Genome-Wide Identification of Nucleotide-Binding Site–Leucine-Rich Repeat Gene Family in Cymbidium ensifolium and Expression Profiles in Response to Fusarium Wilt Infection
Abstract
1. Introduction
2. Materials and Methods
2.1. Identification of the CeNBS-LRR Genes
2.2. Phylogenetic Analysis, Physicochemical Properties, and Chromosome Localization Analysis
2.3. Cis-Acting Elements and Genome Collinearity Analysis of CeNBS-LRR Genes
2.4. Analysis of Gene Structure and Conserved Domains
2.5. Plant Material and Gene Expression Analysis to Fusarium Wilt Infection
2.6. Real-Time Fluorescence Quantitative Experiment
3. Results
3.1. Identification and Classification of NBS-LRR Genes in C. ensifolium
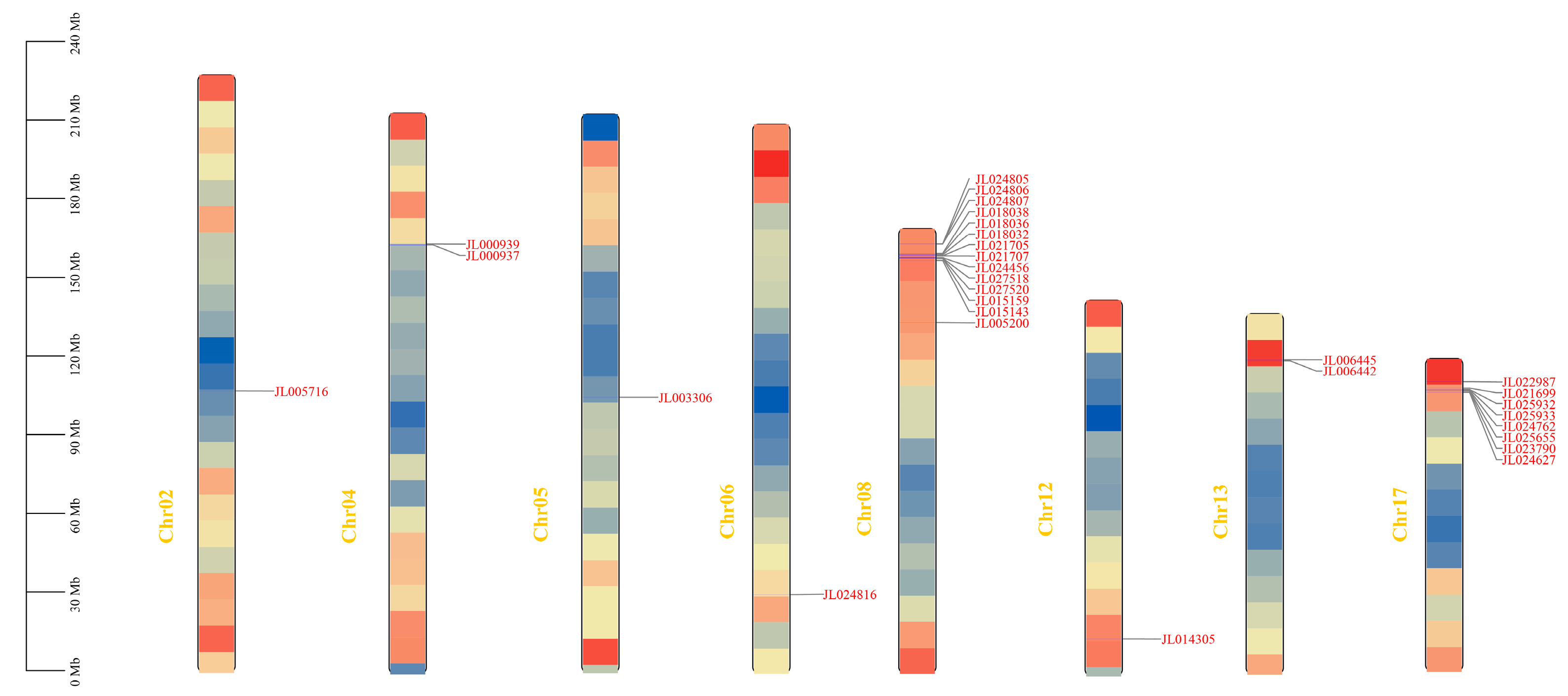
3.2. Phylogenetic Analysis, Physicochemical Properties, and Chromosomal Localization Analysis
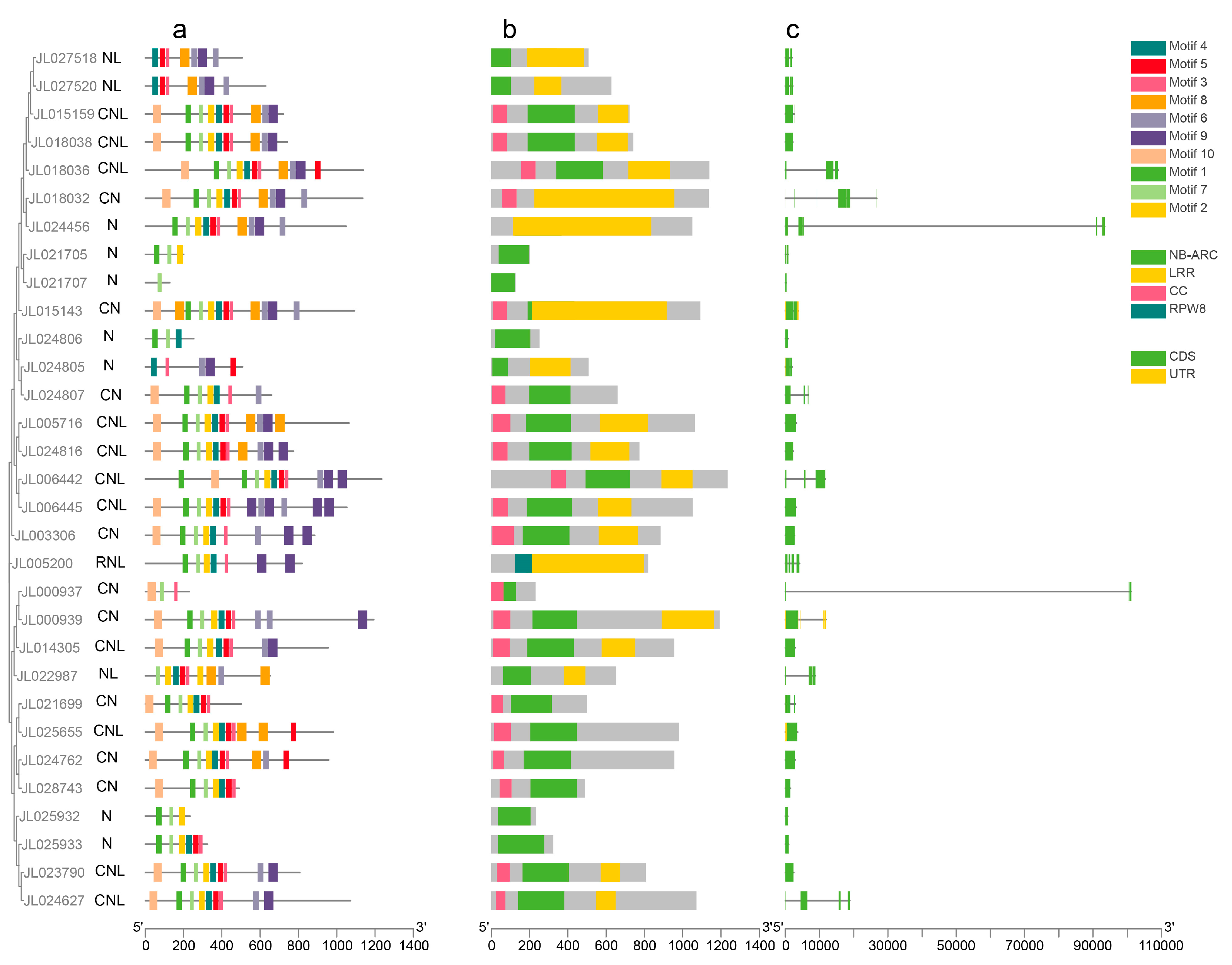
3.3. Gene Structure Analysis, Conserved Motif Analysis, and Conserved Domain Analysis in C. ensifolium
3.4. Collinearity Analysis of CeNBS-LRR Genes
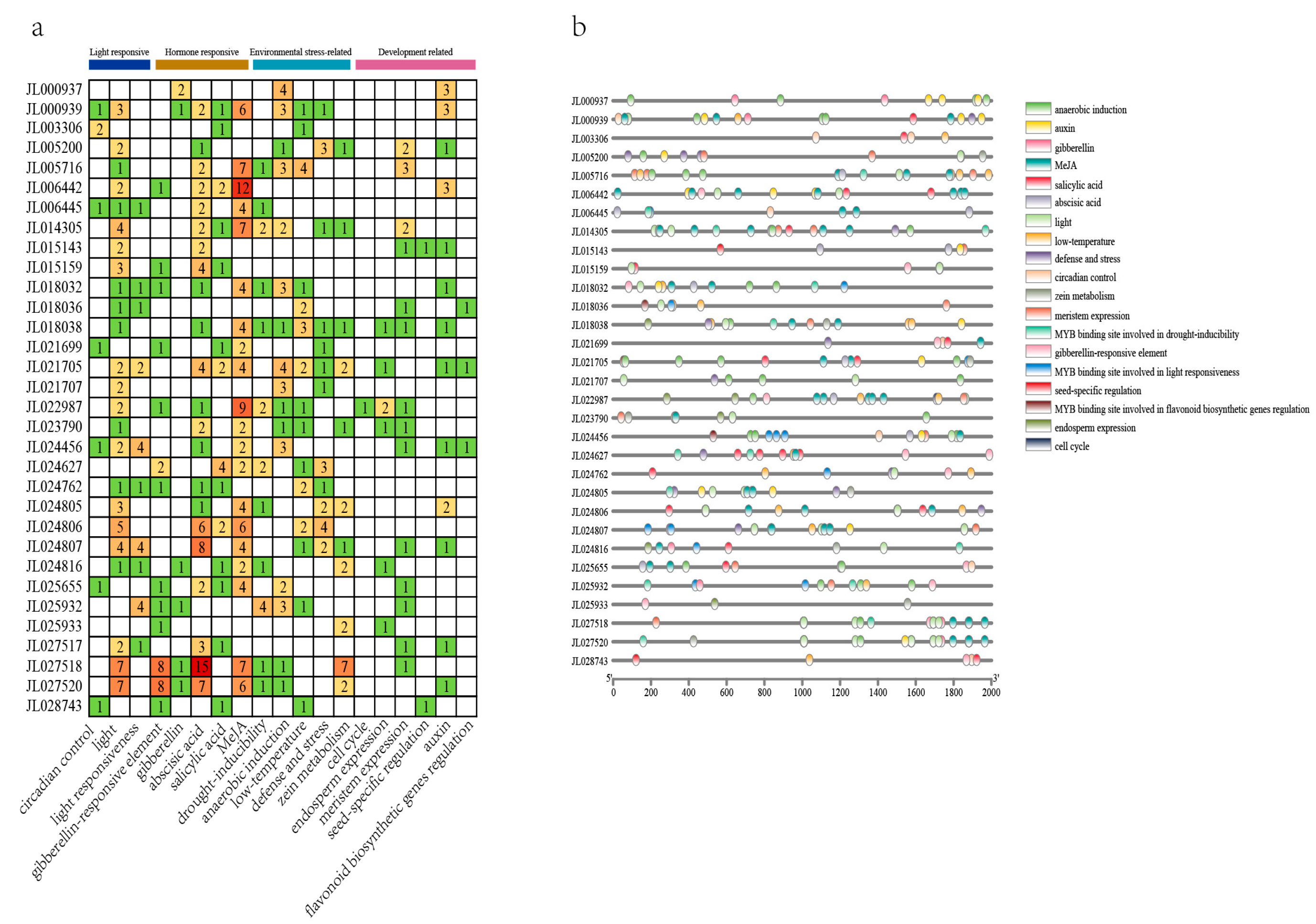
3.5. Analysis of Cis-Acting Elements
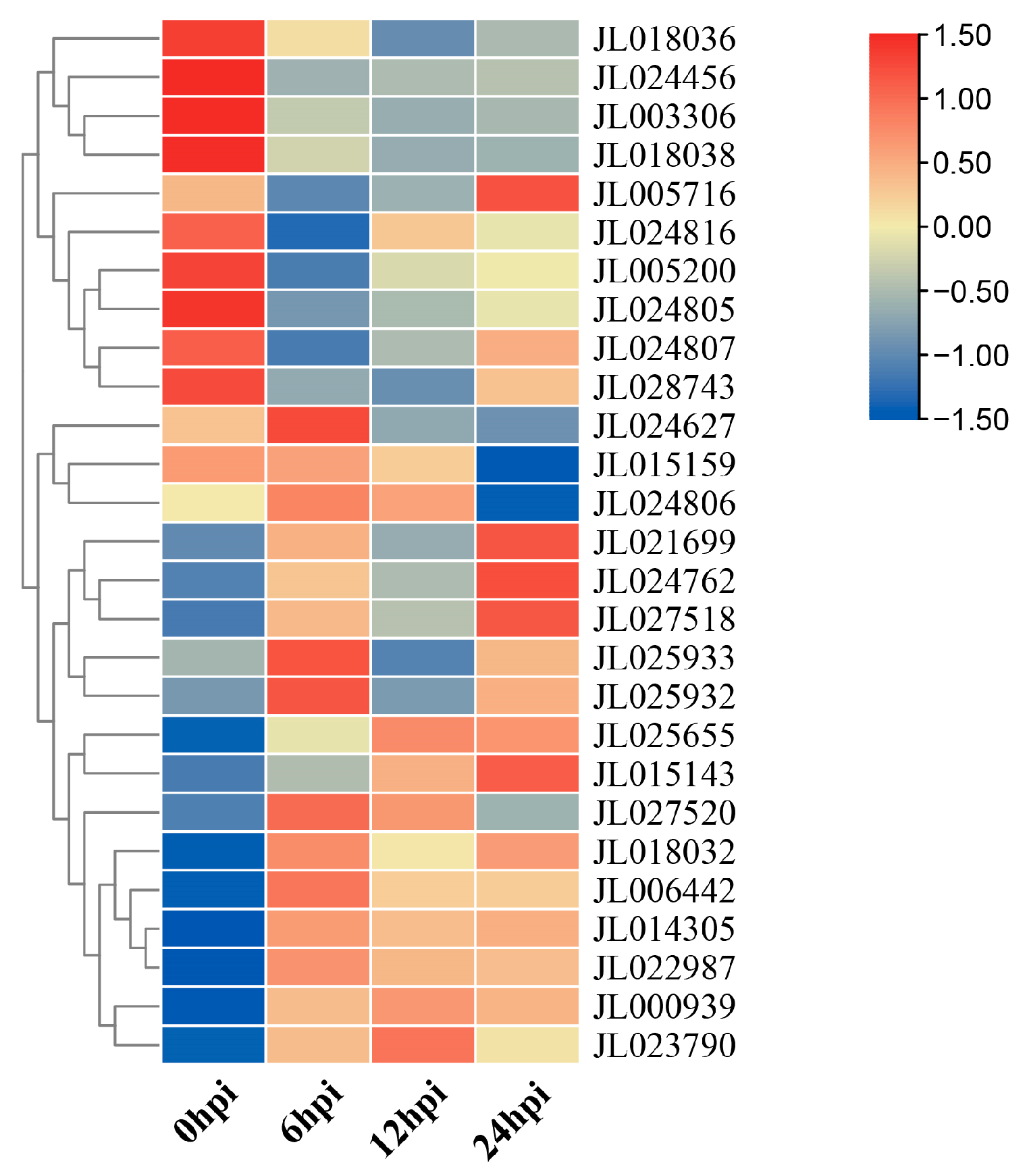
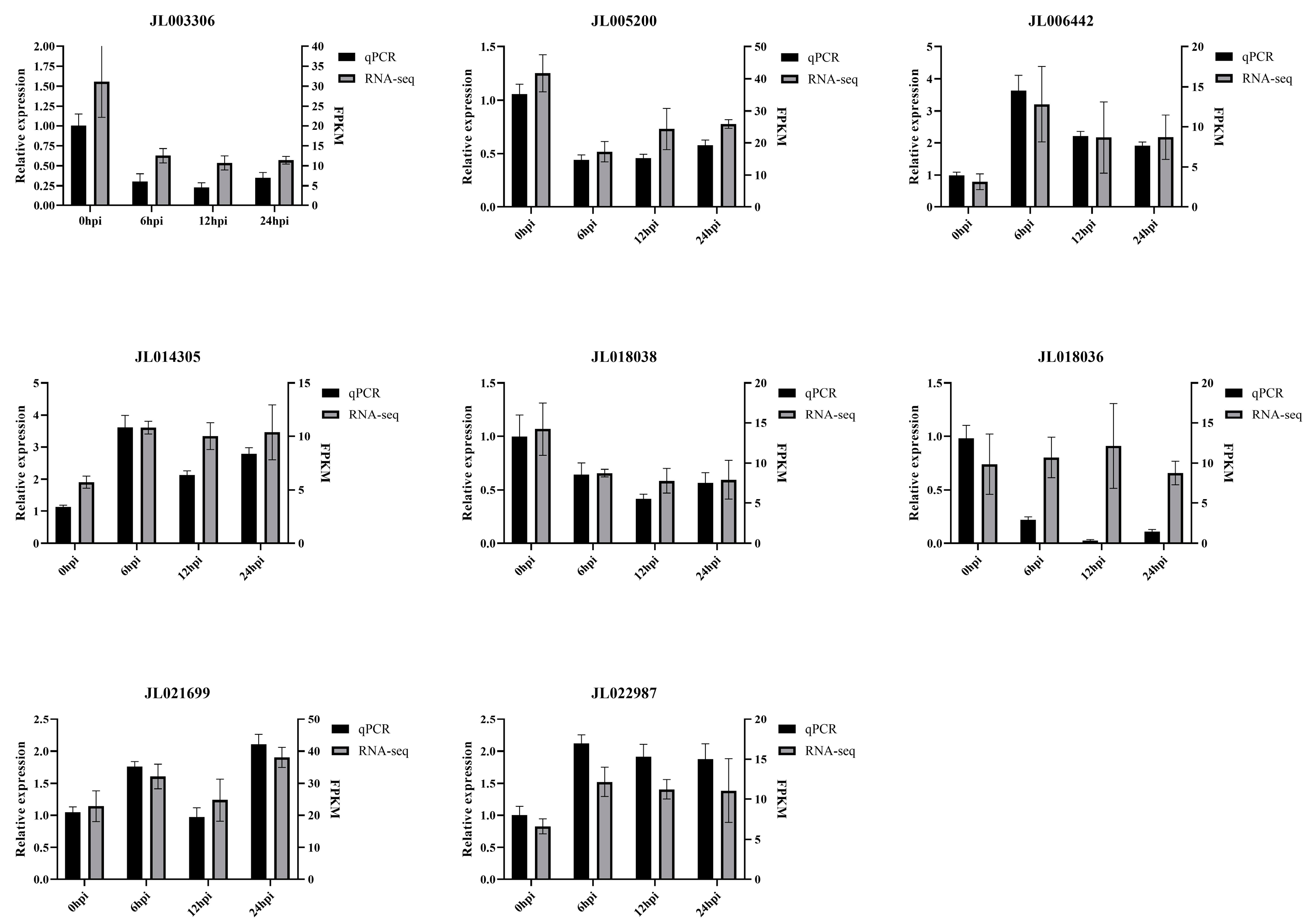
3.6. Expression Patterns of CeNBS-LRR Genes in Response to Fusarium Wilt Infection
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Ai, Y.; Li, Z.; Sun, W.-H.; Chen, J.; Zhang, D.; Ma, L.; Zhang, Q.-H.; Chen, M.-K.; Zheng, Q.-D.; Liu, J.-F.; et al. The Cymbidium genome reveals the evolution of unique morphological traits. Hortic. Res. 2021, 8, 255–264. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Liu, L.; Sun, S.; Li, Y.; Jia, L.; Ye, S.; Yu, Y.; Dossa, K.; Luan, Y. Transcriptome analysis reveals the key pathways and candidate genes involved in salt stress responses in Cymbidium ensifolium leaves. BMC Plant Biol. 2023, 23, 64. [Google Scholar] [CrossRef] [PubMed]
- Paul, S.K.; Mahmud, N.U.; Gupta, D.R.; Alam, M.N.; Chakraborty, M.; Islam, M.T. First Report of Fusarium sacchari Causing Sugarcane Wilt in Bangladesh. Plant Dis. 2022, 106, 319. [Google Scholar] [CrossRef] [PubMed]
- Jin-Ai, Y.; Peng, H.; Cheng-Zhong, L.; De-Yi, Y. Stem rot on Cymbidium ensifolium (Orchidaceae) caused by Fusarium oxysporum in China. Can. J. Plant. Pathol. 2017, 40, 105–108. [Google Scholar] [CrossRef]
- Chung, W.C.; Chen, L.W.; Huang, J.H.; Huang, H.C.; Chung, W.H. A new ‘forma specialis’ of Fusarium solani causing leaf yellowing of Phalaenopsis. Plant Pathol. 2010, 60, 244–252. [Google Scholar] [CrossRef]
- Staskawicz, B.J.; Ausubel, F.M.; Baker, B.J.; Ellis, J.G.; Jones, J.D.G. Molecular Genetics of Plant Disease Resistance. Science 1995, 268, 661–667. [Google Scholar] [CrossRef]
- Wei, W.R.; Yue, H.L.; Nie, Y.X.; Chen, T.X. First Report of Colletotrichum siamense Causing Daylily (Hemerocallis citrina) Anthracnose in China. Plant Dis. 2022, 106, 1061. [Google Scholar] [CrossRef] [PubMed]
- Ajithkumar, K.; Savitha, A.S.; Mahadevakumar, S.; Sujatha, M.; Maharachchikumbura, S.S.N.; Sreenivasa, M.Y.; Renuka, M.; Yenjerappa, S.T. First Report of Powdery Mildew Caused by Erysiphe diffusa on Cluster Bean in India. Plant. Dis. 2022, 106, 3203. [Google Scholar] [CrossRef]
- Ji, H.M.; Mao, H.Y.; Li, S.J.; Feng, T.; Zhang, Z.Y.; Cheng, L.; Luo, S.J.; Borkovich, K.A.; Ouyang, S.Q. Fol-milR1, a pathogenicity factor of Fusarium oxysporum, confers tomato wilt disease resistance by impairing host immune responses. New Phytol. 2021, 232, 705–718. [Google Scholar] [CrossRef]
- Kourelis, J.; van der Hoorn, R.A.L. Defended to the Nines: 25 Years of Resistance Gene Cloning Identifies Nine Mechanisms for R Protein Function. Plant Cell 2018, 30, 285–299. [Google Scholar] [CrossRef]
- Ellis, J.G.; Lawrence, G.J.; Dodds, P.N. Further analysis of gene-for-gene disease resistance specificity in flax. Mol. Plant Pathol. 2007, 8, 103–109. [Google Scholar] [CrossRef] [PubMed]
- DeYoung, B.J.; Innes, R.W. Plant NBS-LRR proteins in pathogen sensing and host defense. Nat. Immunol. 2006, 7, 1243–1249. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.Q.; Xue, J.Y.; Wang, Q.; Wang, B.; Chen, J.Q. Revisiting the Origin of Plant NBS-LRR Genes. Trends Plant Sci. 2019, 24, 9–12. [Google Scholar] [CrossRef] [PubMed]
- McDowell, J.M.; Woffenden, B.J. Plant disease resistance genes: Recent insights and potential applications. Trends Biotechnol. 2003, 21, 178–183. [Google Scholar] [CrossRef] [PubMed]
- Kobe, B.; Kajava, A.V. The leucine-rich repeat as a protein recognition motif. Curr. Opin. Struct. Biol. 2001, 11, 725–732. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhong, S.; Yang, H.; Chen, C.; Chen, W.; Yang, H.; Guan, J.; Fu, P.; Tan, F.; Ren, T.; et al. Identification and Characterization of NBS Resistance Genes in Akebia trifoliata. Front. Plant Sci. 2021, 12, 758559. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.C.; Kaushik, S.; Nandety, R.S. Evolving disease resistance genes. Curr. Opin. Plant Biol. 2005, 8, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Bai, J.; Pennill, L.A.; Ning, J.; Lee, S.W.; Ramalingam, J.; Webb, C.A.; Zhao, B.; Sun, Q.; Nelson, J.C.; Leach, J.E.; et al. Diversity in Nucleotide Binding Site–Leucine-Rich Repeat Genes in Cereals. Genome Res. 2002, 12, 1871–1884. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.C.; Kozik, A.; Griego, A.; Kuang, H.; Michelmore, R.W. Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 2003, 15, 809–834. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Ghouri, F.; Yu, H.; Li, X.; Yu, S.; Shahid, M.Q.; Liu, X. Genome wide re-sequencing of newly developed Rice Lines from common wild rice (Oryza rufipogon Griff.) for the identification of NBS-LRR genes. PLoS ONE 2017, 12, e0180662. [Google Scholar] [CrossRef]
- Liu, J.; Qiao, L.Y.; Zhang, X.J.; Li, X.; Zhan, H.X.; Guo, H.J.; Zheng, J.; Chang, Z.J. Genome-wide identification and resistance expression analysis of the NBS gene family in Triticum urartu. Genes Genom. 2017, 39, 611–621. [Google Scholar] [CrossRef]
- Ameline-Torregrosa, C.; Wang, B.B.; O’Bleness, M.S.; Deshpande, S.; Zhu, H.; Roe, B.; Young, N.D.; Cannon, S.B. Identification and characterization of nucleotide-binding site-leucine-rich repeat genes in the model plant Medicago truncatula. Plant Physiol. 2008, 146, 5–21. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Jia, Z.H.; Zhang, J.Y.; Liu, M.; Guo, Z.R.; Wang, G. Identification and Analysis of NBS-LRR Genes in Actinidia chinensis Genome. Plants 2020, 9, 1350. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Hernandez, M.; Berardini, T.Z.; Chen, G.; Crist, D.; Doyle, A.; Huala, E.; Knee, E.; Lambrecht, M.; Miller, N.; Mueller, L.A.; et al. TAIR: A resource for integrated Arabidopsis data. Funct. Integr. Genomics 2002, 2, 239–253. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Wang, J.; Chitsaz, F.; Derbyshire, M.K.; Geer, R.C.; Gonzales, N.R.; Gwadz, M.; Hurwitz, D.I.; Marchler, G.H.; Song, J.S.; et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 2020, 48, D265–D268. [Google Scholar] [CrossRef] [PubMed]
- Letunic, I.; Khedkar, S.; Bork, P. SMART: Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021, 49, D458–D460. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Xie, J.; Chen, Y.; Cai, G.; Cai, R.; Hu, Z.; Wang, H. Tree Visualization By One Table (tvBOT): A web application for visualizing, modifying and annotating phylogenetic trees. Nucleic Acids Res. 2023, 51, W587–W592. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.C.; Shen, H.B. Cell-PLoc: A package of Web servers for predicting subcellular localization of proteins in various organisms. Nat. Protoc. 2008, 3, 153–162. [Google Scholar] [CrossRef]
- Gasteiger, E.; Gattiker, A.; Hoogland, C.; Ivanyi, I.; Appel, R.D.; Bairoch, A. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Lupas, A.; Van Dyke, M.; Stock, J. Predicting coiled coils from protein sequences. Science 1991, 252, 1162–1164. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.Q.; Xue, J.Y.; Wu, P.; Zhang, Y.M.; Wu, Y.; Hang, Y.Y.; Wang, B.; Chen, J.Q. Large-Scale Analyses of Angiosperm Nucleotide-Binding Site-Leucine-Rich Repeat Genes Reveal Three Anciently Diverged Classes with Distinct Evolutionary Patterns. Plant Physiol. 2016, 170, 2095–2109. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Song, W.; Chai, J. Structure, biochemical function, and signaling mechanism of plant NLRs. Mol. Plant 2023, 16, 75–95. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Chen, C.; Li, L.; Tan, X.; Wei, Z.; Li, Y.; Li, J.; Yan, F.; Chen, J.; Sun, Z. A rice LRR receptor-like protein associates with its adaptor kinase OsSOBIR1 to mediate plant immunity against viral infection. Plant Biotechnol. J. 2021, 19, 2319–2332. [Google Scholar] [CrossRef]
- Ke, Y.-J.; Zheng, Q.-D.; Yao, Y.-H.; Ou, Y.; Chen, J.-Y.; Wang, M.-J.; Lai, H.-P.; Yan, L.; Liu, Z.-J.; Ai, Y. Genome-Wide Identification of the MYB Gene Family in Cymbidium ensifolium and Its Expression Analysis in Different Flower Colors. Int. J. Mol. Sci. 2021, 22, 13245. [Google Scholar] [CrossRef] [PubMed]
- Ahmad, S.; Chen, G.; Huang, J.; Yang, K.; Hao, Y.; Zhou, Y.; Zhao, K.; Lan, S.; Liu, Z.; Peng, D. Beauty and the pathogens: A leaf-less control presents a better image of Cymbidium orchids defense strategy. Front. Plant Sci. 2022, 13, 1001427. [Google Scholar] [CrossRef]
- Saucet, S.B.; Ma, Y.; Sarris, P.F.; Furzer, O.J.; Sohn, K.H.; Jones, J.D.G. Two linked pairs of Arabidopsis TNL resistance genes independently confer recognition of bacterial effector AvrRps4. Nat. Commun. 2015, 6, 6338. [Google Scholar] [CrossRef]
- Pedley, K.F.; Martin, G.B. Molecular Basis of Pto-Mediated Resistance to Bacterial Speck Disease in Tomato. Annu. Rev. Phytopathol. 2003, 41, 215–243. [Google Scholar] [CrossRef]
- Ercoli, M.F.; Luu, D.D.; Rim, E.Y.; Shigenaga, A.; Teixeira de Araujo, A.; Chern, M.; Jain, R.; Ruan, R.; Joe, A.; Stewart, V.; et al. Plant immunity: Rice XA21-mediated resistance to bacterial infection. Proc. Natl. Acad. Sci. USA 2022, 119, e2121568119. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Dong, L.; Li, B.; Wang, Z.; Xie, J.; Qiu, D.; Li, Y.; Shi, W.; Yang, L.; Wu, Q.; et al. A CNL protein in wild emmer wheat confers powdery mildew resistance. New Phytol. 2020, 228, 1027–1037. [Google Scholar] [CrossRef] [PubMed]
- Staal, J.; Kaliff, M.; Dewaele, E.; Persson, M.; Dixelius, C. RLM3, a TIR domain encoding gene involved in broad-range immunity of Arabidopsis to necrotrophic fungal pathogens. Plant J. 2008, 55, 188–200. [Google Scholar] [CrossRef] [PubMed]
- Meyers, B.C.; Dickerman, A.W.; Michelmore, R.W.; Sivaramakrishnan, S.; Sobral, B.W.; Young, N.D. Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J. 1999, 20, 317–332. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Zhao, M.; Qin, H.; Li, S.; Yang, X. Genome-wide analysis of NBS-LRR genes revealed contribution of disease resistance from Saccharum spontaneum to modern sugarcane cultivar. Front. Plant Sci. 2023, 14, 1091567. [Google Scholar] [CrossRef]
- Tetreault, H.M.; Grover, S.; Scully, E.D.; Gries, T.; Palmer, N.A.; Sarath, G.; Louis, J.; Sattler, S.E. Global Responses of Resistant and Susceptible Sorghum (Sorghum bicolor) to Sugarcane Aphid (Melanaphis sacchari). Front. Plant Sci. 2019, 10, 145. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Jiang, X.-M.; Shao, Z.-Q. Genome-Wide Analysis of NLR Disease Resistance Genes in an Updated Reference Genome of Barley. Front. Genet. 2021, 12, 694682. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.-H.; Wang, Y.; Chen, M.; Liu, J.; Lu, R.-S.; Zou, X.; Sun, X.-Q.; Zhang, Y.-M. Genome-wide Identification and Evolutionary Analysis of NBS-LRR Genes From Secale cereale. Front. Genet. 2021, 12, 771814. [Google Scholar] [CrossRef] [PubMed]
- Xiao, S.; Ellwood, S.; Calis, O.; Patrick, E.; Li, T.; Coleman, M.; Turner, J.G. Broad-spectrum mildew resistance in Arabidopsis thaliana mediated by RPW8. Science 2001, 291, 118–120. [Google Scholar] [CrossRef]
- Cannon, S.B.; May, G.D.; Jackson, S.A. Three Sequenced Legume Genomes and Many Crop Species: Rich Opportunities for Translational Genomics. Plant Physiol. 2009, 151, 970–977. [Google Scholar] [CrossRef]
- Xue, J.Y.; Zhao, T.; Liu, Y.; Liu, Y.; Zhang, Y.X.; Zhang, G.Q.; Chen, H.; Zhou, G.C.; Zhang, S.Z.; Shao, Z.Q. Genome- Wide Analysis of the Nucleotide Binding Site Leucine-Rich Repeat Genes of Four Orchids Revealed Extremely Low Numbers of Disease Resistance Genes. Front. Genet. 2019, 10, 1286. [Google Scholar] [CrossRef]
- Loutre, C.; Wicker, T.; Travella, S.; Galli, P.; Scofield, S.; Fahima, T.; Feuillet, C.; Keller, B. Two different CC-NBS-LRR genes are required for Lr10-mediated leaf rust resistance in tetraploid and hexaploid wheat. Plant J. 2009, 60, 1043–1054. [Google Scholar] [CrossRef] [PubMed]
- Rushton, P.J.; Reinstädler, A.; Lipka, V.; Lippok, B.; Somssich, I.E. Synthetic plant promoters containing defined regulatory elements provide novel insights into pathogen- and wound-induced signaling. Plant Cell 2002, 14, 749–762. [Google Scholar] [CrossRef] [PubMed]
- Spoel, S.H.; Koornneef, A.; Claessens, S.M.C.; Korzelius, J.P.; Van Pelt, J.A.; Mueller, M.J.; Buchala, A.J.; Métraux, J.-P.; Brown, R.; Kazan, K.; et al. NPR1 Modulates Cross-Talk between Salicylate- and Jasmonate-Dependent Defense Pathways through a Novel Function in the Cytosol. Plant Cell 2003, 15, 760–770. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.; Zhang, W.; Wang, X. Post-translational control of ABA signalling: The roles of protein phosphorylation and ubiquitination. Plant Biotechnol. J. 2016, 15, 4–14. [Google Scholar] [CrossRef]
- Yang, J.; Xiong, C.; Li, S.; Zhou, C.; Li, L.; Xue, Q.; Liu, W.; Niu, Z.; Ding, X. Evolution patterns of NBS genes in the genus Dendrobium and NBS-LRR gene expression in D. officinale by salicylic acid treatment. BMC Plant Biol. 2022, 22, 529. [Google Scholar]



| Type | Domain | C. ensifolium | A. thaliana | O. sativa |
|---|---|---|---|---|
| CNL | CC NBS LRR | 11 | 18 | 199 |
| CN | CC NBS | 9 | 3 | 61 |
| NL | NBS LRR | 4 | 32 | 123 |
| N | NBS | 6 | 6 | 109 |
| TNL | TIR NBS LRR | 0 | 101 | 0 |
| TN | TIR NBS | 0 | 2 | 3 |
| RNL | RPW8 NBS LRR | 1 | 5 | 0 |
| RN | RPW8 NBS | 0 | 1 | 0 |
| Total | 31 | 168 | 495 |
| Sequence ID | Length/bp | Size/aa | MW/Kda | pI | II | AI | GRAVY | Subcellular Location |
|---|---|---|---|---|---|---|---|---|
| JL000937 | 693 | 230 | 19,641.8 | 5.46 | 48.08 | 99.61 | −0.381 | Cytoplasm. Nucleus. |
| JL000939 | 3579 | 1192 | 136,675.62 | 6.08 | 46.76 | 102.42 | −0.243 | Cytoplasm. |
| JL003306 | 2655 | 884 | 100,476.43 | 9.23 | 57.12 | 103.82 | −0.16 | Cytoplasm. |
| JL005200 | 2457 | 818 | 93,067.12 | 6.32 | 47.07 | 104.82 | −0.145 | Cytoplasm. |
| JL005716 | 3192 | 1063 | 121,322.26 | 5.65 | 54.08 | 106.01 | −0.114 | Cell membrane. Cytoplasm. |
| JL006442 | 3705 | 1234 | 138,792.65 | 7.13 | 44.04 | 95.43 | −0.252 | Cell membrane. Cytoplasm. |
| JL006445 | 3156 | 1051 | 118,860.11 | 5.52 | 45.63 | 103.42 | −0.153 | Cell membrane. Cytoplasm. |
| JL014305 | 2865 | 954 | 109,640.25 | 6.27 | 42.86 | 104.25 | −0.233 | Cytoplasm. |
| JL015143 | 3279 | 1092 | 124,529.05 | 6.22 | 44.61 | 101.77 | −0.187 | Cytoplasm. |
| JL015159 | 2166 | 721 | 82,248.03 | 6.6 | 45.93 | 103.43 | −0.21 | Cytoplasm. |
| JL018032 | 3408 | 1135 | 128,937.11 | 6.45 | 43.25 | 101.26 | −0.2 | Cytoplasm. |
| JL018036 | 3417 | 1138 | 128,968.93 | 6.45 | 45.21 | 100.07 | −0.177 | Cytoplasm. |
| JL018038 | 2226 | 741 | 84,065.55 | 7.26 | 42.34 | 106.67 | −0.119 | Cytoplasm. |
| JL021699 | 1500 | 499 | 58,741.93 | 5.47 | 52.32 | 94.33 | −0.323 | Cytoplasm. |
| JL021705 | 600 | 199 | 22,529.59 | 4.67 | 46.89 | 94.47 | −0.184 | Cytoplasm. Nucleus. |
| JL021707 | 384 | 127 | 14,328.63 | 6.91 | 40.37 | 104.96 | −0.102 | Cytoplasm. |
| JL022987 | 1956 | 651 | 74,820.48 | 7.57 | 39.5 | 99.69 | −0.28 | Cell membrane. Cytoplasm. |
| JL023790 | 2424 | 807 | 93,218.67 | 5.43 | 56.47 | 99.42 | −0.247 | Cytoplasm. |
| JL024456 | 3150 | 1049 | 119,420.27 | 7.99 | 40.32 | 102.76 | −0.202 | Cytoplasm. |
| JL024627 | 3216 | 1071 | 121,917.88 | 5.67 | 51.05 | 97.4 | −0.19 | Cell membrane. Cytoplasm. |
| JL024762 | 2871 | 956 | 110,390.18 | 6.76 | 51.67 | 95.65 | −0.292 | Cytoplasm. |
| JL024805 | 1527 | 508 | 57,241.5 | 8.67 | 59.54 | 95.08 | −0.267 | Cell membrane. Cytoplasm. |
| JL024806 | 756 | 251 | 28,129.13 | 8.86 | 44.4 | 105.62 | 0.063 | Cytoplasm. |
| JL024807 | 1980 | 659 | 74,718.97 | 7.19 | 49.51 | 99.59 | −0.113 | Cytoplasm. |
| JL024816 | 2322 | 773 | 87,897.62 | 6.71 | 52.46 | 105.27 | −0.182 | Cytoplasm. |
| JL025655 | 2943 | 980 | 113,173.42 | 6.47 | 50.31 | 95.3 | −0.252 | Cytoplasm. |
| JL025932 | 702 | 233 | 27,012.08 | 5.3 | 62.72 | 97.04 | −0.141 | Cytoplasm. Nucleus. |
| JL025933 | 972 | 323 | 37,748.46 | 5.37 | 61.94 | 96.56 | −0.218 | Cytoplasm. |
| JL027518 | 1524 | 507 | 57,848.73 | 8.9 | 44.18 | 106.77 | −0.084 | Cell membrane. Cytoplasm. |
| JL027520 | 1884 | 654 | 75,060.62 | 8.7 | 41.99 | 105.7 | −0.1 | Cell membrane. Cytoplasm. |
| JL028743 | 1470 | 489 | 56,510.41 | 5.7 | 47.08 | 85.75 | −0.504 | Chloroplast. Cytoplasm. |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yan, L.; Su, B.-X.; Li, J.-J.; Li, Y.-Y.; Chen, S.-Y.; Feng, C.-Y.; Tian, Y.; Ai, Y.; Zhang, Q.-H. Genome-Wide Identification of Nucleotide-Binding Site–Leucine-Rich Repeat Gene Family in Cymbidium ensifolium and Expression Profiles in Response to Fusarium Wilt Infection. Horticulturae 2024, 10, 634. https://doi.org/10.3390/horticulturae10060634
Yan L, Su B-X, Li J-J, Li Y-Y, Chen S-Y, Feng C-Y, Tian Y, Ai Y, Zhang Q-H. Genome-Wide Identification of Nucleotide-Binding Site–Leucine-Rich Repeat Gene Family in Cymbidium ensifolium and Expression Profiles in Response to Fusarium Wilt Infection. Horticulturae. 2024; 10(6):634. https://doi.org/10.3390/horticulturae10060634
Chicago/Turabian StyleYan, Lu, Bin-Xian Su, Jin-Jin Li, Yu-Yan Li, Shu-Yi Chen, Cai-Yun Feng, Yang Tian, Ye Ai, and Qing-Hua Zhang. 2024. "Genome-Wide Identification of Nucleotide-Binding Site–Leucine-Rich Repeat Gene Family in Cymbidium ensifolium and Expression Profiles in Response to Fusarium Wilt Infection" Horticulturae 10, no. 6: 634. https://doi.org/10.3390/horticulturae10060634
APA StyleYan, L., Su, B.-X., Li, J.-J., Li, Y.-Y., Chen, S.-Y., Feng, C.-Y., Tian, Y., Ai, Y., & Zhang, Q.-H. (2024). Genome-Wide Identification of Nucleotide-Binding Site–Leucine-Rich Repeat Gene Family in Cymbidium ensifolium and Expression Profiles in Response to Fusarium Wilt Infection. Horticulturae, 10(6), 634. https://doi.org/10.3390/horticulturae10060634






