Elucidating Softening Mechanism of Honey Peach (Prunus persica L.) Stored at Ambient Temperature Using Untargeted Metabolomics Based on Liquid Chromatography-Mass Spectrometry
Abstract
:1. Introduction
2. Materials and Methods
2.1. Analytical Standards and Reagents
2.2. Plant Materials and Treatments
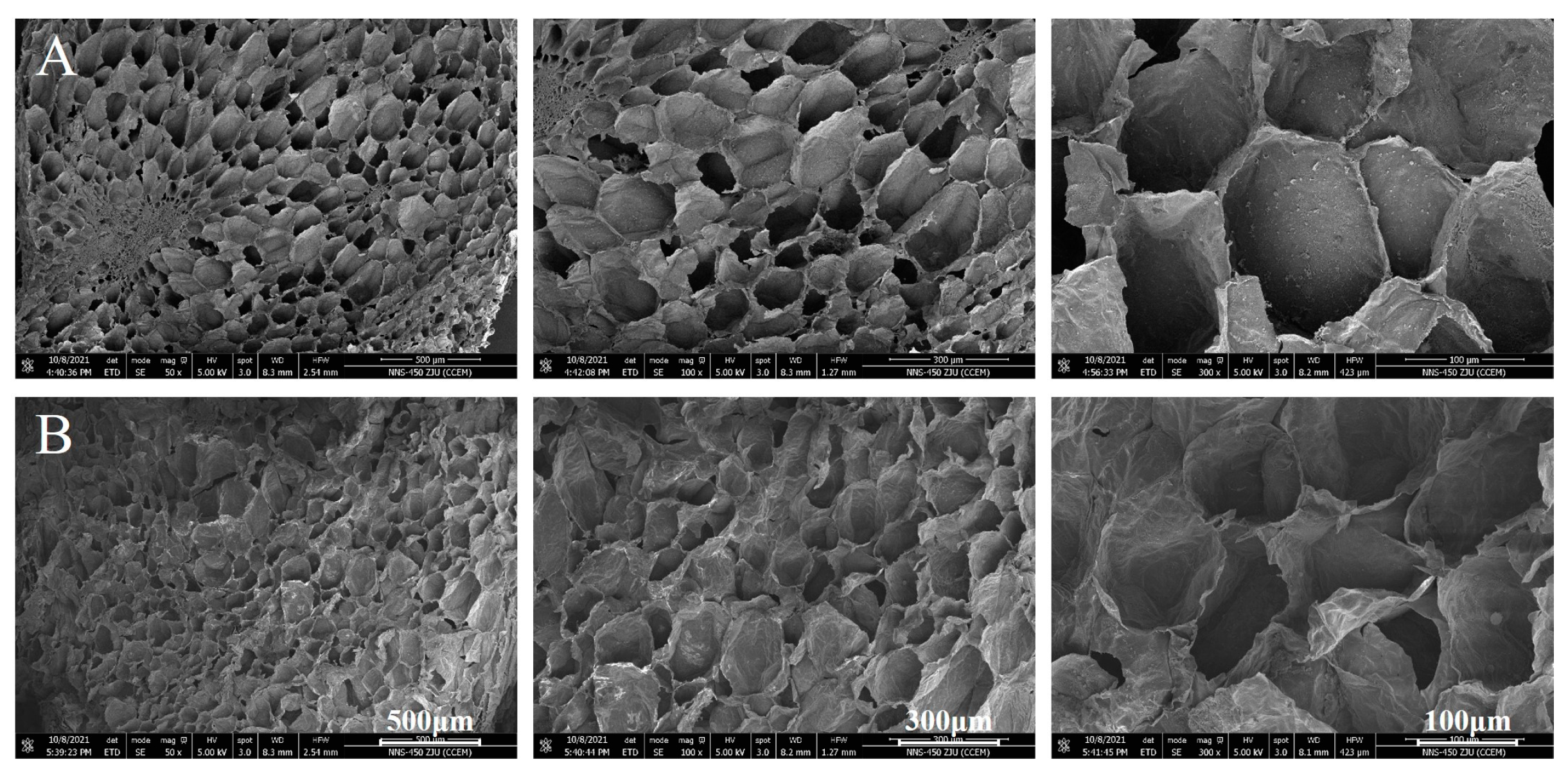
2.3. Visualization of the Ultrastructure
2.4. Sample Preparation for LC-MS
2.5. Ultra-High-Performance LC with Quadrupole Time-of-Flight Mass Spectrometry (UPLC-Q-TOF-MS)
2.6. Metabolome Data Analysis
3. Results
3.1. Cellular Ultrastructure of Peaches before and after Softening
3.2. Metabolite Identification
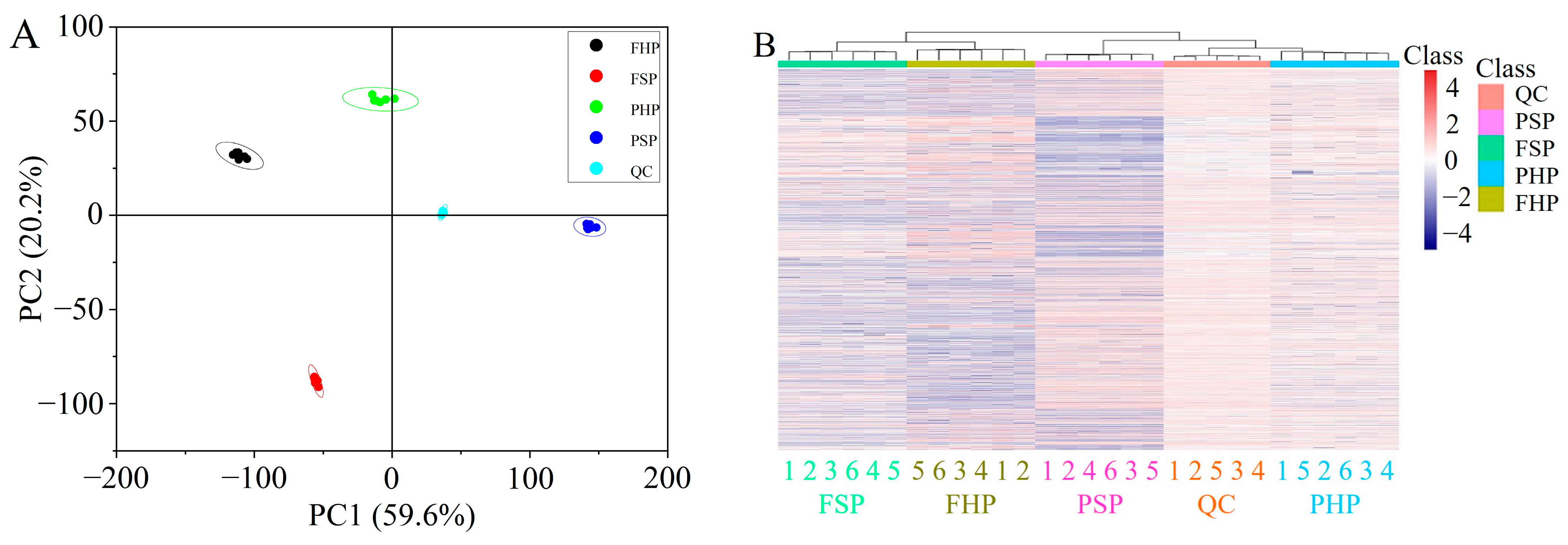
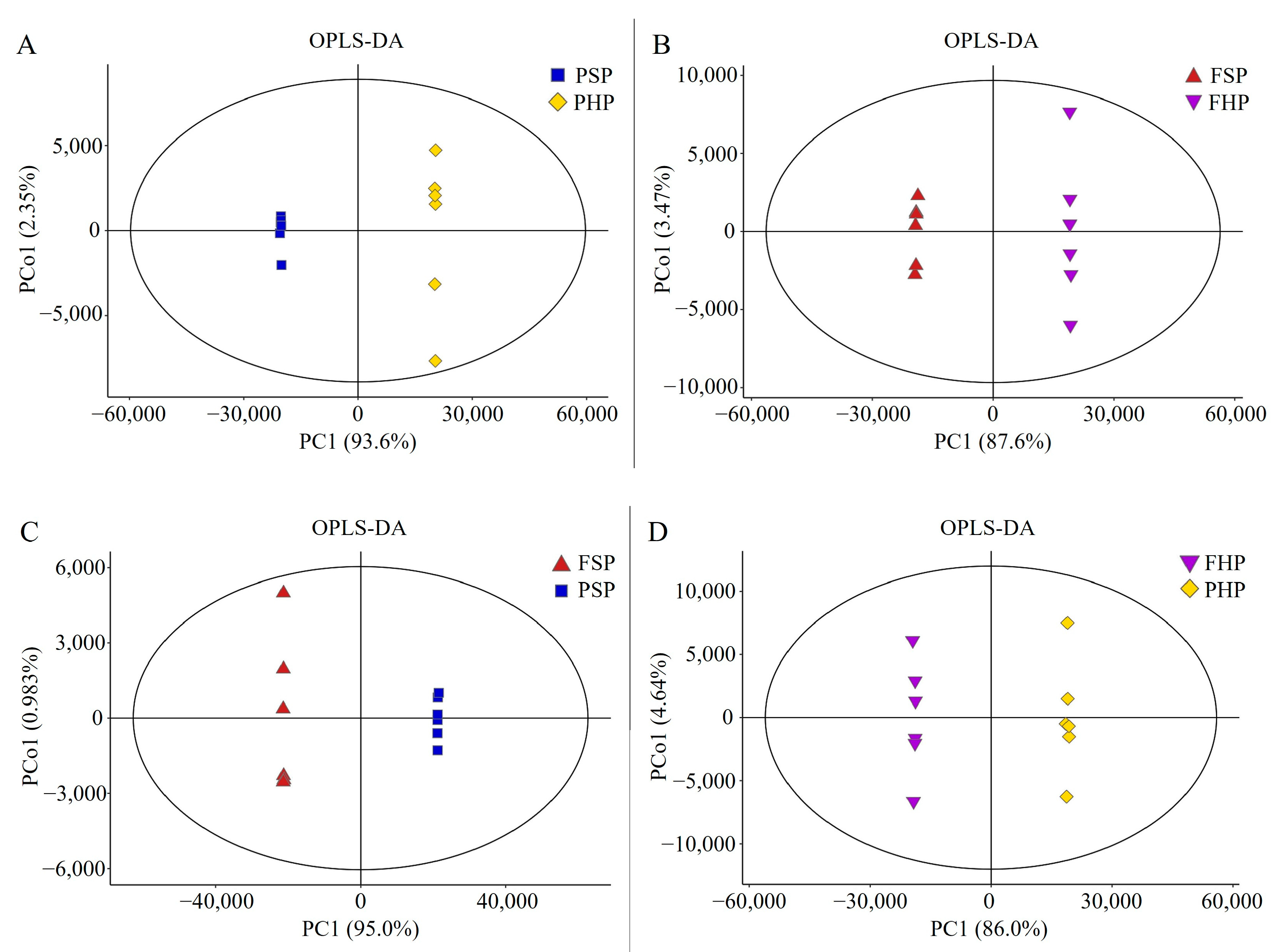
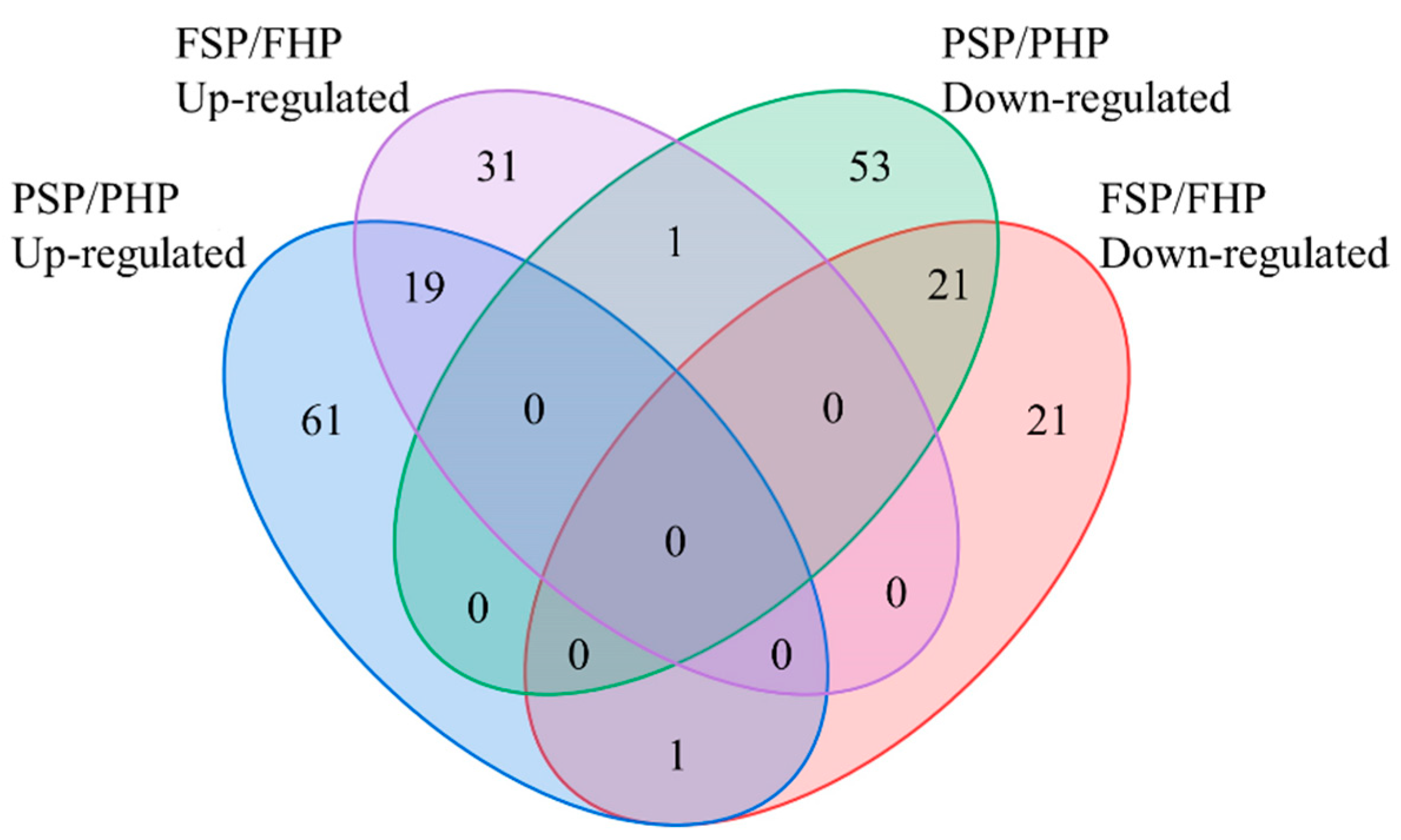
3.3. QC and Identification of Differential Metabolites
3.4. Hierarchical Clustering Analysis (HCA)
3.5. KEGG Annotation and Metabolic Pathway Analysis
4. Discussion
4.1. Degradation of Cell Wall Materials
4.2. Energy Metabolism
4.3. Oxidative Damage
4.4. Plant Hormone Regulation
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Rodriguez, C.A.; Bustamante, C.A.; Budde, C.O.; Müller, G.L.; Drincovich, M.F.; Lara, M.V. Peach fruit development: A comparative proteomic study between endocarp and mesocarp at very early stages underpins the main differential biochemical processes between these tissues. Front. Plant Sci. 2019, 10, 715–734. [Google Scholar] [CrossRef]
- Monti, L.L.; Bustamante, C.A.; Osorio, S.; Gabilondo, J.; Borsani, J.; Lauxmann, M.A.; Maulión, E.; Valentini, G.; Budde, C.O.; Fernie, A.R.; et al. Metabolic profiling of a range of peach fruit varieties reveals high metabolic diversity and commonalities and differences during ripening. Food Chem. 2016, 190, 879–888. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, L.; Zhang, X.P.; Mu, Q.; Tian, J.; Yan, J.; Guo, L.; Wang, Y.; Song, L.X.; Yu, X.Y. Differences in total phenolics, antioxidant activity and metabolic characteristics in peach fruits at different stages of ripening. LWT-Food Sci. Technol. 2023, 178, 114586. [Google Scholar] [CrossRef]
- Kan, J.; Liu, J.; Jin, C. Changes in cell walls during fruit ripening in Chinese ‘honey’ peach. J. Hortic. Sci. Biotech. 2013, 88, 37–46. [Google Scholar] [CrossRef]
- Oliveira, M.G.; Mazorra, L.M.; Souza, A.F.; Silva, G.M.C.; Correa, S.F.; Santos, W.C.; Saraiva, K.D.C.; Teixeira, A.J.; Melo, D.F.; Silva, M.G.; et al. The structure changes of water-soluble polysaccharides in papaya during ripening. Int. J. Biol. Macromol. 2018, 115, 152–156. [Google Scholar] [CrossRef]
- Oliveira, M.G.; Mazorra, L.M.; Souza, A.F.; Silva, G.M.C.; Correa, S.F.; Santos, W.C.; Saraiva, K.D.C.; Teixeira, A.J., Jr.; Melo, D.F.; Silva, M.G.; et al. Involvement of AOX and UCP pathways in the post-harvest ripening of papaya fruits. J. Plant Physiol. 2015, 189, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Feng, L.; Zhang, F.; Luo, H.; Yu, Z. Peach fruit ripening: Proteomic comparative analyses of two cultivars with different flesh texture phenotypes at two ripening stages. Sci. Hortic. 2020, 260, 108610. [Google Scholar] [CrossRef]
- Zhou, H.; Yu, Z.; Ye, Z. Key proteins associated to coloured compounds of peach peel using iTRAQ proteomic techniques during development and postharvest. Sci. Hortic. 2018, 239, 123–132. [Google Scholar] [CrossRef]
- Han, S.; Cai, H.; An, X.; Huan, C.; Wu, X.; Jiang, L.; Yu, M.; Ma, R.; Yu, Z. Effect of nitric oxide on sugar metabolism in peach fruit (cv. Xiahui 6) during cold storage. Postharvest Biol. Technol. 2018, 142, 72–80. [Google Scholar] [CrossRef]
- Huan, C.; Jiang, L.; An, X.; Yu, M.; Xu, Y.; Ma, R.; Yu, Z. Potential role of reactive oxygen species and antioxidant genes in the regulation of peach fruit development and ripening. Plant Physiol. Biochem. 2016, 104, 294–303. [Google Scholar] [CrossRef]
- Li, X.; Peng, S.; Yu, R.; Li, P.; Zhou, C.; Qu, Y.; Li, H.; Luo, H.; Yu, L. Co-application of 1-MCP and laser microporous plastic bag packaging maintains postharvest quality and extends the shelf-life of honey peach fruit. Foods 2022, 11, 1733. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jiang, L.; Shi, Y.; Luo, H.; Kang, R.; Yu, Z. Post-harvest 1-methylcyclopropene and ethephon treatments differently modify protein profiles of peach fruit during ripening. Food Res. Int. 2012, 48, 609–619. [Google Scholar] [CrossRef]
- Wani, A.B.; Chadar, H.; Wani, A.H.; Singh, S.; Upadhyay, N. Salicylic acid to decrease plant stress. Environ. Chem. Lett. 2017, 15, 101–123. [Google Scholar] [CrossRef]
- Li, Z.; Xu, X.; Yang, K.; Zhu, C.; Liu, Y.; Gao, Z. Multifaceted analyses reveal carbohydrate metabolism mainly affecting the quality of postharvest bamboo shoots. Front. Plant Sci. 2022, 13, 1021161. [Google Scholar] [CrossRef]
- Jiang, L.; Kang, R.; Feng, L.; Yu, Z.; Luo, H. iTRAQ-based quantitative proteomic analysis of peach fruit (Prunus persica L.) at different ripening and postharvest storage stages. Postharvest Biol. Technol. 2020, 164, 111137. [Google Scholar] [CrossRef]
- Rajeev, K.V.; Manish, K.P.; Annapurna, C. Metabolomics in plant stress physiology. In Plant Genetics and Molecular Biology; Springer: Cham, Switzerland, 2018; p. 164. ISBN 978-3-319-91312-4. [Google Scholar]
- Liu, J.; Zhang, X.; Tian, J.; Li, Y.; Liu, Q.; Chen, X.; Feng, F.; Yu, X.; Yang, C. Multiomics analysis reveals that peach gum colouring reflects plant defense responses against pathogenic fungi. Food Chem. 2022, 383, 132424. [Google Scholar] [CrossRef]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef]
- Dai, J.Y.; Xu, Z.; Xu, Y.T.; Fang, Z.H.; Shah, K.; Kang, T.Y.; Wu, H.X.; Zhang, D.; Xing, L.B.; Ma, J.J.; et al. A novel NAC transcription factor, PpNAP6, is involved in peach ripening by activating ethylene synthesis. Postharvest Biol. Technol. 2023, 201, 112363. [Google Scholar] [CrossRef]
- Sun, H.; Wang, H.; Zhang, A.; Yan, G.; Zhang, Y.; An, N.; Wang, X. Berberine ameliorates nonbacterial prostatitis via multi-target metabolic network regulation. OMICS 2015, 19, 186–195. [Google Scholar] [CrossRef]
- Zhang, M.; Li, Y.; Mu, Q.; Feng, F.; Yu, X.; Ge, J.; Zhang, Y.; Nie, J. Effects of chlorpyrifos on the metabolic profiling of Bacillus megaterium strain RRB. Chemosphere 2022, 297, 134189. [Google Scholar] [CrossRef]
- Fonseca, T.A.H.; Von, R.C.P.; Araújo, R.; Oliveira, M.C.; Justino, G.C.; Bento, L.; Calado, C.R.C. The impact of the serum extraction protocol on metabolomic profiling ysing UPLC-MS/MS and FTIR spectroscopy. ACS Omega 2023, 8, 20755–20766. [Google Scholar] [CrossRef] [PubMed]
- Emmanuel, A.; Yu, Y.; Chen, C.S.; Lu, L.; Hu, S.K.; Yu, H.P.; Ma, Q.Q.; Kuberan, T.; Rajiv, P.; Anburaj, J.; et al. Untargeted metabolomic analysis using UPLC-MS/MS identifies metabolites involved in shoot growth and development in pruned tea plants (Camellia sinensis (L.) O. Kuntz). Sci. Hortic. 2020, 264, 109164. [Google Scholar] [CrossRef]
- Luo, H.; Zhou, T.; Kong, X.; Tao, M.; Zhang, J.; Wang, W.; Jiang, L.; Yu, L.; Yu, Z. iTRAQ-based mitochondrial proteome analysis of the molecular mechanisms underlying postharvest senescence in Zizania latifolia. J. Food Biochem. 2019, 43, e13053. [Google Scholar] [CrossRef] [PubMed]
- Shi, J.; Xia, C.; Tian, Q.; Zeng, X.; Wu, Z.; Guo, Y.; Pan, D. Untargeted metabolomics based on LC-MS to elucidate the mechanism underlying nitrite degradation by Limosilactobacillus fermentum RC4. LWT-Food Sci. Technol. 2022, 163, 113414. [Google Scholar] [CrossRef]
- Fabrizio, C.; Luca, C.; Marco, F.; Sara, L.; Walter, G.; Pierluigi, M.; Flavia, G.; Franco, B. Texture dynamics during postharvest cold storage ripening in apple (Malus × domestica Borkh.). Postharvest Biol. Technol. 2012, 69, 54–63. [Google Scholar] [CrossRef]
- Wang, S.Y.; Zhou, Q.; Zhou, X.; Zhang, F.; Ji, S.J. Ethylene plays an important role in the softening and sucrose metabolism of blueberries postharvest. Food Chem. 2020, 310, 125965. [Google Scholar] [CrossRef]
- Liu, B.; Wang, K.; Shu, X.; Liang, J.; Fan, X.; Sun, L. Changes in fruit firmness, quality traits and cell wall constituents of two highbush blueberries (Vaccinium corymbosum L.) during postharvest cold storage. Sci. Hortic. 2019, 246, 557–562. [Google Scholar] [CrossRef]
- Zhang, Q.; Tang, F.; Cai, W.; Peng, B.; Ning, M.; Shan, C.; Yang, X. Chitosan treatment reduces softening and chilling injury in cold-stored Hami melon by regulating starch and sucrose metabolism. Front. Plant Sci. 2022, 13, 1096017. [Google Scholar] [CrossRef]
- Chen, Y.; Zhu, C.C.; Zhao, Y.Q.; Zhang, S.J.; Wang, W. Transcriptomics integrated with changes in cell wall material of chestnut (Castanea mollissima Blume) during storage provides a new insight into the “calcification” process. Foods 2022, 11, 1136. [Google Scholar] [CrossRef]
- Favre, L.; Hunter, D.A.; Erridge, Z.A.; Napier, N.J.; Punter, M.; Carr, B.; Tattersall, A.; Johnston, J.W.; Heyes, J.A.; Lill, R.E.; et al. Seasonal differences in softening of early-harvested ‘Royal Gala’ apple fruit are correlated with at-harvest biomarkers indicative of abiotic stress responses. Postharvest Biol. Technol. 2023, 195, 112131. [Google Scholar] [CrossRef]
- Ivan, O.D.; Irene, R.; Dolores, M.A.; Tarradas, R.; Sanchez-Ballesta, M.T.; Escribano, M.I.; Merodio, C. Transcriptomic analysis of CO2-treated strawberries (Fragaria vesca) with enhanced resistance to softening and oxidative stress at consumption. Front. Plant Sci. 2022, 13, 983976. [Google Scholar] [CrossRef]
- Coletta, C.; Botondi, R.; Forniti, R.; Baccelloni, S.; Bellincontro, A.; Mencarelli, F. Alternating temperature in postharvest cooling treatment of ‘Fiano’ and ‘Falanghina’ grapes affects cell wall enzyme rate, berry softening and polyphenols. J. Sci. Food Agric. 2018, 99, 3142–3148. [Google Scholar] [CrossRef]
- Xiang, F.X.; Gao, R.; Chen, Y.; Pang, J.W.; Liu, S.S.; Tian, L.; Zhai, R.; Wang, Z.G.; Xu, L.F. Exogenous putrescine and 1-methylcyclopropene prevent soft scald in ‘Starkrimson’ pear. Postharvest Biol. Technol. 2022, 193, 112035. [Google Scholar] [CrossRef]
- Yao, S.; Cao, Q.; Xie, J.; Deng, L.; Zeng, K. Alteration of sugar and organic acid metabolism in postharvest granulation of Ponkan fruit revealed by transcriptome profiling. Postharvest Biol. Technol. 2018, 139, 2–11. [Google Scholar] [CrossRef]
- Li, D.; Li, L.; Xu, Y.Q.; Wang, L.; Lin, X.Y.; Wang, Y.S.; Luo, Z. Exogenous ATP attenuated fermentative metabolism in postharvest strawberry fruit under elevated CO2 atmosphere by maintaining energy status. Postharvest Biol. Technol. 2021, 182, 111701. [Google Scholar] [CrossRef]
- Aghdam, M.S.; Jannatizadeh, A.; Luo, Z.; Paliyath, G. Ensuring sufficient intracellular ATP supplying and friendly extracellular ATP signaling attenuates stresses, delays senescence and maintains quality in horticultural crops during postharvest life. Trends Food Sci. Technol. 2018, 78, 67–81. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, C.; Chen, C.; Zhang, S.; Zhao, X.; Wu, C.; Kou, X.; Xue, Z. Glycine betaine inhibits postharvest softening and quality decline of winter jujube fruit by regulating energy and antioxidant metabolism. Food Chem. 2023, 410, 135445. [Google Scholar] [CrossRef]
- Chen, H.; Lai, X.; Wang, L.; Li, X.; Chen, W.; Zhu, X.; Song, Z. Ethylene response factor MaERF012 modulates fruit ripening by regulating chlorophyll degradation and softening in banana. Foods 2022, 11, 3882. [Google Scholar] [CrossRef]
- Shan, Y.; Li, F.; Lian, Q.; Xie, L.; Zhu, H.; Li, T.; Zhang, J.; Duan, X.; Jiang, Y. Role of apyrase-mediated eATP signal in chilling injury of postharvest banana fruit during storage. Postharvest Biol. Technol. 2022, 187, 111874. [Google Scholar] [CrossRef]
- Zhang, Y.; Guo, Y.; Song, X. Comprehensive insight into an amino acid metabolic network in postharvest horticultural products: A review. J. Sci. Food Agric. 2023, 103, 5667–5676. [Google Scholar] [CrossRef]
- Lefèvre, P.L.C.; Palin, M.F.; Murphy, B.D. Polyamines on the reproductive landscape. Endocr. Rev. 2011, 32, 694–712. [Google Scholar] [CrossRef] [PubMed]
- Kesarwani, P.; Kant, S.; Prabhu, A.; Chinnaiyan, P. The interplay between metabolic remodeling and immune regulation in glioblastoma. Neuro-Oncology 2017, 19, 1308–1315. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.N.; Li, B.; Sun, Q.; Guo, P.R.; Du, Y.J.; Huang, J.F. Acetolactate synthases regulatory subunit and catalytic subunit genes VdILVs are involved in BCAA biosynthesis, microscletotial and conidial formation and virulence in Verticillium dahlia. Fungal Genet. Biol. 2022, 159, 103667. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.W.; Dissing, M.M.; Agerbirk, N.; Crocoll, C.; Halkier, B.A. Characterization of Arabidopsis CYP79C1 and CYP79C2 by glucosinolate pathway engineering in Nicotiana benthamiana shows substrate specificity toward a range of aliphatic and aromatic amino acids. Front. Plant Sci. 2020, 11, 57–64. [Google Scholar] [CrossRef] [PubMed]
- Kan, J.; Wang, H.M.; Jin, C.H.; Xie, H. Changes of reactive oxygen species and related enzymes in mitochondria respiratory metabolism during the ripening of peach fruit. Agric. Sci. China 2010, 9, 138–146. [Google Scholar] [CrossRef]
- Meitha, K.; Pramesti, Y.; Suhandono, S. Reactive oxygen species and antioxidants in postharvest vegetables and fruits. Int. J. Food Sci. 2020, 2020, 8817778. [Google Scholar] [CrossRef]
- Diaz, E.R.; McCullough, L.D.; Tsvetkov, A.S. The functional role of sphingosine kinase 2. Front. Mol. Biosci. 2021, 8, 683767. [Google Scholar] [CrossRef]
- Toshikazu, H.; Toshihiko, S.; Shigeo, K.; Takamitsu, S.; Takayuki, K.; Yasuyuki, I. Synthesis of fluorescence-Labeled sphingosine and sphingosine 1-phosphate; effective tools for sphingosine and sphingosine 1-phosphate behavior. Bioorg. Med. Chem. Lett. 2003, 13, 661–664. [Google Scholar] [CrossRef]
- Dong, B.; Yao, Q.; Zhu, D.; Han, H.; Tang, H.; Ding, X. Exogenous melatonin maintains quality of postharvest Rosa roxburghii fruitby modulating reactive oxygen species metabolism and energy status. Sci. Hortic. 2022, 304, 111346. [Google Scholar] [CrossRef]
- Wang, C.; Huang, D.; Tian, W.; Zhu, S. Nitric oxide alleviates mitochondrial oxidative damage and maintains mitochondrial functions in peach fruit during cold storage. Sci. Hortic. 2021, 287, 110249. [Google Scholar] [CrossRef]
- Romero, I.; Escribano, M.I.; Merodio, C.; Sanchez, B.M.T. Postharvest high-CO2 treatments on the quality of soft fruit berries: An integrated transcriptomic, proteomic, and metabolomic approach. J. Agric. Food Chem. 2022, 70, 8593–8597. [Google Scholar] [CrossRef] [PubMed]
- Delphine, M.P.; Vallarino, J.G.; Osorio, S. Review: Metabolite changes during postharvest storage: Effects on fruit quality traits. Metabolites 2020, 187, 10050187. [Google Scholar] [CrossRef]
- Springsteen, G.; Yerabolu, J.R.; Nelson, J.; Rhea, C.J.; Krishnamurthy, R. Linked cycles of oxidative decarboxylation of glyoxylate as protometabolic analogs of the citric acid cycle. Nat. Commun. 2018, 9, 91–100. [Google Scholar] [CrossRef]
- Alves, V.E. Exogenous abscisic acid (ABA) and jasmonate (JA) promote metabolic regulation in Jacarandá-Pardo (Machaerium villosum Vog.) seedlings under PEG-induced water deficit. Plant Stress 2023, 9, 100174. [Google Scholar] [CrossRef]
- Elisabeth, P.; Olivier, R.; Claudie, R.; Stéphanie, B.M.; Alessandra, M.G.; Limami, A.M. Nitrogen metabolism responses to water deficit act through both abscisic acid (ABA)-dependent and independent pathways in Medicago truncatula during post-germination. J. Exp. Bot. 2011, 62, 605–615. [Google Scholar] [CrossRef]
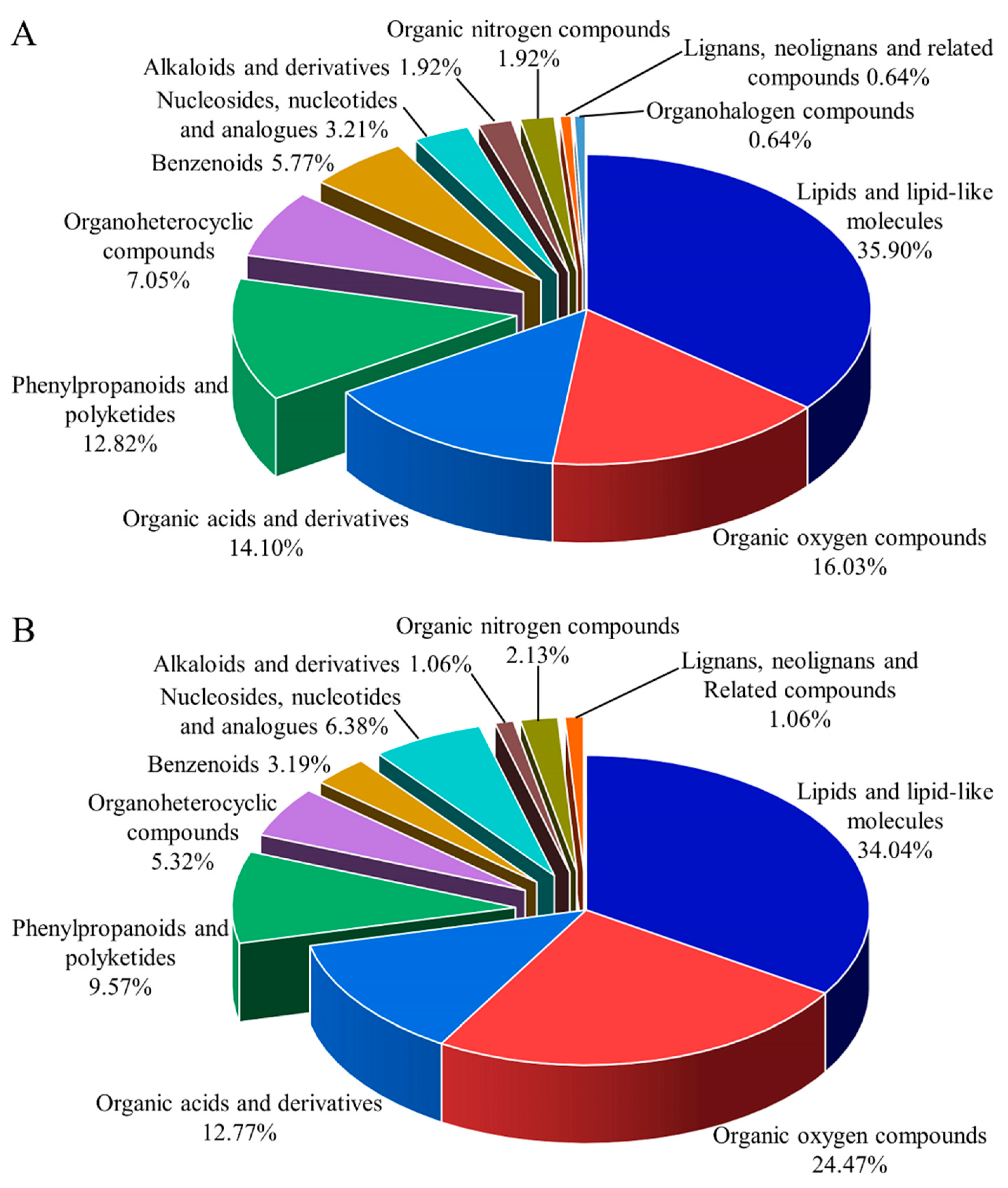
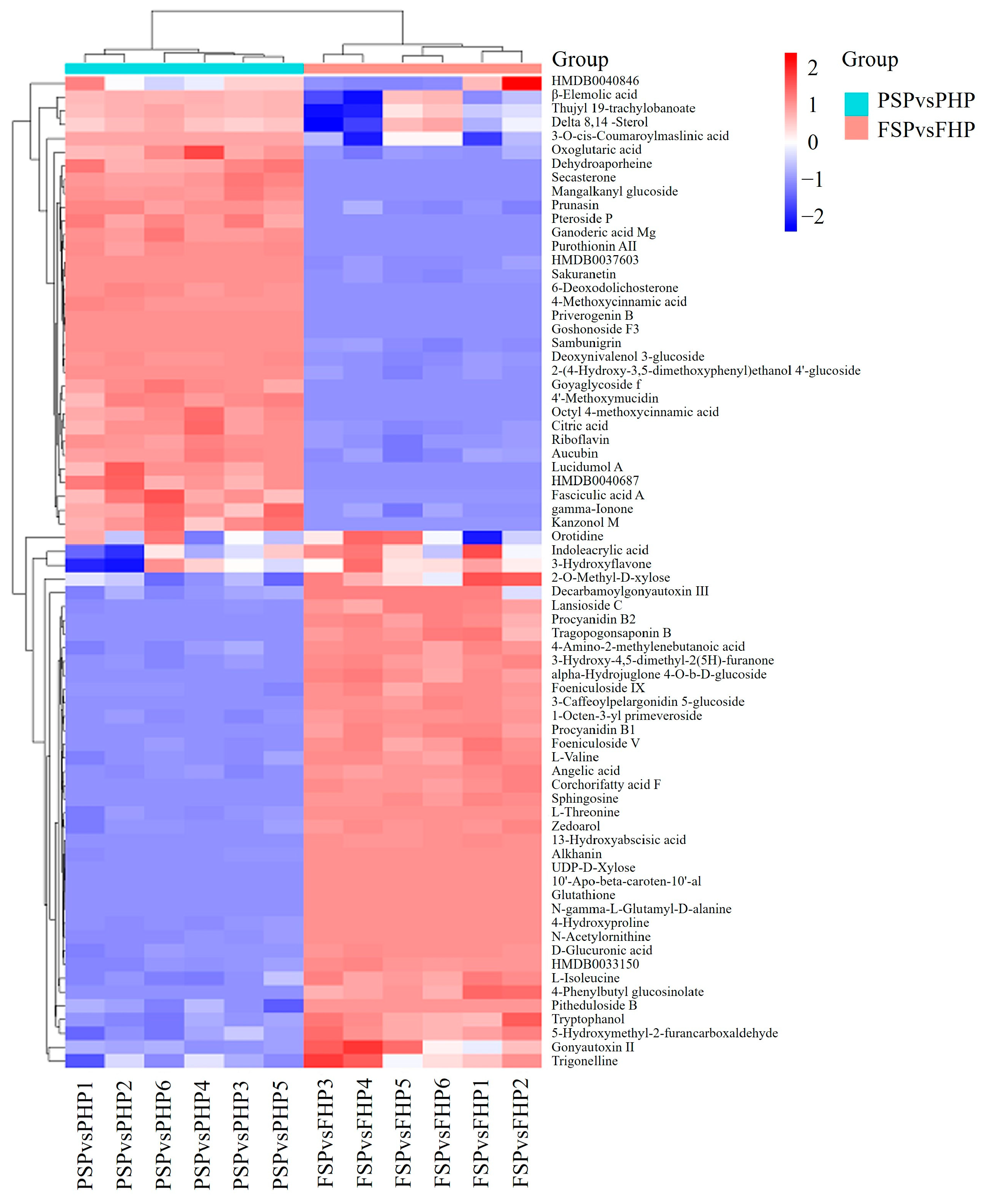
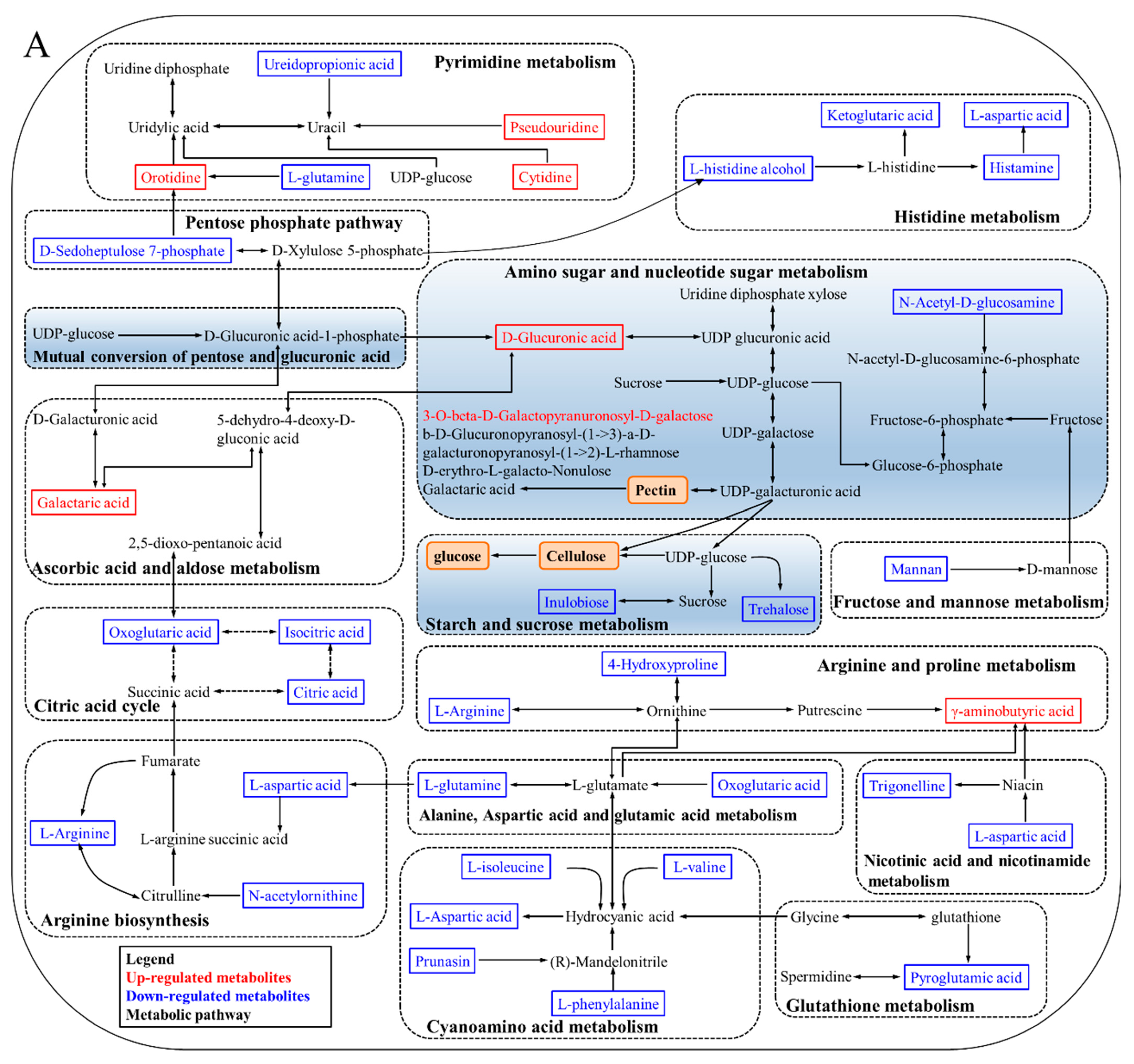
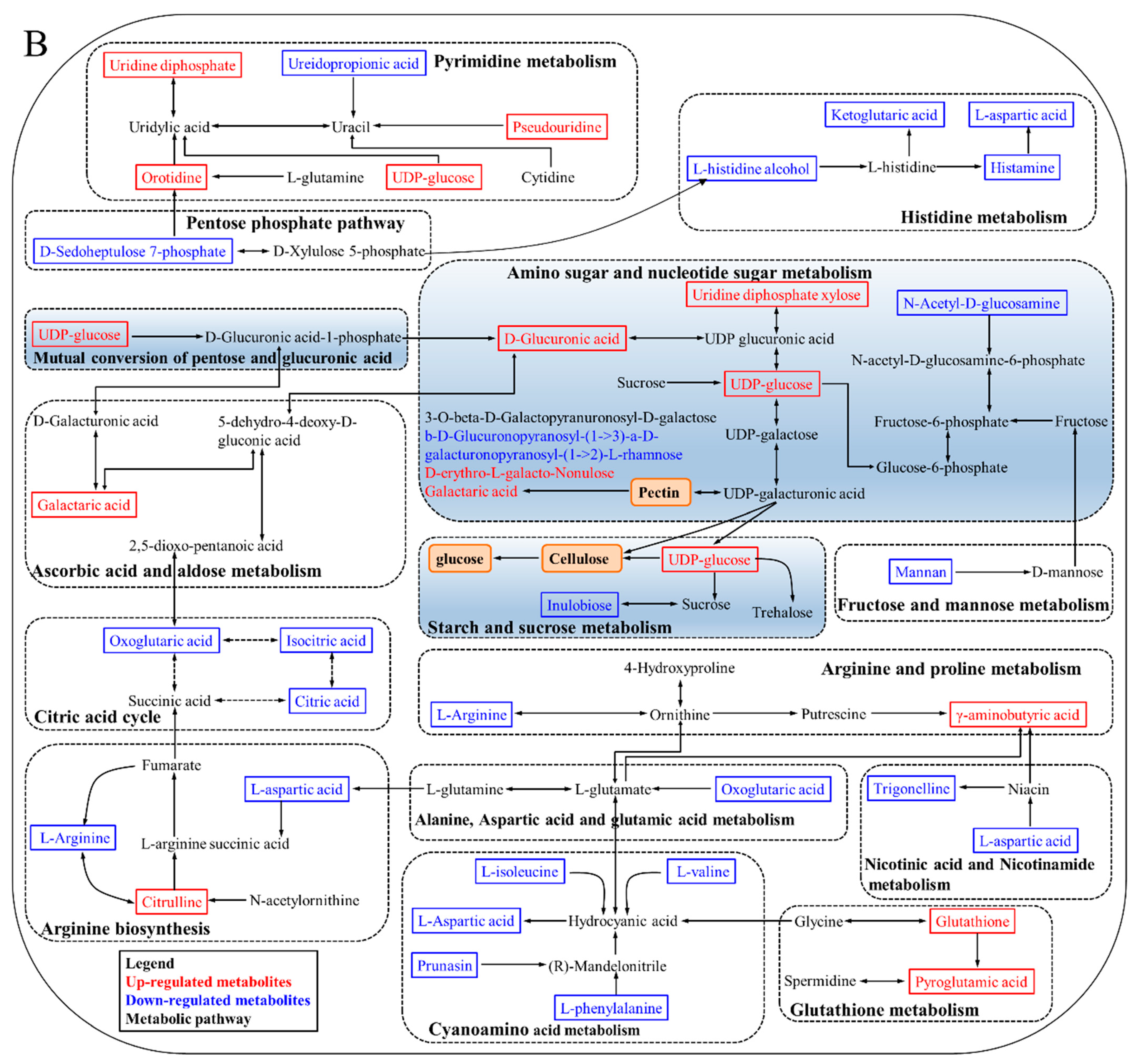
| No. | ID | m/z | Retention Time (min) | Ion Mode | Metabolites | Compound ID | PSP/PHP | FSP/FHP |
|---|---|---|---|---|---|---|---|---|
| Alkaloids and derivatives | ||||||||
| 1 | 5.03_553.2138m/z | 553.2138 | 5.0349167 | neg | Dehydroaporheine | HMDB0033355 | 5.5528348 | |
| 2 | 0.91_675.0976m/z | 675.09764 | 0.9134167 | neg | Prebetanin | HMDB0029411 | −1.1931527 | −0.2097159 |
| 3 | 0.82_137.0476n | 160.0368 | 0.8212167 | pos | Trigonelline | HMDB0000875 | −6.296828 | −3.4098136 |
| Benzenoids | ||||||||
| 4 | 12.05_333.1354m/z | 333.13543 | 12.045667 | neg | 4′-Methoxymucidin | HMDB0030019 | 3.3352022 | |
| 5 | 12.59_501.2238m/z | 501.2238 | 12.591167 | neg | Purothionin AII | HMDB0039001 | 2.8072538 | |
| 6 | 10.98_292.2037n | 293.21085 | 10.978267 | pos | [7]-Paradol | HMDB0040806 | 2.5051899 | |
| 7 | 5.16_437.2030m/z | 437.20305 | 5.15925 | neg | N-Phenyl-2-naphthylamine | HMDB0032865 | 2.0844425 | 2.2331908 |
| 8 | 5.03_524.1345m/z | 524.13452 | 5.0319 | pos | Protohypericin | HMDB0034180 | −1.2897264 | |
| 9 | 15.27_150.1277m/z | 150.12768 | 15.2738 | pos | p-Mentha-1,3,5,8-tetraene | HMDB0029641 | −1.422287 | −0.6103573 |
| 10 | 0.91_521.1087m/z | 521.10873 | 0.9064 | pos | Isomelitric acid A | HMDB0039523 | −1.580731 | −0.6180271 |
| 11 | 4.72_493.1289m/z | 493.12889 | 4.7210667 | pos | Palmidin A | HMDB0034038 | −1.5901031 | −1.6091476 |
| 12 | 1.83_278.1516n | 301.14084 | 1.83035 | pos | Dibutyl phthalate | HMDB0033244 | −1.5974966 | −0.7392945 |
| 13 | 4.72_583.1255m/z | 583.12551 | 4.7247833 | neg | Rheidin C | HMDB0038508 | −1.2387168 | |
| Lignans, neolignans, and related compounds | ||||||||
| 14 | 5.39_522.2105n | 567.20854 | 5.3948167 | neg | Isolariciresinol 4′-O-beta-D-glucoside | HMDB0040471 | 1.220256 | |
| 15 | 5.18_567.2084m/z | 567.20838 | 5.1766 | neg | Isolariciresinol 9-O-beta-D-glucoside | HMDB0032907 | 0.793055 | −1.1147439 |
| Lipids and lipid-like molecules | ||||||||
| 16 | 15.26_474.3706n | 497.35981 | 15.256067 | pos | Priverogenin B | HMDB0034644 | 37.261839 | |
| 17 | 12.40_781.4695m/z | 781.46946 | 12.4023 | pos | Goyaglycoside f | HMDB0037124 | 15.036405 | |
| 18 | 15.10_472.3550n | 495.34426 | 15.096133 | pos | Lucidumol A | HMDB0033233 | 8.9189368 | |
| 19 | 14.51_446.3394n | 469.32866 | 14.510817 | pos | Secasterone | HMDB0040999 | 5.2741667 | |
| 20 | 15.13_448.3551n | 471.3443 | 15.131617 | pos | 6-Deoxodolichosterone | HMDB0034332 | 5.1783877 | |
| 21 | 14.07_643.4173m/z | 643.41728 | 14.06755 | pos | Fasciculic acid A | HMDB0036439 | 3.6356945 | |
| 22 | 13.89_585.3757m/z | 585.3757 | 13.894517 | pos | Ganoderic acid Mg | HMDB0035999 | 2.9360617 | |
| 23 | 5.31_192.1514n | 175.14811 | 5.3125 | pos | gamma-Ionone | HMDB0034979 | 2.7246443 | 2.3180287 |
| 24 | 8.73_518.3244n | 563.32257 | 8.7302167 | neg | Ganolucidic acid C | HMDB0039691 | 2.462305 | |
| 25 | 14.21_508.3764n | 531.36563 | 14.208733 | pos | Fasciculol C | HMDB0035853 | 2.2688443 | |
| 26 | 9.66_535.2879m/z | 535.28789 | 9.6555167 | pos | Corchoroside A | HMDB0033846 | 2.0353167 | |
| 27 | 11.96_633.3968m/z | 633.39684 | 11.955867 | pos | Calenduloside E | HMDB0040851 | 1.9493374 | |
| 28 | 8.28_518.3234n | 563.32252 | 8.2771333 | neg | Ganoderic acid C2 | HMDB0035304 | 1.9198447 | |
| 29 | 5.16_415.1936m/z | 415.19356 | 5.1589 | pos | S-Furanopetasitin | HMDB0036131 | 1.852255 | 1.1850532 |
| 30 | 5.50_415.1975m/z | 415.1975 | 5.5021833 | neg | (3S,7E,9S)-9-Hydroxy-4,7-megastigmadien-3-one 9-glucoside | HMDB0036822 | 1.7683917 | 1.4457104 |
| 31 | 5.29_377.1817m/z | 377.18167 | 5.2873333 | neg | 6Z-8-Hydroxygeraniol 8-O-glucoside | HMDB0035025 | 1.7644214 | 1.3697986 |
| 32 | 5.58_373.1868m/z | 373.1868 | 5.5755667 | neg | 6-Epi-7-isocucurbic acid glucoside | HMDB0029782 | 1.755631 | |
| 33 | 4.74_379.1610m/z | 379.161 | 4.7429333 | neg | Prenyl arabinosyl-(1->6)-glucoside | HMDB0041360 | 1.6820333 | |
| 34 | 5.47_282.1467n | 281.13938 | 5.4670833 | neg | Epidihydrophaseic acid | HMDB0038661 | 1.6585013 | 2.5180264 |
| 35 | 11.35_294.2193n | 295.22654 | 11.354317 | pos | 2-Hydroxylinolenic acid | HMDB0031103 | 1.6570269 | −1.0014807 |
| 36 | 9.15_502.3297n | 547.32789 | 9.1458 | neg | Ganolucidic acid B | HMDB0035751 | 1.6085416 | |
| 37 | 5.41_441.1978m/z | 441.1978 | 5.4130167 | neg | 1-Hexanol arabinosylglucoside | HMDB0031689 | 1.6013305 | |
| 38 | 5.36_471.1872m/z | 471.18718 | 5.35895 | neg | 11,13-Dihydrotaraxinic acid glucosyl ester | HMDB0035867 | 1.5551523 | 1.0746529 |
| 39 | 8.75_500.3135n | 483.3102 | 8.7520667 | pos | Ganolucidic acid A | HMDB0035302 | 1.5458326 | |
| 40 | 7.71_695.4014m/z | 695.40141 | 7.7110833 | neg | Momordicoside E | HMDB0035697 | 1.531432 | |
| 41 | 5.36_433.2079m/z | 433.20795 | 5.35895 | neg | Dihydroroseoside | HMDB0040614 | 1.4165233 | |
| 42 | 9.55_502.3292n | 503.33642 | 9.5534167 | pos | Medicagenic acid | HMDB0034551 | 1.3474913 | |
| 43 | 5.39_194.1670n | 177.16372 | 5.3880333 | pos | 5-Isopropyl-2-(2-methylpropyl)-2-cyclohexen-1-one | HMDB0038216 | 1.3416272 | |
| 44 | 9.57_410.3181n | 433.30983 | 9.5741 | pos | (6alpha,22E)-6-Hydroxy-4,7,22-ergostatrien-3-one | HMDB0037380 | 1.3000743 | |
| 45 | 5.02_433.2080m/z | 433.20803 | 5.0172833 | neg | 9,13-Dihydroxy-4-megastigmen-3-one 9-glucoside | HMDB0036318 | 1.2650463 | |
| 46 | 5.34_393.1768m/z | 393.17677 | 5.3405333 | neg | Nepetariaside | HMDB0039014 | 1.2443563 | 0.5806051 |
| 47 | 4.12_451.2187m/z | 451.21868 | 4.12275 | neg | Kiwiionoside | HMDB0038691 | 1.2365472 | |
| 48 | 5.09_427.1938m/z | 427.19381 | 5.0859667 | pos | Pisumionoside | HMDB0039947 | 1.2300592 | |
| 49 | 4.91_282.1466n | 281.13946 | 4.9062833 | neg | Pisumic acid | HMDB0039241 | 1.1927666 | 2.1976093 |
| 50 | 5.29_332.1832n | 355.17243 | 5.2941167 | pos | (2E,4E,7R)-2,7-Dimethyl-2,4-octadiene-1,8-diol 8-O-b-D-glucopyranoside | HMDB0038747 | 1.16536 | 0.727943 |
| 51 | 5.19_439.1822m/z | 439.18219 | 5.1942167 | neg | cis-3-Hexenyl b-primeveroside | HMDB0031690 | 1.1648099 | |
| 52 | 4.85_386.1940n | 431.19223 | 4.8532667 | neg | Citroside A | HMDB0030370 | 1.1464168 | 0.5008677 |
| 53 | 9.57_468.3237n | 469.33097 | 9.5741 | pos | Uralenolide | HMDB0038797 | 1.1359664 | |
| 54 | 9.15_502.3291n | 503.33641 | 9.14545 | pos | Esculentic acid (Phytolacca) | HMDB0034639 | 1.119195 | |
| 55 | 5.03_348.1781n | 371.16731 | 5.0319 | pos | Foeniculoside V | HMDB0034874 | 1.1036398 | 2.628105 |
| 56 | 5.95_421.2081m/z | 421.20813 | 5.94505 | neg | 1-Octen-3-yl primeveroside | HMDB0032960 | 1.0953733 | 2.8884386 |
| 57 | 4.80_433.2080m/z | 433.20798 | 4.7979 | neg | Icariside B8 | HMDB0036846 | 1.0514936 | |
| 58 | 5.48_280.1311n | 279.1238 | 5.4847667 | neg | Nigellic acid | HMDB0036094 | 1.0289409 | 1.9319225 |
| 59 | 10.99_679.3853m/z | 679.38531 | 10.9895 | neg | 2alpha-Hydroxypyracrenic acid | HMDB0029780 | 1.0173618 | |
| 60 | 4.91_264.1360n | 265.14329 | 4.90995 | pos | 3-Epiarmefolin | HMDB0036135 | 0.6841163 | 1.4082933 |
| 61 | 11.35_454.3443n | 455.35163 | 11.354317 | pos | Ursonic acid | HMDB0036007 | −0.4947675 | −1.8832459 |
| 62 | 5.57_458.1786n | 481.16784 | 5.56605 | pos | Deoxynivalenol 3-glucoside | HMDB0039852 | −0.5041028 | −3.9033631 |
| 63 | 11.34_473.3624m/z | 473.36241 | 11.336667 | pos | 27-Hydroxyisomangiferolic acid | HMDB0036064 | −0.6364165 | −1.9997725 |
| 64 | 0.81_344.1316n | 389.12984 | 0.81115 | neg | Lactitol | HMDB0040937 | −0.8733643 | −1.2932778 |
| 65 | 12.98_438.3496n | 439.35686 | 12.97695 | pos | Thujyl 19-trachylobanoate | HMDB0036840 | −0.9976602 | −2.3387444 |
| 66 | 0.79_207.0503m/z | 207.05031 | 0.7941333 | neg | 3-Hydroxymethylglutaric acid | HMDB0000355 | −1.032756 | −0.6809323 |
| 67 | 2.14_346.1261n | 369.11535 | 2.1447 | pos | Aucubin | HMDB0036562 | −1.057286 | −2.7078305 |
| 68 | 12.99_457.3672m/z | 457.36724 | 12.99425 | pos | beta-Elemolic acid | HMDB0034961 | −1.2980053 | −3.2925127 |
| 69 | 13.01_410.3545n | 411.36175 | 13.011333 | pos | Delta 8,14 -Sterol | HMDB0006928 | −1.3317368 | −2.134609 |
| 70 | 6.98_292.1883n | 315.17763 | 6.9816333 | pos | (S)-3-Octanol glucoside | HMDB0032958 | −1.3828306 | −0.5888407 |
| 71 | 11.39_277.1797m/z | 277.1797 | 11.38935 | pos | Phytuberin | HMDB0035754 | −1.4766195 | −0.5807962 |
| 72 | 14.14_310.3102m/z | 310.31019 | 14.137933 | pos | Geranylcitronellol | HMDB0032147 | −1.5039805 | |
| 73 | 1.13_118.0865m/z | 118.08646 | 1.1279 | pos | Angelic acid | HMDB0029608 | −2.0222781 | −0.4844698 |
| 74 | 7.07_414.2252n | 437.21441 | 7.0656833 | pos | (4R,5S,7R,11S)-11,12-Dihydroxy-1(10)-spirovetiven-2-one 11-glucoside | HMDB0033150 | −2.3741167 | −0.7120234 |
| 75 | 6.47_264.1362n | 263.12892 | 6.4695167 | neg | Alkhanin | HMDB0036202 | −2.8303188 | |
| 76 | 6.48_246.1255n | 247.13273 | 6.4793333 | pos | Zedoarol | HMDB0038202 | −3.1160144 | −1.5618091 |
| 77 | 13.03_883.5013m/z | 883.50126 | 13.028717 | pos | Pitheduloside B | HMDB0034865 | −5.0778196 | |
| 78 | 14.99_377.2835m/z | 377.2835 | 14.989867 | pos | 10′-Apo-beta-caroten-10′-al | HMDB0036887 | 35.192181 | |
| 79 | 7.28_327.2176m/z | 327.21764 | 7.2776667 | neg | Corchorifatty acid F | HMDB0035919 | 4.5032417 | |
| 80 | 4.54_926.4697n | 927.47698 | 4.54215 | pos | Tragopogonsaponin B | HMDB0037911 | 3.7664956 | |
| 81 | 14.81_395.3670m/z | 395.36697 | 14.812417 | pos | Stigmasterol | HMDB0000937 | 3.149023 | |
| 82 | 6.00_280.1311n | 279.12379 | 6.00005 | neg | 13-Hydroxyabscisic acid | HMDB0036095 | 3.0563838 | |
| 83 | 7.73_329.2334m/z | 329.23337 | 7.7299667 | neg | 9,10,13-TriHOME | HMDB0004710 | 2.4227629 | |
| 84 | 5.95_197.1536m/z | 197.15357 | 5.9453333 | pos | alpha-Terpineol acetate | HMDB0032051 | 1.9997694 | |
| 85 | 5.48_280.1311n | 279.1238 | 5.4847667 | neg | Nigellic acid | HMDB0036094 | 1.932 | |
| 86 | 5.49_280.1309n | 263.12764 | 5.4853167 | pos | Crispolide | HMDB0036695 | 1.3681446 | |
| 87 | 12.19_618.3915n | 619.39874 | 12.190483 | pos | 3-O-cis-Coumaroylmaslinic acid | HMDB0034539 | −4.4936192 | |
| 88 | 8.41_644.3399n | 667.32928 | 8.40645 | pos | Goshonoside F3 | HMDB0038376 | −35.34355 | |
| Nucleosides, nucleotides, and analogues | ||||||||
| 89 | 0.79_575.1100m/z | 575.10997 | 0.7941333 | neg | Orotidine | HMDB0000788 | 36.14114 | 36.162397 |
| 90 | 5.58_485.1643m/z | 485.16426 | 5.5755667 | neg | Cytidine | HMDB0000089 | 1.7393751 | |
| 91 | 0.84_244.0926m/z | 244.09257 | 0.8382333 | pos | Cytarabine | HMDB0015122 | 1.3531597 | |
| 92 | 1.19_244.0693n | 243.06197 | 1.1876833 | neg | Pseudouridine | HMDB0000767 | 1.3475125 | 2.4172629 |
| 93 | 1.98_267.0722m/z | 267.07222 | 1.9836833 | neg | Inosine | HMDB0000195 | −1.5091056 | |
| 94 | 0.81_535.0369m/z | 535.0369 | 0.81115 | neg | UDP-D-Xylose | HMDB0001018 | 35.862012 | |
| 95 | 0.82_405.0089m/z | 405.0089 | 0.8212167 | pos | Uridine 5′-diphosphate | HMDB0000295 | 2.3748643 | |
| 96 | 0.81_565.0474m/z | 565.04744 | 0.81115 | neg | Uridine diphosphate glucose | HMDB0000286 | 1.8838349 | |
| 97 | 2.16_283.0915n | 284.09878 | 2.1632167 | pos | Guanosine | HMDB0000133 | 1.0393267 | |
| Organic acids and derivatives | ||||||||
| 98 | 5.59_627.2407m/z | 627.24074 | 5.5859833 | pos | 6-Hydroxysandoricin | HMDB0037556 | 1.1439601 | |
| 99 | 0.75_104.0710m/z | 104.07099 | 0.7531167 | pos | gamma-Aminobutyric acid | HMDB0000112 | 1.022634 | 1.8454808 |
| 100 | 1.12_192.0261n | 191.01882 | 1.1193833 | neg | Isocitric acid | HMDB0000193 | −0.6188259 | −1.6664215 |
| 101 | 1.13_147.0896n | 130.0863 | 1.1279 | pos | (2R,3R,4R)-2-Amino-4-hydroxy-3-methylpentanoic acid | HMDB0029449 | −1.0198727 | |
| 102 | 1.11_146.0216n | 129.0183 | 1.1108833 | pos | Oxoglutaric acid | HMDB0000208 | −1.1202847 | −2.1185923 |
| 103 | 1.11_192.0271n | 215.01603 | 1.1108833 | pos | Citric acid | HMDB0000094 | −1.1723789 | −2.4103012 |
| 104 | 0.92_324.2166m/z | 324.21664 | 0.9234167 | pos | N-Jasmonoylisoleucine | HMDB0029391 | −1.2033973 | −0.5569567 |
| 105 | 1.98_141.0182m/z | 141.01819 | 1.9789 | pos | 2-Methylene-4-oxopentanedioic acid | HMDB0037759 | −1.4134892 | −0.5977763 |
| 106 | 0.74_147.0763m/z | 147.07632 | 0.7360833 | pos | L-Glutamine | HMDB0000641 | −1.4326922 | |
| 107 | 15.27_115.0505m/z | 115.05045 | 15.2738 | pos | Ureidopropionic acid | HMDB0000026 | −1.4409771 | −0.6352039 |
| 108 | 0.55_143.0339m/z | 143.03386 | 0.5475833 | pos | 2-Methyl-4-oxopentanedioic acid | HMDB0039447 | −1.4770497 | −0.4252457 |
| 109 | 1.11_143.0339m/z | 143.03388 | 1.1108833 | pos | Oxoadipic acid | HMDB0000225 | −1.5351415 | −0.717166 |
| 110 | 4.18_202.0441m/z | 202.0441 | 4.1786667 | pos | L-Oxalylalbizziine | HMDB0039164 | −1.6272134 | −1.0362134 |
| 111 | 0.75_130.0500m/z | 130.04995 | 0.7531167 | pos | Pyroglutamic acid | HMDB0000267 | −1.6855014 | 0.2421316 |
| 112 | 0.70_175.1189m/z | 175.11886 | 0.7020333 | pos | L-Arginine | HMDB0000517 | −1.7061716 | −1.3389484 |
| 113 | 0.72_134.0447m/z | 134.04468 | 0.7190667 | pos | L-Aspartic acid | HMDB0000191 | −1.7547972 | −0.9691886 |
| 114 | 2.84_166.0862m/z | 166.08623 | 2.8391 | pos | L-Phenylalanine | HMDB0000159 | −1.8175168 | −1.0099049 |
| 115 | 0.75_119.0586n | 120.06569 | 0.7531167 | pos | L-Threonine | HMDB0000167 | −1.8452429 | |
| 116 | 0.89_118.0864m/z | 118.08643 | 0.88935 | pos | L-Valine | HMDB0000883 | −1.9042841 | −0.2565016 |
| 117 | 2.07_132.1020m/z | 132.10196 | 2.0707667 | pos | L-Isoleucine | HMDB0000172 | −1.9205672 | −1.2438406 |
| 118 | 0.84_116.0708m/z | 116.07082 | 0.8382333 | pos | 4-Amino-2-methylenebutanoic acid | HMDB0030409 | −2.1902771 | −0.7244668 |
| 119 | 0.75_132.0656m/z | 132.06558 | 0.7531167 | pos | 4-Hydroxyproline | HMDB0000725 | −2.5643691 | |
| 120 | 0.84_175.1076m/z | 175.10763 | 0.8382333 | pos | N-Acetylornithine | HMDB0003357 | −2.7493487 | |
| 121 | 1.13_307.0835n | 308.09078 | 1.1279 | pos | Glutathione | HMDB0000125 | 37.25281 | |
| 122 | 0.86_218.0902n | 219.09738 | 0.8552667 | pos | N-gamma-L-Glutamyl-D-alanine | HMDB0036301 | 35.156087 | |
| 123 | 0.77_176.1028m/z | 176.10284 | 0.7701333 | pos | Citrulline | HMDB0000904 | 2.4286486 | |
| 124 | 0.74_244.0224m/z | 244.02236 | 0.74305 | neg | O-Phosphohomoserine | HMDB0003484 | 1.9696354 | |
| Organic nitrogen compounds | ||||||||
| 125 | 15.27_124.0871m/z | 124.08706 | 15.2738 | pos | L-Histidinol | HMDB0003431 | −1.4183979 | −0.6265774 |
| 126 | 15.27_122.0966m/z | 122.0966 | 15.2738 | pos | N,N-Dimethylaniline | HMDB0001020 | −1.4320285 | −0.6158732 |
| 127 | 15.29_112.0872m/z | 112.0872 | 15.291383 | pos | Histamine | HMDB0000870 | −1.4439979 | −0.6506019 |
| 128 | 12.39_300.2895m/z | 300.28947 | 12.385033 | pos | Sphingosine | HMDB0000252 | 3.8095576 | |
| 129 | 2.39_124.0395m/z | 124.03947 | 2.3868833 | pos | 2-Hydroxy-4-imino-2,5-cyclohexadienone | HMDB0031713 | −1.8874874 | |
| Organic oxygen compounds | ||||||||
| 130 | 4.85_817.3868m/z | 817.38677 | 4.8532667 | neg | (3x,5x,10x)-9,10-Didehydroisohumbertiol O-[rhamnosyl-(1->4)-rhamnosyl-(1->2)-[rhamnosyl-(1->6)]-glucoside] | HMDB0040687 | 3.9229994 | |
| 131 | 10.96_369.2633m/z | 369.26334 | 10.959167 | pos | Mangalkanyl glucoside | HMDB0036015 | 3.3342658 | |
| 132 | 5.16_441.1765m/z | 441.17651 | 5.15925 | neg | Pteroside P | HMDB0036608 | 3.2783919 | |
| 133 | 10.14_676.3662n | 699.35529 | 10.144617 | pos | (S)-Nerolidol 3-O-[a-L-rhamnopyranosyl-(1->4)-a-L-rhamnopyranosyl- (1->6)-b-D-glucopyranoside] | HMDB0040846 | 3.1660281 | 3.0346403 |
| 134 | 5.16_359.1349m/z | 359.13486 | 5.15925 | neg | 2′-Methoxy-3-(2,4-dihydroxyphenyl)-1,2-propanediol 4′-glucoside | HMDB0039473 | 1.9883008 | |
| 135 | 5.45_357.1192m/z | 357.11919 | 5.4494 | neg | Moringyne | HMDB0031724 | 1.8337359 | 1.9333416 |
| 136 | 0.76_194.0418n | 193.03453 | 0.7600833 | neg | D-Glucuronic acid | HMDB0000127 | 1.7667359 | 3.7537263 |
| 137 | 0.75_356.0951n | 379.08431 | 0.7531167 | pos | 3-O-beta-D-Galactopyranuronosyl-D-galactose | HMDB0039726 | 1.7473063 | |
| 138 | 5.03_393.1767m/z | 393.17668 | 5.0349167 | neg | Foeniculoside IX | HMDB0033011 | 1.3872272 | 3.323785 |
| 139 | 5.19_463.0885m/z | 463.0885 | 5.1942167 | neg | 3′-(2″-Galloylglucosyl)-phloroacetophenone | HMDB0040622 | 1.3004351 | |
| 140 | 5.36_539.1745m/z | 539.17454 | 5.35895 | neg | Torachrysone 8-(2-apiosylglucoside) | HMDB0034612 | 1.2716972 | |
| 141 | 5.18_509.2238m/z | 509.2238 | 5.1766 | neg | Linalool 3,6-oxide primeveroside | HMDB0035489 | 1.0205958 | |
| 142 | 5.14_377.1817m/z | 377.18167 | 5.1415833 | neg | 7-Hydroxyterpineol 8-glucoside | HMDB0033019 | 0.6037439 | 1.8587819 |
| 143 | 0.76_209.0296m/z | 209.02957 | 0.7600833 | neg | Galactaric acid | HMDB0000639 | 0.4372531 | 1.7316785 |
| 144 | 5.14_355.1724m/z | 355.17236 | 5.1416 | pos | (1S,2S,4R)-1,8-Epoxy-p-menthan-2-ol glucoside | HMDB0033110 | 0.1965645 | 1.3746685 |
| 145 | 4.72_402.1525n | 447.15077 | 4.7247833 | neg | Benzyl O-[arabinofuranosyl-(1->6)-glucoside] | HMDB0041514 | −0.8390669 | −1.2836963 |
| 146 | 0.86_504.1687n | 527.15791 | 0.8552667 | pos | Gentiotriose | HMDB0029910 | −1.1018444 | −0.5817183 |
| 147 | 5.00_295.1057n | 340.10362 | 4.99905 | neg | Prunasin | HMDB0034934 | −1.1644387 | −3.8543421 |
| 148 | 9.86_329.0049m/z | 329.00487 | 9.86165 | pos | D-Sedoheptulose 7-phosphate | HMDB0001068 | −1.3360925 | −0.552754 |
| 149 | 0.79_204.0866m/z | 204.08657 | 0.7871667 | pos | N-Acetyl-D-glucosamine | HMDB0000215 | −1.3540415 | −0.6775015 |
| 150 | 0.86_522.2025m/z | 522.20253 | 0.8552667 | pos | 6-Kestose | HMDB0033673 | −1.4496564 | −0.6952694 |
| 151 | 0.87_342.1158n | 365.10504 | 0.8722833 | pos | Allolactose | HMDB0038489 | −1.6033747 | −0.5975339 |
| 152 | 0.85_342.1160n | 387.11424 | 0.8452333 | neg | Trehalose | HMDB0000975 | −1.6106083 | |
| 153 | 0.86_689.2101m/z | 689.21012 | 0.8552667 | pos | Mannan | HMDB0029931 | −1.6263271 | −0.6079286 |
| 154 | 0.84_288.0843n | 289.09139 | 0.8382333 | pos | Phlorin | HMDB0035589 | −1.6994496 | −0.670122 |
| 155 | 0.86_342.1158n | 360.14975 | 0.8552667 | pos | Inulobiose | HMDB0029898 | −1.7089782 | −0.7158346 |
| 156 | 14.21_589.4072m/z | 589.40716 | 14.208733 | pos | Lansioside C | HMDB0035103 | −1.8987114 | 2.4629941 |
| 157 | 0.77_144.0655m/z | 144.06547 | 0.7701333 | pos | 5-Hydroxymethyl-2-furancarboxaldehyde | HMDB0034355 | −2.2634989 | −1.3366433 |
| 158 | 0.77_164.0684n | 147.06512 | 0.7701333 | pos | 2-O-Methyl-D-xylose | HMDB0033821 | −3.5640663 | −3.2664454 |
| 159 | 4.78_469.1318m/z | 469.13181 | 4.77945 | pos | 4-Phenylbutyl glucosinolate | HMDB0038415 | 3.9823775 | |
| 160 | 0.76_383.1000m/z | 383.09996 | 0.7600833 | neg | alpha-Hydrojuglone 4-O-b-D-glucoside | HMDB0034242 | 2.8118682 | |
| 161 | 1.32_231.0838m/z | 231.08378 | 1.3224 | pos | Ethyl beta-D-glucopyranoside | HMDB0029968 | 2.3002148 | |
| 162 | 0.79_315.0933m/z | 315.09329 | 0.7941333 | neg | D-erythro-L-galacto-Nonulose | HMDB0029955 | 1.6214985 | |
| 163 | 0.81_479.1617m/z | 479.16172 | 0.81115 | neg | D-glycero-L-galacto-Octulose | HMDB0029954 | 1.4373274 | |
| 164 | 4.35_342.1311n | 365.1203 | 4.3501167 | pos | Sphalleroside A | HMDB0032767 | −1.506083 | |
| 165 | 1.13_305.0840m/z | 305.08405 | 1.1279 | pos | Arabinopyranobiose | HMDB0029619 | −1.684668 | |
| 166 | 1.13_539.1214m/z | 539.12143 | 1.1279 | pos | b-D-Glucuronopyranosyl-(1->3)-a-D-galacturonopyranosyl-(1->2)-L-rhamnose | HMDB0039728 | −2.2164514 | |
| 167 | 2.54_360.1417n | 383.13095 | 2.5369167 | pos | 2-(4-Hydroxy-3,5-dimethoxyphenyl) ethanol 4′-glucoside | HMDB0038381 | −2.5423974 | |
| 168 | 5.56_458.1789n | 503.17718 | 5.5570667 | neg | Eugenol O-[a-L-Arabinofuranosyl-(1->6)-b-D-glucopyranoside] | HMDB0037603 | −3.5179227 | |
| 169 | 4.99_295.1054n | 318.09467 | 4.9916 | pos | Sambunigrin | HMDB0034981 | −4.9839447 | |
| Organohalogen compounds | ||||||||
| 170 | 13.77_226.9513m/z | 226.95127 | 13.77355 | pos | Perflutren | HMDB0014696 | −1.5064546 | −0.654059 |
| Organoheterocyclic compounds | ||||||||
| 171 | 2.42_376.1367n | 399.12588 | 2.424 | pos | Riboflavin | HMDB0000244 | −1.0930927 | −2.5176871 |
| 172 | 0.77_118.0865m/z | 118.08645 | 0.7701333 | pos | 2-Methyltetrahydrofuran-3-one | HMDB0031178 | −1.231741 | |
| 173 | 15.17_175.1229m/z | 175.12292 | 15.167433 | pos | 3-(Dimethylaminomethyl)indole | HMDB0035762 | −1.4142639 | −0.6390608 |
| 174 | 15.29_147.0916m/z | 147.09158 | 15.291383 | pos | 1H-Indole-3-methanamine | HMDB0029740 | −1.425459 | −0.6368287 |
| 175 | 15.27_108.0811m/z | 108.0811 | 15.2738 | pos | 6-Acetyl-1,2,3,4-tetrahydropyridine | HMDB0030345 | −1.441196 | −0.5951493 |
| 176 | 1.79_125.0235m/z | 125.02348 | 1.7935667 | pos | 5-Hydroxymaltol | HMDB0032988 | −1.489562 | −0.594498 |
| 177 | 0.55_127.0390m/z | 127.03905 | 0.5475833 | pos | Maltol | HMDB0030776 | −1.5004781 | −0.6021647 |
| 178 | 0.86_163.0600m/z | 163.05997 | 0.8552667 | pos | D-1,5-Anhydrofructose | HMDB0041561 | −1.7541497 | −0.9841154 |
| 179 | 0.72_184.0732m/z | 184.07321 | 0.7190667 | pos | Tryptophanol | HMDB0003447 | −2.2213504 | −1.5218705 |
| 180 | 4.12_187.0633n | 188.07057 | 4.12355 | pos | Indoleacrylic acid | HMDB0000734 | −2.3607275 | −2.2746153 |
| 181 | 0.77_128.0474n | 129.05468 | 0.7701333 | pos | 3-Hydroxy-4,5-dimethyl-2(5H)-furanone | HMDB0031306 | −2.8330093 | −1.9165285 |
| 182 | 3.87_271.1150m/z | 271.11503 | 3.8704833 | pos | Neopterin | HMDB0000845 | −1.7434786 | |
| Phenylpropanoids and polyketides | ||||||||
| 183 | 12.96_291.1952m/z | 291.19521 | 12.959667 | pos | Octyl 4-methoxycinnamic acid | HMDB0061861 | 8.2831723 | |
| 184 | 12.96_178.0629n | 179.07014 | 12.959667 | pos | 4-Methoxycinnamic acid | HMDB0002040 | 4.7751364 | |
| 185 | 0.76_397.0791m/z | 397.07908 | 0.7600833 | neg | Decarbamoylgonyautoxin III | HMDB0040137 | 3.5373353 | 7.5091867 |
| 186 | 14.20_379.1561m/z | 379.15614 | 14.2023 | neg | Kanzonol M | HMDB0041101 | 3.014213 | |
| 187 | 7.26_565.2866m/z | 565.28665 | 7.2588167 | neg | Hordatine A | HMDB0030461 | 2.6440336 | |
| 188 | 14.19_357.1467m/z | 357.14673 | 14.19105 | pos | [8]-Dehydrogingerdione | HMDB0039277 | 2.4931618 | |
| 189 | 0.85_695.2246m/z | 695.22456 | 0.8452333 | neg | 5-Hydroxy-7,3′,4′-trimethoxy-8-methylisoflavone 5-neohesperidoside | HMDB0030627 | 2.094353 | |
| 190 | 10.08_488.3504n | 975.69367 | 10.076433 | neg | 16beta-Hydroxystellatogenin | HMDB0040391 | 1.6017311 | |
| 191 | 5.31_624.1690n | 623.16171 | 5.305 | neg | Isorhamnetin 3-O-[b-D-glucopyranosyl-(1->2)-a-L-rhamnopyranoside] | HMDB0037085 | 1.4061339 | |
| 192 | 4.46_384.1057n | 383.09847 | 4.4620167 | neg | Eleutheroside B1 | HMDB0029549 | 1.2791289 | |
| 193 | 5.27_593.1512m/z | 593.15122 | 5.2687167 | neg | Kaempferol 3-neohesperidoside | HMDB0037573 | 1.1632915 | |
| 194 | 4.85_421.1637m/z | 421.1637 | 4.8532667 | neg | Mulberrin | HMDB0029507 | 1.0887678 | |
| 195 | 5.31_624.1684n | 625.1757 | 5.3125 | pos | Azaleatin 3-rutinoside | HMDB0037361 | 1.01554 | |
| 196 | 10.20_460.2690m/z | 460.26903 | 10.204233 | pos | Pectachol | HMDB0039064 | −1.0181638 | −0.6769391 |
| 197 | 9.44_432.2378m/z | 432.23776 | 9.4353667 | pos | Clausarinol | HMDB0041407 | −1.127634 | −0.6728349 |
| 198 | 1.30_164.0474n | 182.08123 | 1.3049 | pos | 2-Hydroxycinnamic acid | HMDB0002641 | −1.4332254 | −0.6687264 |
| 199 | 0.92_520.1013n | 543.09055 | 0.9234167 | pos | Melitric acid B | HMDB0040680 | −1.5110783 | −0.5940138 |
| 200 | 0.86_252.0633n | 253.07042 | 0.8552667 | pos | 2-O-(Z-p-Hydroxycinnamoyl)-(x)-glyceric acid | HMDB0041195 | −1.7930666 | −0.5375964 |
| 201 | 0.76_219.0449m/z | 219.04493 | 0.7600833 | neg | 3-Hydroxyflavone | HMDB0031816 | −3.0443569 | −2.8643833 |
| 202 | 0.77_418.0763m/z | 418.07634 | 0.7701333 | pos | Gonyautoxin II | HMDB0033507 | −6.0958687 | −5.450026 |
| 203 | 4.23_578.1420n | 579.14932 | 4.23295 | pos | Procyanidin B1 | HMDB0029754 | 9.5256207 | |
| 204 | 4.24_577.1352m/z | 577.13516 | 4.2357167 | neg | Procyanidin B2 | HMDB0033973 | 8.6781467 | |
| 205 | 4.16_595.1465n | 596.1538 | 4.16075 | pos | 3-Caffeoylpelargonidin 5-glucoside | HMDB0038087 | 5.5068457 | |
| 206 | 5.95_467.1864m/z | 467.18638 | 5.9453333 | pos | Thamnosin | HMDB0030550 | 2.4912899 | |
| 207 | 5.29_475.1161m/z | 475.1161 | 5.2941167 | pos | Albanin B | HMDB0034143 | −1.0039232 | |
| 208 | 5.56_571.1644m/z | 571.16438 | 5.5570667 | neg | Sakuranetin | HMDB0030090 | −3.6529585 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kong, X.; Luo, H.; Chen, Y.; Shen, H.; Shi, P.; Yang, F.; Li, H.; Yu, L. Elucidating Softening Mechanism of Honey Peach (Prunus persica L.) Stored at Ambient Temperature Using Untargeted Metabolomics Based on Liquid Chromatography-Mass Spectrometry. Horticulturae 2023, 9, 1210. https://doi.org/10.3390/horticulturae9111210
Kong X, Luo H, Chen Y, Shen H, Shi P, Yang F, Li H, Yu L. Elucidating Softening Mechanism of Honey Peach (Prunus persica L.) Stored at Ambient Temperature Using Untargeted Metabolomics Based on Liquid Chromatography-Mass Spectrometry. Horticulturae. 2023; 9(11):1210. https://doi.org/10.3390/horticulturae9111210
Chicago/Turabian StyleKong, Xiaoxue, Haibo Luo, Yanan Chen, Hui Shen, Pingping Shi, Fang Yang, Hong Li, and Lijuan Yu. 2023. "Elucidating Softening Mechanism of Honey Peach (Prunus persica L.) Stored at Ambient Temperature Using Untargeted Metabolomics Based on Liquid Chromatography-Mass Spectrometry" Horticulturae 9, no. 11: 1210. https://doi.org/10.3390/horticulturae9111210
APA StyleKong, X., Luo, H., Chen, Y., Shen, H., Shi, P., Yang, F., Li, H., & Yu, L. (2023). Elucidating Softening Mechanism of Honey Peach (Prunus persica L.) Stored at Ambient Temperature Using Untargeted Metabolomics Based on Liquid Chromatography-Mass Spectrometry. Horticulturae, 9(11), 1210. https://doi.org/10.3390/horticulturae9111210







