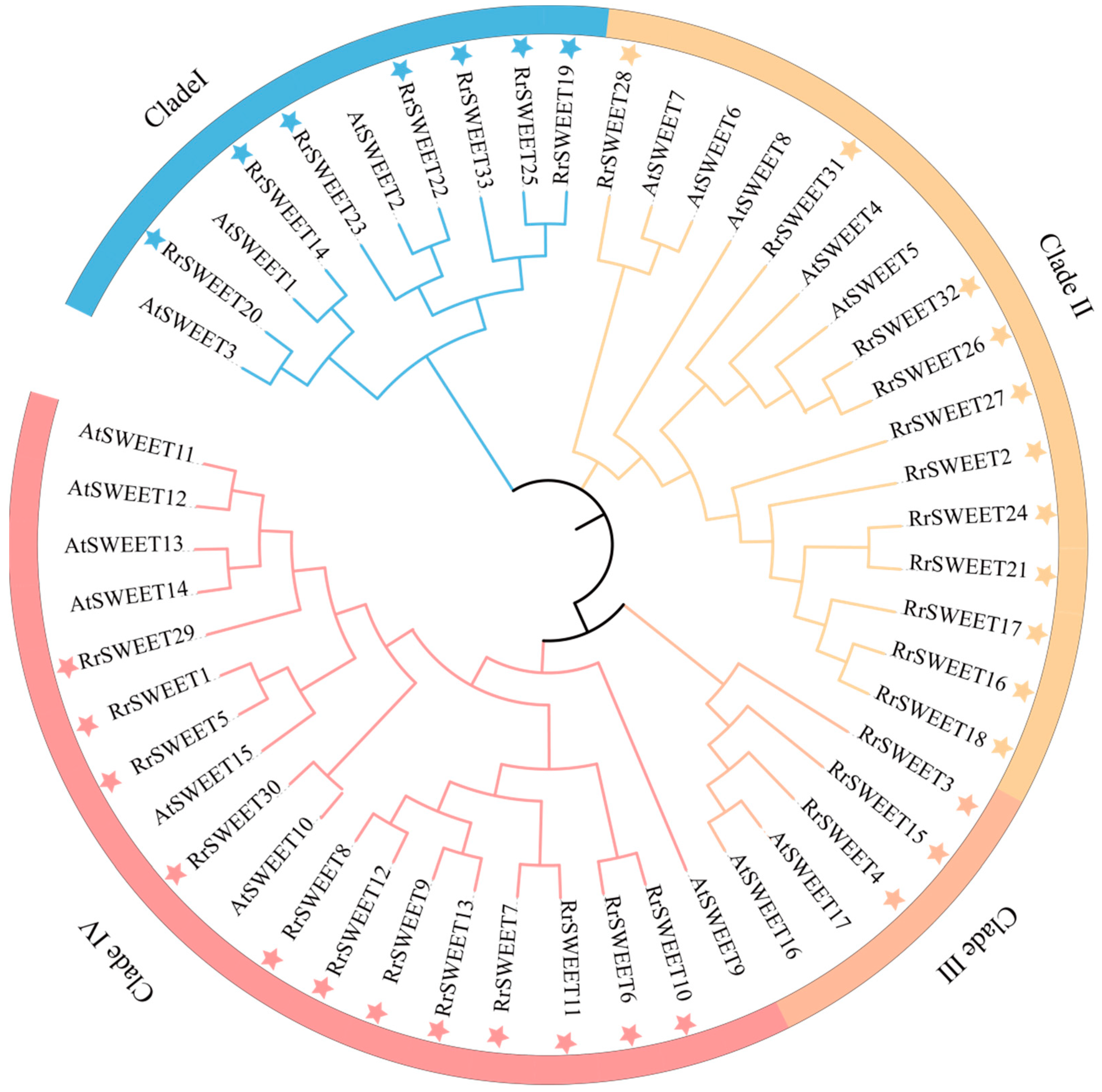
3.2. Phylogenetic Analysis, Conserved Motifs, Gene Structure Compositions and Cis-Regulatory Element Analysis of RrSWEETs
3.3. Chromosomal Distribution and Duplication Analysis of RrSWEET Genes
3.4. Collinearity Analysis
3.5. Specific-Tissue Expression Analysis of RrSWEETs
3.6. Expression Patterns of RrSWEETs under Cold Treatment
3.7. Subcellular Localization Analysis of RrSWEETs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wei, L.; Li, J.J.; Yang, Y.; Zhu, M.; Zhao, M.K.; Yang, J.H.; Yang, Z.Y.; Zhou, L.Y.; Zhou, S.Y.; Gong, J.J.; et al. Characterization and potential bioactivity of polyphenols of Rosa Rugosa. Food Biosci. 2022, 50, 102108. [Google Scholar] [CrossRef]
- Cetinbas-Genc, A.; Cai, G.; Del Duca, S. Treatment with spermidine alleviates the effects of concomitantly applied cold stress by modulating Ca2+, pH and ROS homeostasis, actin filament organization and cell wall deposition in pollen tubes of Camellia sinensis. Plant Physiol. Biochem. 2020, 156, 578–590. [Google Scholar] [CrossRef]
- Nishida, I.; Murata, N. Chilling sensitivity in plants and cyanobateria: The Crucial Contribution of Membrane Lipids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 541–568. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, F.; Kopka, J.; Haskell, D.W.; Zhao, W.; Schiller, K.C.; Gatzke, N.; Sung, D.Y.; Guy, C.L. Exploring the temperature-stress metabolome of Arabidopsis. Plant Physiol. 2004, 136, 4159–4168. [Google Scholar] [CrossRef] [PubMed]
- Bittner, A.; van Buer, J.; Baier, M. Cold priming uncouples light- and cold-regulation of gene expression in Arab Thaliana. BMC Plant Biol. 2020, 20, 281. [Google Scholar] [CrossRef]
- Buttner, M.; Sauer, N. Monosaccharide transporters in plants: Structure, function and physiology. Biochim. Biophys. Acta 2000, 1465, 263–274. [Google Scholar] [CrossRef]
- Chen, L.Q.; Cheung, L.S.; Feng, L.; Tanner, W.; Frommer, W.B. Transport of Sugars. Annu. Rev. Biochem. 2015, 84, 865–894. [Google Scholar] [CrossRef]
- Eom, J.S.; Chen, L.Q.; Sosso, D.; Julius, B.T.; Lin, I.W.; Qu, X.Q.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef]
- Feng, L.; Frommer, W.B. Structure and function of SemiSWEET and SWEET sugar transporters. Trends Biochem. Sci. 2015, 40, 480–486. [Google Scholar] [CrossRef]
- Feng, L.; Frommer, W.B. Evolution of Transporters: The Relationship of SWEETs, PQ-loop, and PnuC Transporters. Trends Biochem. Sci. 2016, 41, 118–119. [Google Scholar] [CrossRef]
- Chen, L.Q.; Hou, B.H.; Lalonde, S.; Takanaga, H.; Hartung, M.L.; Qu, X.Q.; Guo, W.J.; Kim, J.G.; Underwood, W.; Chaudhuri, B.; et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature 2010, 468, 527–532. [Google Scholar] [CrossRef]
- Chen, L.Q.; Qu, X.Q.; Hou, B.H.; Sosso, D.; Osorio, S.; Fernie, A.R.; Frommer, W.B. Sucrose Efflux Mediated by SWEET Proteins as a Key Step for Phloem Transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Doidy, J.; Grace, E.; Kuhn, C.; Simon-Plas, F.; Casieri, L.; Wipf, D. Sugar transporters in plants and in their interactions with fungi. Trends Plant Sci. 2012, 17, 413–422. [Google Scholar] [CrossRef]
- Chen, L.Q.; Lin, I.W.N.; Qu, X.Q.; Sosso, D.; McFarlane, H.E.; Londono, A.; Samuels, A.L.; Frommer, W.B. A Cascade of Sequentially Expressed Sucrose Transporters in the Seed Coat and Endosperm Provides Nutrition for the Arabidopsis Embryo. Plant Cell 2015, 27, 607–619. [Google Scholar] [CrossRef] [PubMed]
- Sosso, D.; Luo, D.P.; Li, Q.B.; Sasse, J.; Yang, J.L.; Gendrot, G.; Suzuki, M.; Koch, K.E.; McCarty, D.R.; Chourey, P.S.; et al. Seed filling in domesticated maize and rice depends on SWEET-mediated hexose transport. Nat. Genet. 2015, 47, 1489–1493. [Google Scholar] [CrossRef] [PubMed]
- Guan, Y.F.; Huang, X.Y.; Zhu, J.; Gao, J.F.; Zhang, H.X.; Yang, Z.N. Ruptured pollen grain1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspores in Arabidopsis. Plant Physiol. 2008, 147, 852–863. [Google Scholar] [CrossRef] [PubMed]
- Sun, M.X.; Huang, X.Y.; Yang, J.; Guan, Y.F.; Yang, Z.N. Arabidopsis RPG1 is important for primexine deposition and functions redundantly with RPG2 for plant fertility at the late reproductive stage. Plant Reprod. 2013, 26, 83–91. [Google Scholar] [CrossRef]
- Wang, L.; Yao, L.N.; Hao, X.Y.; Li, N.N.; Wang, Y.C.; Ding, C.Q.; Lei, L.; Qian, W.J.; Zeng, J.M.; Yang, Y.J.; et al. Transcriptional and physiological analyses reveal the association of ROS metabolism with cold tolerance in tea plant. Environ. And. Exp. Bot. 2019, 160, 45–58. [Google Scholar] [CrossRef]
- Valifard, M.; Le Hir, R.; Muller, J.; Scheuring, D.; Neuhaus, H.E.; Pommerrenig, B. Vacuolar fructose transporter SWEET17 is critical for root development and drought tolerance. Plant Physiol. 2021, 187, 2716–2730. [Google Scholar] [CrossRef]
- Wang, L.; Yao, L.; Hao, X.; Li, N.; Qian, W.; Yue, C.; Ding, C.; Zeng, J.; Yang, Y.; Wang, X. Tea plant SWEET transporters: Expression profiling, sugar transport, and the involvement of CsSWEET16 in modifying cold tolerance in Arabidopsis. Plant Mol. Biol. 2018, 96, 577–592. [Google Scholar] [CrossRef]
- Klemens, P.A.W.; Patzke, K.; Deitmer, J.; Spinner, L.; Le Hir, R.; Bellini, C.; Bedu, M.; Chardon, F.; Krapp, A.; Neuhaus, H.E. Overexpression of the Vacuolar Sugar Carrier AtSWEET16 Modifies Germination, Growth, and Stress Tolerance in Arabidopsis. Plant Physiol. 2013, 163, 1338–1352. [Google Scholar] [CrossRef]
- Yang, G.; Xu, H.; Zou, Q.; Zhang, J.; Chen, X.J.P.C.T.; Culture, O. The vacuolar membrane sucrose transporter MdSWEET16 plays essential roles in the cold tolerance of apple. Plant Cell Tissue Organ Cult. (PCTOC) 2020, 140, 129–142. [Google Scholar] [CrossRef]
- Yao, L.N.; Ding, C.Q.; Hao, X.Y.; Zeng, J.M.; Yang, Y.J.; Wang, X.C.; Wang, L. CsSWEET1a and CsSWEET17 Mediate Growth and Freezing Tolerance by Promoting Sugar Transport across the Plasma Membrane. Plant And. Cell Physiol. 2020, 61, 1669–1682. [Google Scholar] [CrossRef] [PubMed]
- Rouina, H.; Tseng, Y.H.; Nataraja, K.N.; Shaanker, R.U.; Oelmueller, R. Arabidopsis Restricts Sugar Loss to a Colonizing Trichoderma harzianum Strain by Downregulating SWEET11 and -12 and Upregulation of SUC1 and SWEET2 in the Roots. Microorganisms 2021, 9, 1246. [Google Scholar] [CrossRef]
- Chen, H.Y.; Huh, J.H.; Yu, Y.C.; Ho, L.H.; Chen, L.Q.; Tholl, D.; Frommer, W.B.; Guo, W.J. The Arabidopsis vacuolar sugar transporter SWEET2 limits carbon sequestration from roots and restricts Pythium infection. Plant J. 2015, 83, 1046–1058. [Google Scholar] [CrossRef]
- Wang, X.; Zhang, L.; Ji, H.; Mo, X.; Li, P.; Wang, J.; Dong, H. Hpa1 is a type III translocator in Xanthomonas oryzae pv. oryzae. BMC Microbiol. 2018, 18, 105. [Google Scholar] [CrossRef]
- Wu, Y.; Lee, S.-K.; Yoo, Y.; Wei, J.; Kwon, S.-Y.; Lee, S.-W.; Jeon, J.-S.; An, G. Rice Transcription Factor OsDOF11 Modulates Sugar Transport by Promoting Expression of Sucrose Transporter and SWEET Genes. Mol. Plant 2018, 11, 833–845. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Q.; Mao, W.; Xie, W.; Liu, Q.; Cao, J.; Yuan, M.; Zhang, Q.; Li, X.; Wang, S. Characterization of a disease susceptibility locus for exploring an efficient way to improve rice resistance against bacterial blight. Sci. China Life Sci. 2017, 60, 298–306. [Google Scholar] [CrossRef]
- Meteier, E.; La Camera, S.; Goddard, M.-L.; Laloue, H.; Mestre, P.; Chong, J. Overexpression of the VvSWEET4 Transporter in Grapevine Hairy Roots Increases Sugar Transport and Contents and Enhances Resistance to Pythium irregulare, a Soilborne Pathogen. Front. Plant Sci. 2019, 10, 884. [Google Scholar] [CrossRef] [PubMed]
- Jung, S.; Lee, T.; Cheng, C.H.; Buble, K.; Zheng, P.; Yu, J.; Humann, J.; Ficklin, S.P.; Gasic, K.; Scott, K.; et al. 15 years of GDR: New data and functionality in the Genome Database for Rosaceae. Nucleic Acids Res. 2019, 47, D1137–D1145. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER web server: 2018 update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed]
- Liu, Q.; Zhu, Y.S.; Wang, B.H.; Li, Y.X. A HMM-based method to predict the transmembrane regions of beta-barrel membrane proteins. Comput. Biol. Chem. 2003, 27, 69–76. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Howe, K.L.; Achuthan, P.; Allen, J.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Azov, A.G.; Bennett, R.; Bhai, J.; et al. Ensembl 2021. Nucleic Acids Res. 2021, 49, D884–D891. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proceedings. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Baek, J.-H.; Kim, J.; Kim, C.-K.; Sohn, S.-H.; Choi, D.; Ratnaparkhe, M.B.; Kim, D.-W.; Lee, T.-H. MultiSyn: A Webtool for Multiple Synteny Detection and Visualization of User’s Sequence of Interest Compared to Public Plant Species. Evol. Bioinform. Online 2016, 12, 193–199. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Singh, V.K.; Mangalam, A.K.; Dwivedi, S.; Naik, S. Primer premier: Program for design of degenerate primers from a protein sequence. BioTechniques 1998, 24, 318–319. [Google Scholar] [CrossRef]
- Zhang, Q.; Zhang, W.J.; Yin, Z.G.; Li, W.J.; Zhao, H.H.; Zhang, S.; Zhuang, L.; Wang, Y.X.; Zhang, W.H.; Du, J.D. Genome- and Transcriptome-Wide Identification of C3Hs in Common Bean (Phaseolus vulgaris L.) and Structural and Expression-Based Analyses of Their Functions During the Sprout Stage Under Salt-Stress Conditions. Front. Genet. 2020, 11, 564607. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- An, X.H.; Tian, Y.; Chen, K.Q.; Liu, X.J.; Liu, D.D.; Xie, X.B.; Cheng, C.G.; Cong, P.H.; Hao, Y.J. MdMYB9 and MdMYB11 are Involved in the Regulation of the JA-Induced Biosynthesis of Anthocyanin and Proanthocyanidin in Apples. Plant Cell Physiol. 2015, 56, 650–662. [Google Scholar] [CrossRef]
- Feng, C.Y.; Han, J.X.; Han, X.X.; Jiang, J. Genome-wide identification, phylogeny, and expression analysis of the SWEET gene family in tomato. Gene 2015, 573, 261–272. [Google Scholar] [CrossRef]
- Zhang, W.; Wang, S.Y.; Yu, F.W.; Tang, J.; Shan, X.; Bao, K.; Yu, L.; Wang, H.; Fei, Z.J.; Li, J.B. Genome-wide characterization and expression profiling of SWEET genes in cabbage (Brassica oleracea var. capitata L.) reveal their roles in chilling and clubroot disease responses. BMC Genom. 2019, 20, 93. [Google Scholar] [CrossRef]
- Manck-Gotzenberger, J.; Requena, N. Arbuscular mycorrhiza Symbiosis Induces a Major Transcriptional Reprogramming of the Potato SWEET Sugar Transporter Family. Front. Plant Sci. 2016, 7, 487. [Google Scholar] [CrossRef]
- Sui, J.L.; Xiao, X.H.; Qi, J.Y.; Fang, Y.J.; Tang, C.R. The SWEET gene family in Hevea brasiliensis—Its evolution and expression compared with four other plant species. FEBS Open Bio. 2017, 7, 1943–1959. [Google Scholar] [CrossRef]
- Miao, H.X.; Sun, P.G.; Liu, Q.; Miao, Y.L.; Liu, J.H.; Zhang, K.X.; Hu, W.; Zhang, J.B.; Wang, J.Y.; Wang, Z.; et al. Genome-wide analyses of SWEET family proteins reveal involvement in fruit development and abiotic/biotic stress responses in banana. Sci. Rep. 2017, 7, 3536. [Google Scholar] [CrossRef]
- Filyushin, M.A.; Slugina, M.A.; Shchennikova, A.V.; Kochieva, E.Z. Differential Expression of Sugar Uniporter Genes of the SWEET Family in the Regulation of Qualitative Fruit Traits in Tomato Species (Solanum Section Lycopersicon). Russ. J. Plant Physiol. 2023, 70, 70. [Google Scholar] [CrossRef]
- Sharma, H.; Sharma, A.; Rajput, R.; Sidhu, S.; Dhillon, H.; Verma, P.C.; Pandey, A.; Upadhyay, S.K. Molecular Characterization, Evolutionary Analysis, and Expression Profiling of BOR Genes in Important Cereals. Plants 2022, 11, 911. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.M.; Lv, Y.X.; Kong, L.J.; Chen, Q.Z.; Chen, C.Q.; Li, J.; Zeng, F.H.; Wang, S.Y.; Li, J.B.; Huang, L.; et al. Genome-wide identification, phylogeny, evolution, and expression patterns of MtN3/saliva/SWEET genes and functional analysis of BcNS in Brassica rapa. BMC Genom. 2018, 19, 174. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Xue, X.Y.; Zeng, H.Q.; Li, J.K.; Chen, L.Q. Sucrose rather than GA transported by AtSWEET13 and AtSWEET14 supports pollen fitness at late anther development stages. New Phytol. 2022, 236, 525–537. [Google Scholar] [CrossRef]
- Liu, X.Z.; Zhang, Y.; Yang, C.; Tian, Z.H.; Li, J.X. AtSWEET4, a hexose facilitator, mediates sugar transport to axial sinks and affects plant development. Sci. Rep. 2016, 6, 24563. [Google Scholar] [CrossRef] [PubMed]
- Chardon, F.; Bedu, M.; Calenge, F.; Klemens, P.A.W.; Spinner, L.; Clement, G.; Chietera, G.; Leran, S.; Ferrand, M.; Lacombe, B.; et al. Leaf Fructose Content Is Controlled by the Vacuolar Transporter SWEET17 in Arabidopsis. Curr. Biol. 2013, 23, 697–702. [Google Scholar] [CrossRef]
- Engel, M.L.; Holmes-Davis, R.; McCormick, S. Green sperm. Identification of male gamete promoters in Arabidopsis. Plant Physiology 2005, 138, 2124–2133. [Google Scholar] [CrossRef]
- Wen, Z.; Li, M.; Meng, J.; Li, P.; Cheng, T.; Zhang, Q.; Sun, L. Genome-wide identification of the SWEET gene family mediating the cold stress response in Prunus mume. PeerJ 2022, 10, e13273. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, Z.Y.; Kumar, V.; Xu, X.F.; Yuan, D.P.; Zhu, X.F.; Li, T.Y.; Jia, B.; Xuan, Y.H. Genome-wide identification of the SWEET gene family in wheat. Gene 2018, 642, 284–292. [Google Scholar] [CrossRef]
- Hu, L.P.; Zhang, F.; Song, S.H.; Yu, X.L.; Ren, Y.; Zhao, X.Z.; Liu, H.; Liu, G.M.; Wang, Y.Q.; He, H.J. CsSWEET2, a Hexose Transporter from Cucumber (Cucumis sativus L.), Affects Sugar Metabolism and Improves Cold Tolerance in Arabidopsis. Int. J. Mol. Sci. 2022, 23, 3886. [Google Scholar] [CrossRef] [PubMed]
- Le Hir, R.; Spinner, L.; Klemens, P.A.W.; Chakraborti, D.; de Marco, F.; Vilaine, F.; Wolff, N.; Lemoine, R.; Porcheron, B.; Gery, C.; et al. Disruption of the Sugar Transporters AtSWEET11 and AtSWEET12 Affects Vascular Development and Freezing Tolerance in Arabidopsis. Mol. Plant 2015, 8, 1687–1690. [Google Scholar] [CrossRef] [PubMed]







| Gene Name | Gene Locus | Stand | AA(aa) | PI | MW (kDa) | TM |
|---|---|---|---|---|---|---|
| RrSWEET1 | evm.model.Chr1.60 | − | 127 | 9.28 | 14.30 | 3 |
| RrSWEET2 | evm.model.Chr1.1695 | + | 243 | 8.94 | 27.14 | 7 |
| RrSWEET3 | evm.model.Chr1.2350 | − | 181 | 4.63 | 20.21 | 4 |
| RrSWEET4 | evm.model.Chr1.2452 | − | 241 | 5.85 | 26.98 | 7 |
| RrSWEET5 | evm.model.Chr1.3067 | + | 281 | 7.7 | 31.53 | 7 |
| RrSWEET6 | evm.model.Chr2.1813 | + | 294 | 6.73 | 32.81 | 7 |
| RrSWEET7 | evm.model.Chr2.1816 | + | 309 | 9.68 | 34.48 | 7 |
| RrSWEET8 | evm.model.Chr2.1817 | + | 249 | 9.39 | 27.83 | 7 |
| RrSWEET9 | evm.model.Chr2.1819 | + | 273 | 8.72 | 30.23 | 7 |
| RrSWEET10 | evm.model.Chr2.1921 | + | 293 | 5.48 | 32.70 | 7 |
| RrSWEET11 | evm.model.Chr2.1926 | + | 304 | 9.5 | 33.92 | 7 |
| RrSWEET12 | evm.model.Chr2.1927 | + | 296 | 9.46 | 32.77 | 7 |
| RrSWEET13 | evm.model.Chr2.1933 | + | 273 | 8.72 | 30.23 | 7 |
| RrSWEET14 | evm.model.Chr2.3499 | − | 247 | 9.28 | 27.28 | 7 |
| RrSWEET15 | evm.model.Chr3.2084 | − | 243 | 8.91 | 26.89 | 7 |
| RrSWEET16 | evm.model.Chr3.3219 | + | 243 | 9.02 | 27.29 | 6 |
| RrSWEET17 | evm.model.Chr3.3233 | − | 243 | 8.91 | 27.29 | 7 |
| RrSWEET18 | evm.model.Chr3.3287 | + | 239 | 9.08 | 26.89 | 6 |
| RrSWEET19 | evm.model.Chr3.3681 | − | 136 | 5.42 | 15.68 | 1 |
| RrSWEET20 | evm.model.Chr3.4823 | + | 251 | 8.92 | 28.32 | 7 |
| RrSWEET21 | evm.model.Chr4.1820 | + | 192 | 7.53 | 21.54 | 4 |
| RrSWEET22 | evm.model.Chr5.4695 | + | 235 | 8.61 | 26.30 | 7 |
| RrSWEET23 | evm.model.Chr5.5986 | + | 235 | 9.33 | 26.42 | 7 |
| RrSWEET24 | evm.model.Chr6.3424 | + | 225 | 8.53 | 25.44 | 6 |
| RrSWEET25 | evm.model.Chr6.3661 | + | 170 | 6.55 | 19.08 | 1 |
| RrSWEET26 | evm.model.Chr6.4775 | + | 235 | 9.02 | 26.31 | 6 |
| RrSWEET27 | evm.model.Chr6.6650 | + | 243 | 9.38 | 26.99 | 6 |
| RrSWEET28 | evm.model.Chr7.760 | − | 253 | 9.28 | 28.13 | 7 |
| RrSWEET29 | evm.model.Chr7.1004 | + | 311 | 6.65 | 34.85 | 7 |
| RrSWEET30 | evm.model.Chr7.1007 | + | 288 | 8.15 | 32.70 | 7 |
| RrSWEET31 | evm.model.Chr7.1376 | − | 238 | 8.89 | 26.97 | 7 |
| RrSWEET32 | evm.model.Chr7.1378 | − | 237 | 9.32 | 26.57 | 7 |
| RrSWEET33 | evm.model.Chr7.1609 | + | 91 | 9.76 | 11.02 | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, R.; Gao, P.; Yang, T.; Dong, J.; Chen, Y.; Xie, Y.; Yang, Y.; Liu, C.; Zhang, J.; Che, D. Genome-Wide Analysis of the SWEET Transporters and Their Potential Role in Response to Cold Stress in Rosa rugosa. Horticulturae 2023, 9, 1212. https://doi.org/10.3390/horticulturae9111212
Li R, Gao P, Yang T, Dong J, Chen Y, Xie Y, Yang Y, Liu C, Zhang J, Che D. Genome-Wide Analysis of the SWEET Transporters and Their Potential Role in Response to Cold Stress in Rosa rugosa. Horticulturae. 2023; 9(11):1212. https://doi.org/10.3390/horticulturae9111212
Chicago/Turabian StyleLi, Ronghui, Peng Gao, Tao Yang, Jie Dong, Yunting Chen, Yangyang Xie, Yvtong Yang, Chengzhi Liu, Jinzhu Zhang, and Daidi Che. 2023. "Genome-Wide Analysis of the SWEET Transporters and Their Potential Role in Response to Cold Stress in Rosa rugosa" Horticulturae 9, no. 11: 1212. https://doi.org/10.3390/horticulturae9111212
APA StyleLi, R., Gao, P., Yang, T., Dong, J., Chen, Y., Xie, Y., Yang, Y., Liu, C., Zhang, J., & Che, D. (2023). Genome-Wide Analysis of the SWEET Transporters and Their Potential Role in Response to Cold Stress in Rosa rugosa. Horticulturae, 9(11), 1212. https://doi.org/10.3390/horticulturae9111212








