Proteomic Analysis of the Effect of Accelerated Ageing on Allium mongolicum Seeds
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Methods
2.2.1. Total Protein Extraction from Allium mongolicum Seeds
2.2.2. TMT Labeling and Peptide Separation
2.2.3. LC-MS/MS Analysis and Protein Identification
2.2.4. Data Analysis
3. Results
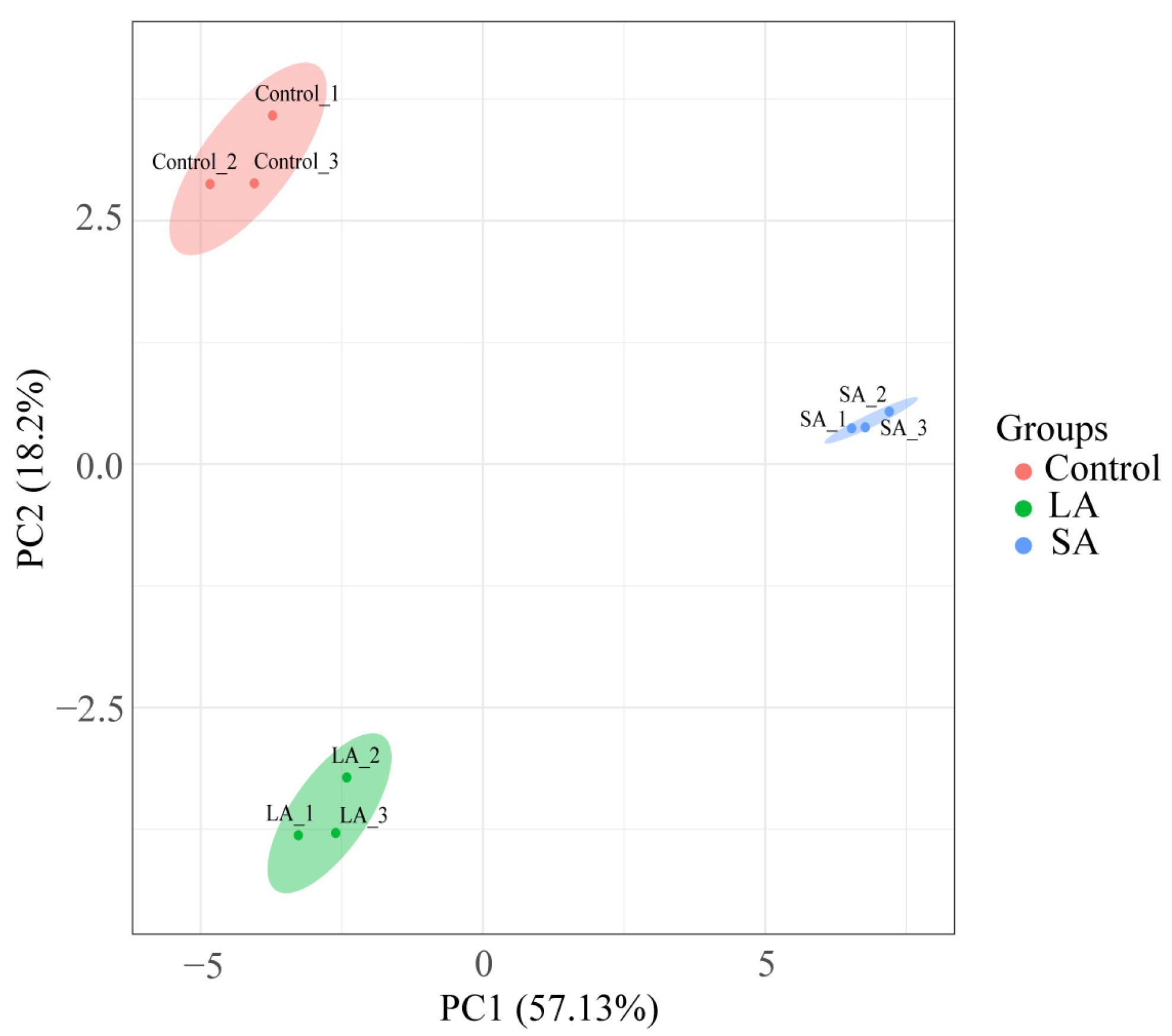
3.1. A General Overview of Protein Identification in Allium mongolicum Seeds under Accelerated Ageing
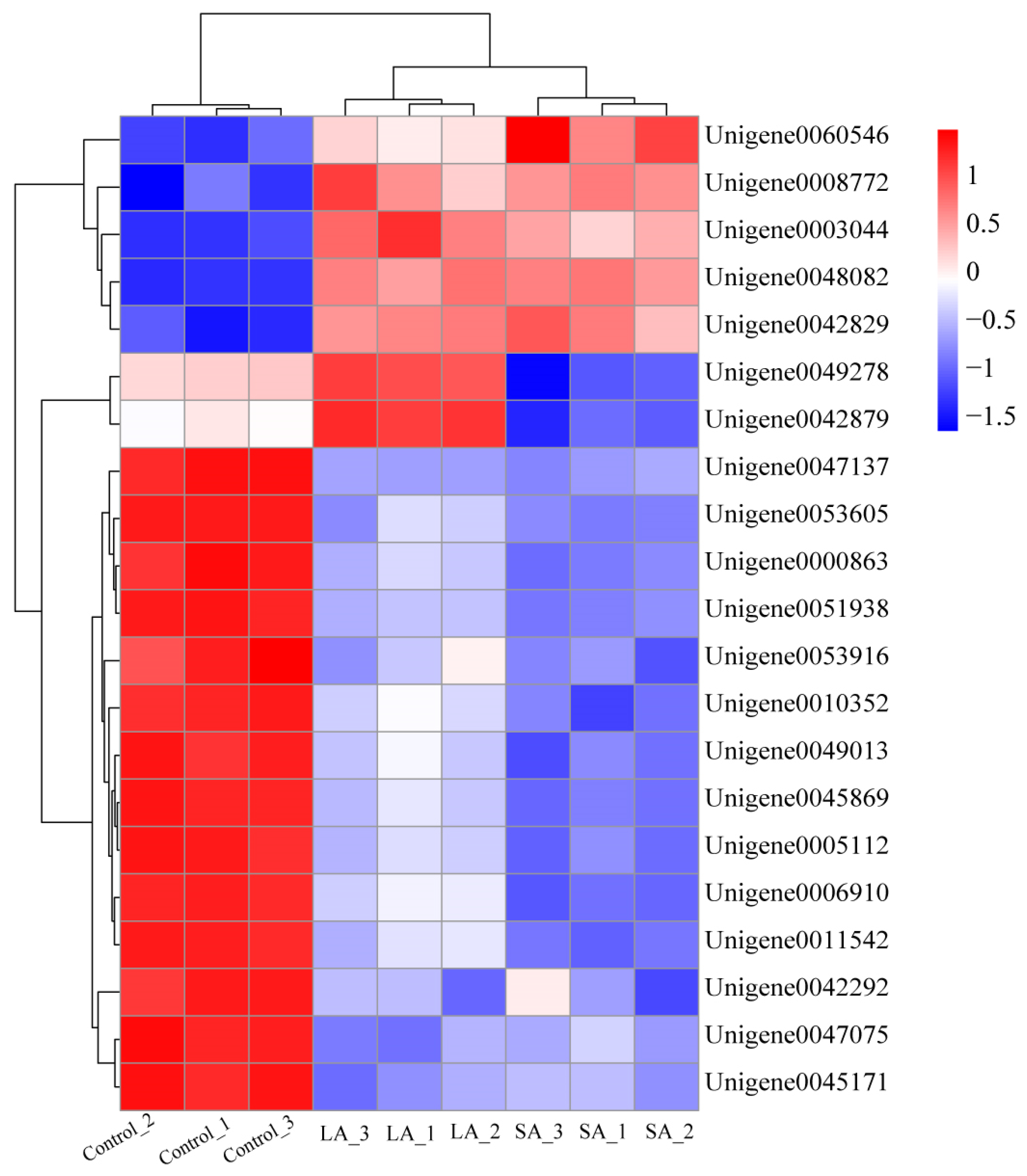
3.2. Quantification and Annotation of Differentially-Expressed Proteins (DEPs) in Allium mongolicum Seeds under Accelerated Ageing
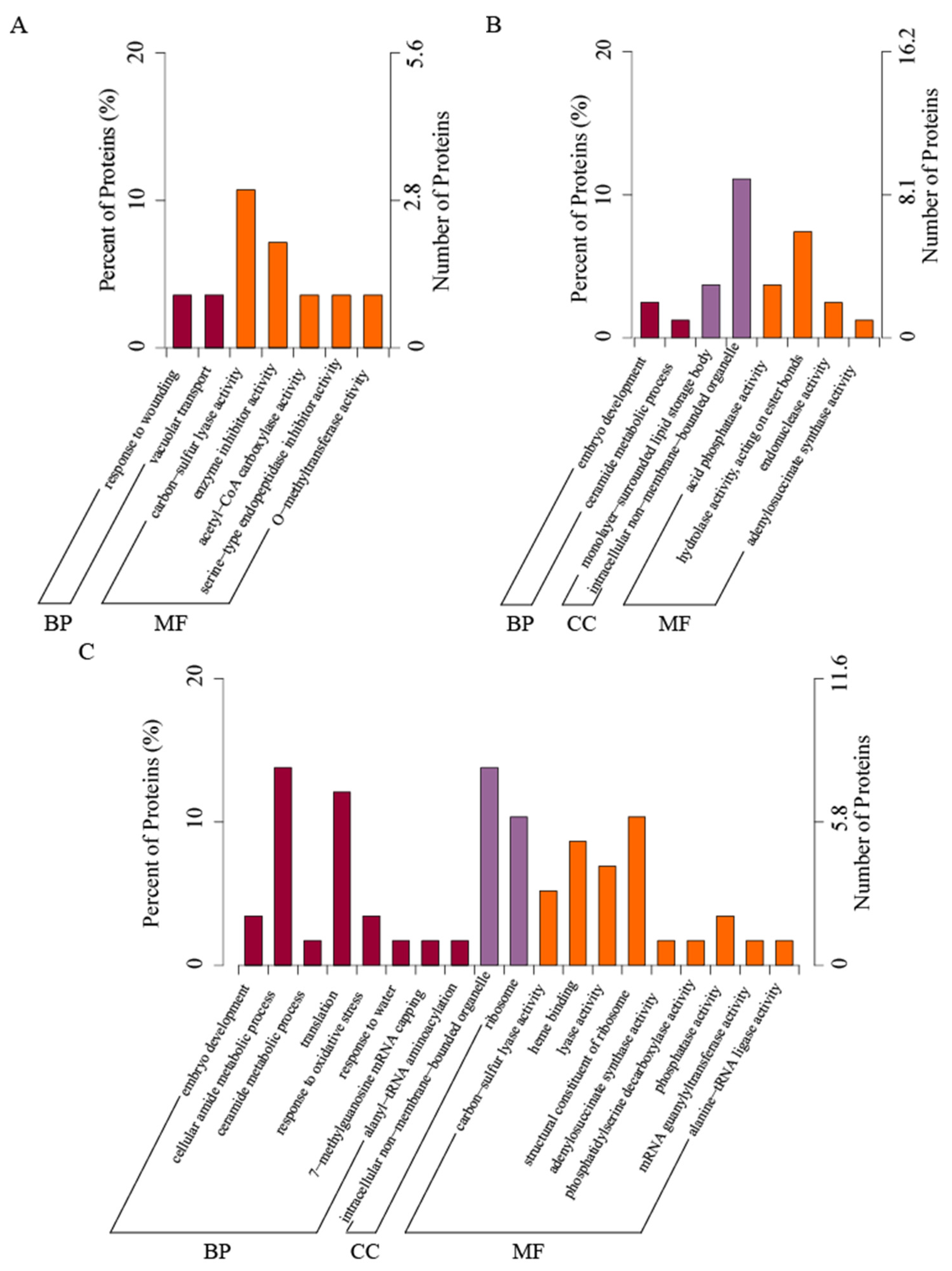
3.3. GO Enrichment Analysis of DEPs in Allium mongolicum Seeds under Accelerated Ageing
3.4. KEGG Pathway Enrichment of DEPs in Allium mongolicum Seeds under Accelerated Ageing
3.5. Analysis of Candidate Differential Proteins of Allium mongolicum Seeds after Accelerated Ageing
4. Discussion
4.1. Effect of Accelerated Ageing Treatment on the Viability of Allium mongolicum Seeds
4.2. Effects of Accelerated Ageing Treatments on Starch and Sucrose Metabolism in Allium mongolicum Seeds
4.3. Effects of Accelerated Ageing Treatment on Proteins Associated with Energy Metabolism in Allium mongolicum Seeds
4.4. Effect of Accelerated Ageing Treatment on Glutathione Metabolism-Related Proteins in Allium mongolicum Seeds
4.5. Effects of Accelerated Ageing Treatments on LEA and HSP Proteins in Allium mongolicum Seeds
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhang, D.; Zhang, F.L.; Yang, Z.R.; Hao, L.Z.; Wang, J.Q.; Ren, X.Y. Effects of Ce3+ and La3+ soaking on seed vigor and physiological characteristics of artificially aged Allium mongolicum seeds. Acta Bot. Boreali-Occident. Sin. 2020, 40, 87–94. [Google Scholar]
- Chang, H.W.; Zhang, F.L.; Yang, Z.R.; Kong, D.J.; Zheng, Q.L.; Hao, L.Z. Physiological and biochemical responses of Allium mongolicum seeds to storage ageing. Plant Physiol. J. 2015, 51, 1075–1081. [Google Scholar]
- Liu, Y.; He, J.W.; Yan, Y.T.; Liu, A.M.; Zhang, H.Q. Comparative Transcriptomic Analysis of Two Rice (Oryza sativa L.) Male Sterile Line Seed Embryos Under Accelerated Aging. Plant Mol. Biol. Rep. 2020, 38, 282–293. [Google Scholar] [CrossRef]
- Lakshmi, C.J.; Jijeesh, C.M.; Seethalakshmi, K.K. Impact of accelerated aging process on seed quality and biochemical changes of Dendrocalamus sikkimensis Gamble. Acta Physiol. Plant 2021, 43, 34. [Google Scholar] [CrossRef]
- Rajjou, L.; Lovigny, Y.; Groot, S.P.C.; Maya, B.; Claudette, J.; Dominique, J. Proteome-wide characterization of seed aging in Arabidopsis: A comparison between artificial and natural aging protocols. Plant Physiol. 2008, 148, 620–641. [Google Scholar] [CrossRef]
- Kaur, H.; Petla, B.P.; Kamble, N.U.; Singh, A.; Rao, V.; Salvi, P.; Ghosh, S.; Majee, M. Differentially expressed seed aging responsive heat shock protein OsHSP18.2 implicates in seed vigor, longevity and improves germination and seedling establishment under abiotic stress. Front. Plant Sci. 2015, 6, 713–725. [Google Scholar] [CrossRef]
- Moraes, C.E.; Lopes, J.C.; Farias, C.C.M.; Maciel, K.S. Physiological quality of Tabernaemontana fuchsiaefolia A. DC seeds due to the accelerated aging test. Cienc. Florest. 2016, 26, 213–223. [Google Scholar] [CrossRef]
- Chen, B.X.; Fu, H.; Gao, J.D.; Zhang, Y.X.; Huang, W.J.; Chen, Z.J.; Zhang, Q.; Yan, S.J.; Liu, J. Identification of Metabolomic Biomarkers of Seed Vigor and Aging in Hybrid Rice. Rice 2022, 15, 7. [Google Scholar] [CrossRef]
- Liu, D.M.; Han, C.X.; Deng, X.; Liu, Y.; Liu, N.N.; Yan, Y.M. Integrated physiological and proteomic analysis of embryo and endosperm reveals central salt stress response proteins during seed germination of winter wheat cultivar Zhengmai 366. BMC Plant Biol. 2019, 19, 406. [Google Scholar] [CrossRef]
- Chatelain, E.; Hundertmark, M.; Leprince, O.; Le, G.S.; Satour, P.; Deligny-Penninck, S.; Rogniaux, H.; Buitink, J. Temporal profiling of the heat-stable proteome during late maturation of Medicago truncatula seeds identifies a restricted subset of late embryogenesis abundant proteins associated with longevity. Plant Cell Environ. 2012, 35, 1440–1455. [Google Scholar] [CrossRef]
- Nogueira, F.C.S.; Palmisano, G.; Schwammle, V.; Soares, E.L.; Soares, A.A.; Roepstorff, P.; Domont, G.B.; Wang, W.Q.; Liu, S.J.; Song, S.Q.; et al. Proteomics of seed development, desiccation tolerance, germination and vigor. Plant Physiol. Biochem. 2015, 86, 1–15. [Google Scholar]
- Chu, P.; Chen, H.H.; Zhou, Y.L.; Li, Y.; Ding, Y.; Jiang, L.W.; Tsang Edward, W.T.; Wu, K.Q.; Huang, S.Z. Proteomic and functional analyses of Nelumbo nucifera annexins involved in seed thermotolerance and germination vigor. Planta 2012, 235, 1271–1288. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Qu, H.B.; Zhu, P.Y.; Su, K.M.; Zhang, C.Q. Comparative Proteomics Reveals the Mechanisms Underlying Variations in Seed Vigor Based on Maize (Zea mays L.) Ear Positions. Plant Mol. Biol. Rep. 2018, 36, 738–749. [Google Scholar] [CrossRef]
- Yan, M.K.; Zheng, L.; Li, B.J.; Shen, R.F.; Lan, P. Comparative proteomics reveals new insights into the endosperm responses to drought, salinity and submergence in germinating wheat seeds. Plant Mol. Biol. 2021, 105, 287–302. [Google Scholar] [CrossRef]
- Li, M.; Chen, X.; He, D.L.; Yang, P.F. Proteomic analysis reveals that calcium channel blockers affect radicle protrusion during rice seed germination. Plant Plant Growth Regul. 2020, 90, 393–407. [Google Scholar] [CrossRef]
- Tian, Q.; Wang, D.; Zhang, W.L.; Duan, N.B.; Li, Q.; Yan, T.J.; Dai, S.; Ding, H.F. Effect of artificial aging on soybean seed vigor and ascorbate-glutathione cycle in mitochondria. Plant Physiol. J. 2016, 52, 543–550. [Google Scholar]
- Niu, L.; Zhang, H.; Wu, Z.; Wang, Y.; Liu, H.; Wu, X.; Wang, W.; Thierry, C. Modifed TCA/acetone precipitation of plant proteins for proteomic analysis. PLoS ONE 2018, 13, e0202238. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, T.; Zhang, Z. Integrated Proteogenomic Characterization of Human High-Grade Serous Ovarian Cancer. Cell 2016, 166, 755. [Google Scholar] [CrossRef]
- Zhang, D.; Yang, Z.R.; Song, X.Q.; Zhang, F.L.; Liu, Y. TMT-based proteomic analysis of liquorice root in response to drought stress. BMC Genom. 2022, 23, 524. [Google Scholar] [CrossRef]
- Guo, F. Study on the Response Mechanism of Allium mongolicum Seeds to Storage Ageing. Master’s Thesis, Inner Mongolia Agricultural University, Hohhot, China, 2017. [Google Scholar]
- Moulder, R.; Lönnberg, T.; Elo, L.L.; Filén, J.J.; Rainio, E.; Corthals, G.; Oresic, M.; Nyman, T.A.; Aittokallio, T.; Lahesmaa, R. Quantitative proteomics analysis of the nuclear fraction of human CD4+ cells in the early phases of IL-4-induced Th2 differentiation. Mol. Cell Proteom. 2010, 9, 1937–1953. [Google Scholar] [CrossRef]
- Jones, P.; Binns, D.; Chang, H.Y.; Fraser, M.; Li, W.Z.; McAnulla, C. InterProScan 5: Genome-scale protein function classifcation. Bioinformatics 2014, 30, 1236–1240. [Google Scholar] [CrossRef] [PubMed]
- Ashburner, M.; Ball, C.A.; Blake, J.A.; Botstein, D.; Butler, H.; Cherry, J.M.; Ringwald, M.; Rubin, G.M.; Sherlock, G. Gene ontology: Tool for the unifcation of biology. The Gene Ontology Consortium. Nat. Genet. 2000, 25, 25–29. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Goto, S.; Hattori, M.; Aoki-Kinoshita, K.F.; Itoh, M.; Kawashima, S.; Katayama, T.; Araki, M.; Hirakawa, M. From genomics to chemical genomics: New developments in KEGG. Nucleic Acids Res. 2006, 34, 354–357. [Google Scholar] [CrossRef] [PubMed]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Bioinformatics enrichment tools: Paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009, 37, 1–13. [Google Scholar] [CrossRef]
- Sveinsdottir, H.; Zhu, Y.Y.; Schubert, S. Seed ageing-induced inhibition of germination and post-germination root growth is related to lower activity of plasma membrane H+-ATPase in maize roots. J. Plant Physiol. 2009, 166, 128–135. [Google Scholar] [CrossRef]
- Liu, H.; Zhu, Y.F.; Liu, X.; Jiang, Y.; Deng, S.M.; Ai, X.R.; Deng, Z.J. Effect of artificially accelerated aging on the vigor of Metasequoia glyptostroboides seeds. J. For. Res. 2020, 31, 769–779. [Google Scholar] [CrossRef]
- Van Treuren, R.; de Groot, E.C.; van Hintum, J.L. Preservation of seed viability during 25 years of storage under standard genebank conditions. Genet. Resour. Crop Evol. 2013, 60, 1407–1421. [Google Scholar] [CrossRef]
- McDonald, M.B. Orthodox seed deterioration and its repair. In Handbook of Seed Physiology: Applications to Agriculture; Benech-Arnold, R.L., Sanchez, R.A., Eds.; The Haworth Press: New York, NY, USA, 2004; pp. 273–304. [Google Scholar]
- Hilhorst, H.W.M.; Toorop, P. Review on dormancy, germinability, and germination in crop and weed seeds. Adv. Agron. 1997, 61, 111–165. [Google Scholar]
- Qin, J.; Zhang, J.N.; Liu, D.; Yin, C.C.; Wang, F.M.; Chen, P.Y.; Chen, H.; Ma, J.B.; Zhang, B.; Xu, J.; et al. iTRAQ-based analysis of developmental dynamics in the soybean leaf proteome reveals pathways associated with leaf photosynthetic rate. Mol. Genet. Genom. 2016, 291, 1595. [Google Scholar] [CrossRef]
- Liu, Y.H.; Lu, S.; Liu, K.F.; Wang, S.; Huang, L.Q.; Guo, L.P. Proteomics: A powerful tool to study plant responses to biotic stress. Plant Methods 2019, 15, 135. [Google Scholar] [CrossRef]
- Angelovici, R.; Fait, A.; Fernie, A.R.; Galili, G. A seed high-lysine trait is negatively associated with the TCA cycle and slows down Arabidopsis seed germination. New Phytol. 2011, 189, 148–159. [Google Scholar] [CrossRef] [PubMed]
- Nonogaki, H.; Nomaguchi, M.; Okumoto, N.; Kaneko, Y.; Matsushima, H.; Morohashi, Y. Temporal and spatial pattern of the biochemical activation of the endosperm during and following imbibition of tomato seeds. Physiol. Plant. 1998, 102, 236–242. [Google Scholar] [CrossRef]
- Li, S.F.; Wei, X.J.; Ren, Y.L.; Qiu, J.H.; Jiao, G.A.; Guo, X.P.; Tang, S.Q.; Wan, J.M.; Hu, P.S. OsBT1 encodes an ADP-glucose transporter involved in starch synthesis and compound granule formation in rice endosperm. Sci. Rep. 2017, 7, 40124. [Google Scholar] [CrossRef]
- Smith, A.M.; Zeeman, S.C.; Smith, S.M. Starch degradation. Annu. Rev. Plant Biol. 2005, 56, 73–98. [Google Scholar] [CrossRef] [PubMed]
- Dong, K.; Zhen, S.M.; Cheng, Z.W.; Cao, H.; Ge, P.; Yan, Y.M. Proteomic analysis reveals key proteins and phosphoproteins upon seed germination of wheat (Triticum aestivum L.). Front. Plant Sci. 2015, 6, 1017–1025. [Google Scholar] [CrossRef]
- Ruan, Y.L. Signaling role of sucrose metabolism in development. Mol. Plant 2012, 5, 763–765. [Google Scholar] [CrossRef] [PubMed]
- Baier, M.C.; Keck, M.; Gdde, V.; Niehaus, K.; Kuster, H.; Hohnjec, N. Knockdown of the symbiotic sucrose synthase Mt SucS1 affects arbuscule maturation and maintenance in mycorrhizal roots of Medicago truncatula. Plant Physiol. 2010, 152, 1000–1014. [Google Scholar] [CrossRef]
- Wang, W.Q.; Ye, J.Q.; Rogowska-Wrzesinska, A.; Wojdyla-Katarzyna, I.; Jensen, O.N.; Møller, I.M.; Song, S.Q. Proteomic comparison between maturation drying and prematurely imposed drying of Zea mays seeds reveals a potential role of maturation drying in preparing proteins for seed germination, seedling vigor, and pathogen resistance. J. Proteome Res. 2013, 13, 606–626. [Google Scholar] [CrossRef]
- Xin, X.; Lin, X.H.; Zhou, Y.C.; Chen, X.L.; Liu, X.; Lu, X.X. Proteome analysis of maize seeds: The effect of artificial ageing. Physiol. Plant. 2011, 143, 126–138. [Google Scholar] [CrossRef]
- Zhu, H.F.; Qian, W.Q.; Lu, X.Z.; Li, D.P.; Liu, X.; Liu, K.F.; Wang, D.W. Expression patterns of purple acid phosphatase genes in Arabidopsis organs and functional analysis of AtPAP23 predominantly transcribed in flower. Plant Mol. Biol. 2005, 59, 581–594. [Google Scholar] [CrossRef]
- Kuang, R.B.; Chan, K.H.; Yeung, E.; Lim, B.L. Molecular and biochemical characterization of AtPAP15, a purple acid phosphatase with phytase activity, in Arabidopsis. Plant Physiol. 2009, 151, 199–209. [Google Scholar] [CrossRef]
- Zamani, K.; Lohrasebi, T.; Sabet, M.S.; Malboobi, M.A.; Mousavi, A. Expression pattern and subcellular localization of Arabidopsis purple acid phosphatase AtPAP9. Gene Expr. Patterns 2014, 14, 9–18. [Google Scholar] [CrossRef]
- Sun, F.; Suen, P.K.; Zhang, Y.J.; Liang, C.; Carrie, C.; Whelan, J.; Ward, J.L.; Hawkins, N.D.; Jiang, L.W.; Rajjou, L.; et al. Seed germination and vigor. Annu. Rev. Plant Biol. 2012, 63, 507–533. [Google Scholar]
- Song, W.; Zhou, F.K.; Shan, C.H.; Zhang, Q.; Ning, M.; Liu, X.M.; Zhao, X.X.; Cai, W.C.; Yang, X.Q.; Hao, G.F.; et al. Identification of glutathione S-transferase genes in Hami melon (Cucumis melo var. saccharinus) and their expression analysis under cold stress. Front. Plant Sci. 2021, 12, 672017. [Google Scholar] [CrossRef]
- Milla, M.A.; Maurer, A.; Huete, A.R.; Gustafson, J.P. Glutathione peroxidase genes in Arabidopsis are ubiquitous and regulated by abiotic stresses through diverse signaling pathways. Plant J. 2003, 36, 602–615. [Google Scholar] [CrossRef]
- Moons, A. Regulatory and functional interactions of plant growth regulators and plant glutathione S-transferases (GSTs). Vitam. Horm. 2005, 72, 155. [Google Scholar]
- Xu, J.; Tian, Y.S.; Xing, X.J.; Peng, R.H.; Zhu, B.; Gao, J.J.; Yao, H. Over-expression of AtGSTUl9 provides tolerance to salt, drought and methyl viologen stresses in Arabidopsis. Physiol. Plant. 2016, 156, 164–175. [Google Scholar] [CrossRef]
- Espelund, M.; Saeboe-Larssen, S.; Hughes, D.W.; Galan, G.A.; Larsen, F.; Jakobsen, K.S. Late embryogenesis-abundant genes encoding proteins with different numbers of hydrophilic repeats are regulated differentially by abscisic acid and osmotic stress. Plant J. 1992, 2, 241–252. [Google Scholar] [CrossRef]
- Grelet, J.; Benamar, A.; Teyssier, E.; Avelange-Macherel, M.H.; Grunwald, D.; Macherel, D. Identification in pea seed mitochondria of a late-embryogenesis abundant protein able to protect enzymes from drying. Plant Physiol. 2005, 137, 157–167. [Google Scholar] [CrossRef]
- Cao, J.; Li, X. Identification and phylogenetic analysis of late embryogenesis abundant proteins family in tomato (Solanum lycopersicum). Planta 2015, 241, 757–772. [Google Scholar] [CrossRef]
- Pedrosa, A.M.; Martins, C.D.P.S.; Gonçalves, L.P.; Costa, M.G.C. Late embryogenesis abundant (LEA) constitutes a large and diverse family of proteins involved in development and abiotic stress responses in sweet orange (Citrus sinensis L. Osb.). PLoS ONE 2015, 10, 785–790. [Google Scholar] [CrossRef] [PubMed]
- Juszczak, I.; Bartels, D. LEA gene expression, RNA stability and pigment accumulation in three closely related linderniaceae species differing in desiccation tolerance. Plant Sci. 2017, 255, 59–71. [Google Scholar] [CrossRef] [PubMed]
- Su, P.; Li, H.M. Arabidopsis stromal 70-kD heat shock proteins are essential for plant development and important for thermos tolerance of germinating seeds. Plant Physiol. 2008, 146, 1231–1241. [Google Scholar] [CrossRef] [PubMed]
- Forward, B.S.; Osusky, M.; Misra, S. The Douglas-fir BiP promoter is functional in Arabidopsis and responds to wounding. Planta 2002, 215, 569–576. [Google Scholar] [CrossRef]
- Carpentier, S.C.; Witters, E.; Laukens, K.; Deckers, P.; Swennen, R.; Panis, B. Preparation of protein extracts from recalcitrant plant tissues:an evaluation of different methods for two-dimensional gel electrophoresis analysis. Proteomics 2005, 5, 2497–2507. [Google Scholar] [CrossRef]
- Shen-Miller, J.; Lindner, P.; Xie, Y.; Villa, S.; Wooding, K.; Clarke, S.G.; Loo, R.R.O.; Loo, J.A. Thermal–Stable Proteins of Fruit of Long–Living Sacred Lotus Nelumbo nucifera Gaertn var. China Antique. Trop. Plant Biol. 2013, 6, 69–84. [Google Scholar] [CrossRef]
- Maikova, A.; Zalutskaya, Z.; Lapina, T.; Ermilova, E. The HSP70 chaperone machines of Chlamydomonas are induced by cold stress. J. Plant Physiol. 2016, 204, 85–91. [Google Scholar] [CrossRef]
- Zhang, X.H.; Hong, W.; Tang, S.; Li, Q.N.; Bao, E.D. Apoptosis in response to heat stress is positively associated with heat-shock protein 90 expression in chicken myocardial cells in vitro. J. Vet. Sci. 2017, 18, 129–140. [Google Scholar] [CrossRef]





| Time (min) | Flow Rate (nL/min) | Mobile Phase A (%) | Mobile Phase B (%) |
|---|---|---|---|
| 0 | 1 | 97 | 3 |
| 10 | 1 | 95 | 5 |
| 30 | 1 | 80 | 20 |
| 48 | 1 | 60 | 40 |
| 50 | 1 | 50 | 50 |
| 53 | 1 | 30 | 70 |
| 54 | 1 | 0 | 100 |
| Total Spectra | Matched Spectrum | Peptide | Identified Protein | ALL |
|---|---|---|---|---|
| 401,747 | 29,560 | 19,076 | 4336 | 4318 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, X.; Yang, Z.; Zhang, D.; Zhang, X.; Zhang, F.; Liu, J.; Yu, C. Proteomic Analysis of the Effect of Accelerated Ageing on Allium mongolicum Seeds. Horticulturae 2023, 9, 1155. https://doi.org/10.3390/horticulturae9101155
Song X, Yang Z, Zhang D, Zhang X, Zhang F, Liu J, Yu C. Proteomic Analysis of the Effect of Accelerated Ageing on Allium mongolicum Seeds. Horticulturae. 2023; 9(10):1155. https://doi.org/10.3390/horticulturae9101155
Chicago/Turabian StyleSong, Xiaoqing, Zhongren Yang, Dong Zhang, Xiaoyan Zhang, Fenglan Zhang, Jiecai Liu, and Chuanzong Yu. 2023. "Proteomic Analysis of the Effect of Accelerated Ageing on Allium mongolicum Seeds" Horticulturae 9, no. 10: 1155. https://doi.org/10.3390/horticulturae9101155
APA StyleSong, X., Yang, Z., Zhang, D., Zhang, X., Zhang, F., Liu, J., & Yu, C. (2023). Proteomic Analysis of the Effect of Accelerated Ageing on Allium mongolicum Seeds. Horticulturae, 9(10), 1155. https://doi.org/10.3390/horticulturae9101155





