BcAMT1;5 Mediates Nitrogen Uptake and Assimilation in Flowering Chinese Cabbage and Improves Plant Growth When Overexpressed in Arabidopsis
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Culture Conditions
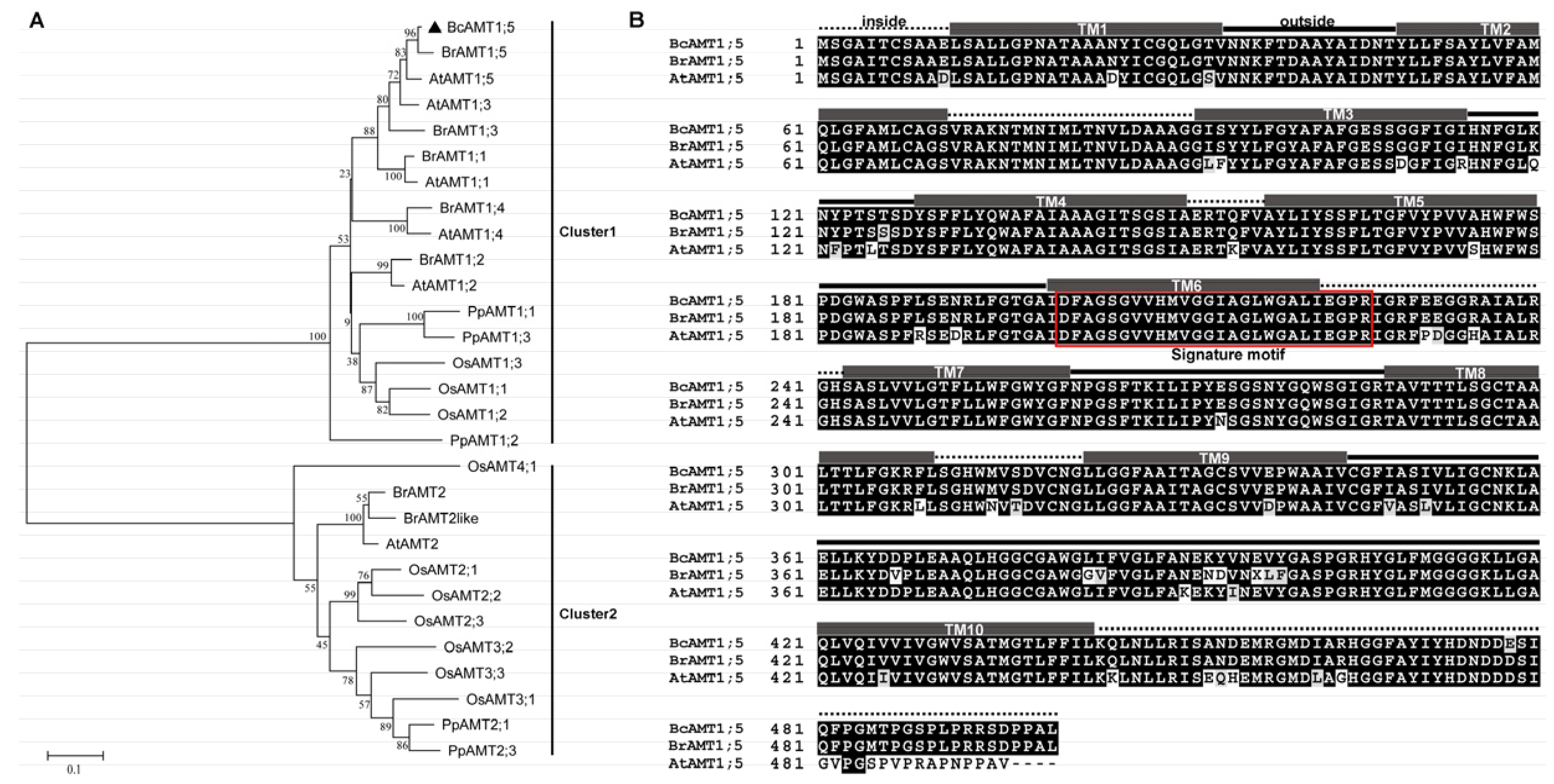
2.2. Cloning of BcAMT1;5 and Bioinformatics Analysis
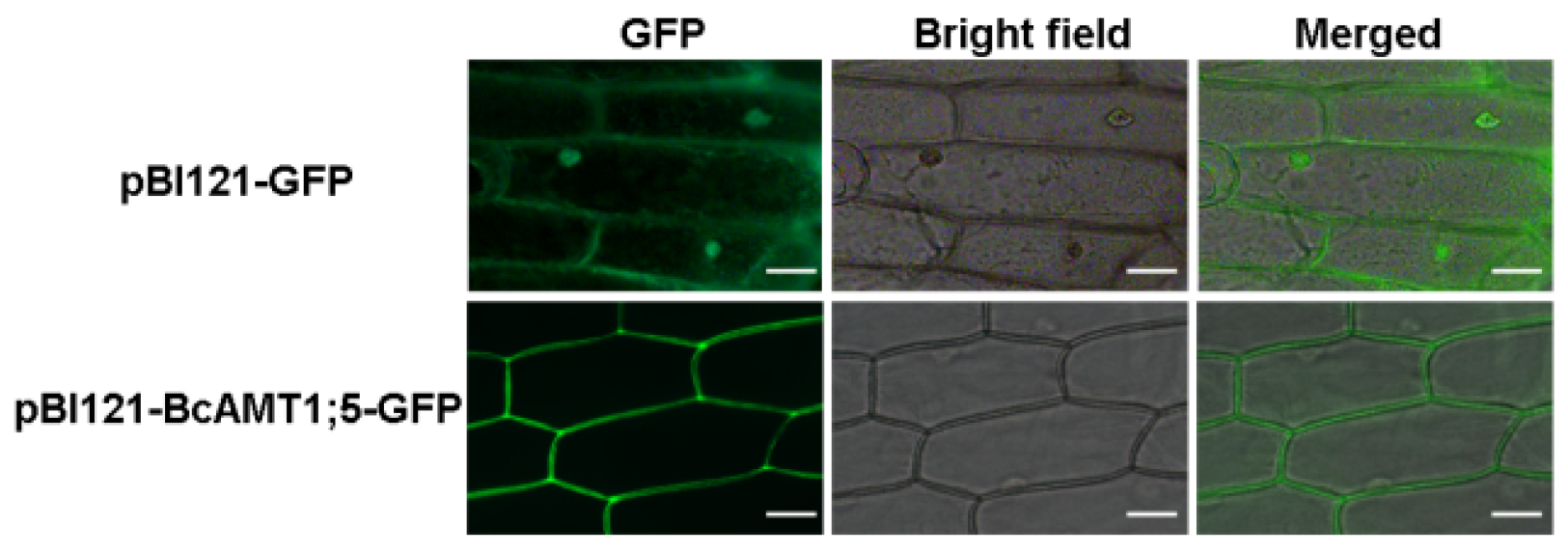
2.3. Sub-Cellular Location
2.4. qPCR
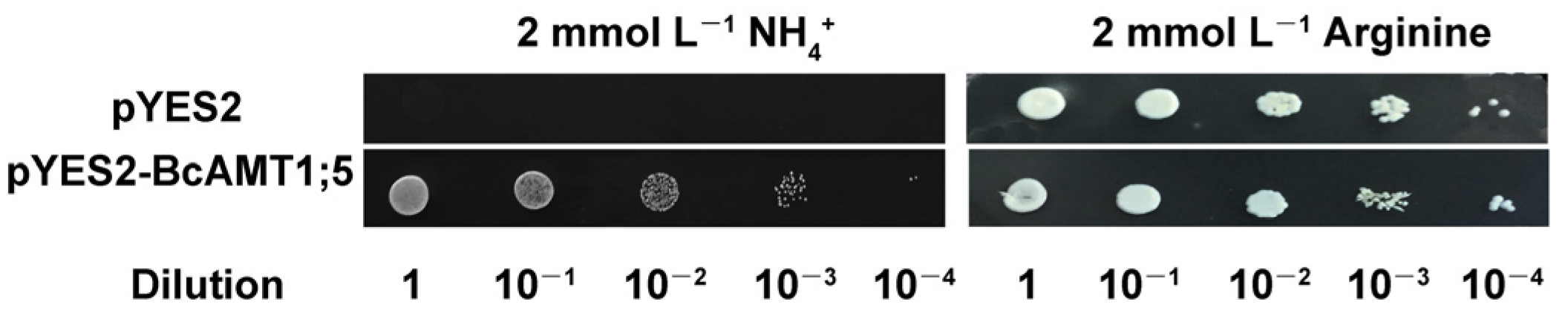
2.5. Functional Complementation in Yeast Mutant 31019b
2.6. Construction of pCAMBIA1391-BcAMT1;5pro::GUS for Arabidopsis Transformation and Glucorinidase (GUS) Assays
2.7. Construction of BcAMT1;5-Overexpressing Lines in Arabidopsis
2.8. Plant Culture for Growth Test, NH4+ Uptake, Ion Fluxes and Gene Expression
2.9. Statistical Analysis
3. Results
3.1. Molecular Identification of AMT1;5 Homolog from Flowering Chinese Cabbage
3.2. Subcellular Localization of BcAMT1;5 and its Functional Complementation Analysis in Yeast Mutant Cells
3.3. Expression Profiles for BcAMT1;5 Gene in Different Tissues of Flowering Chinese Cabbage
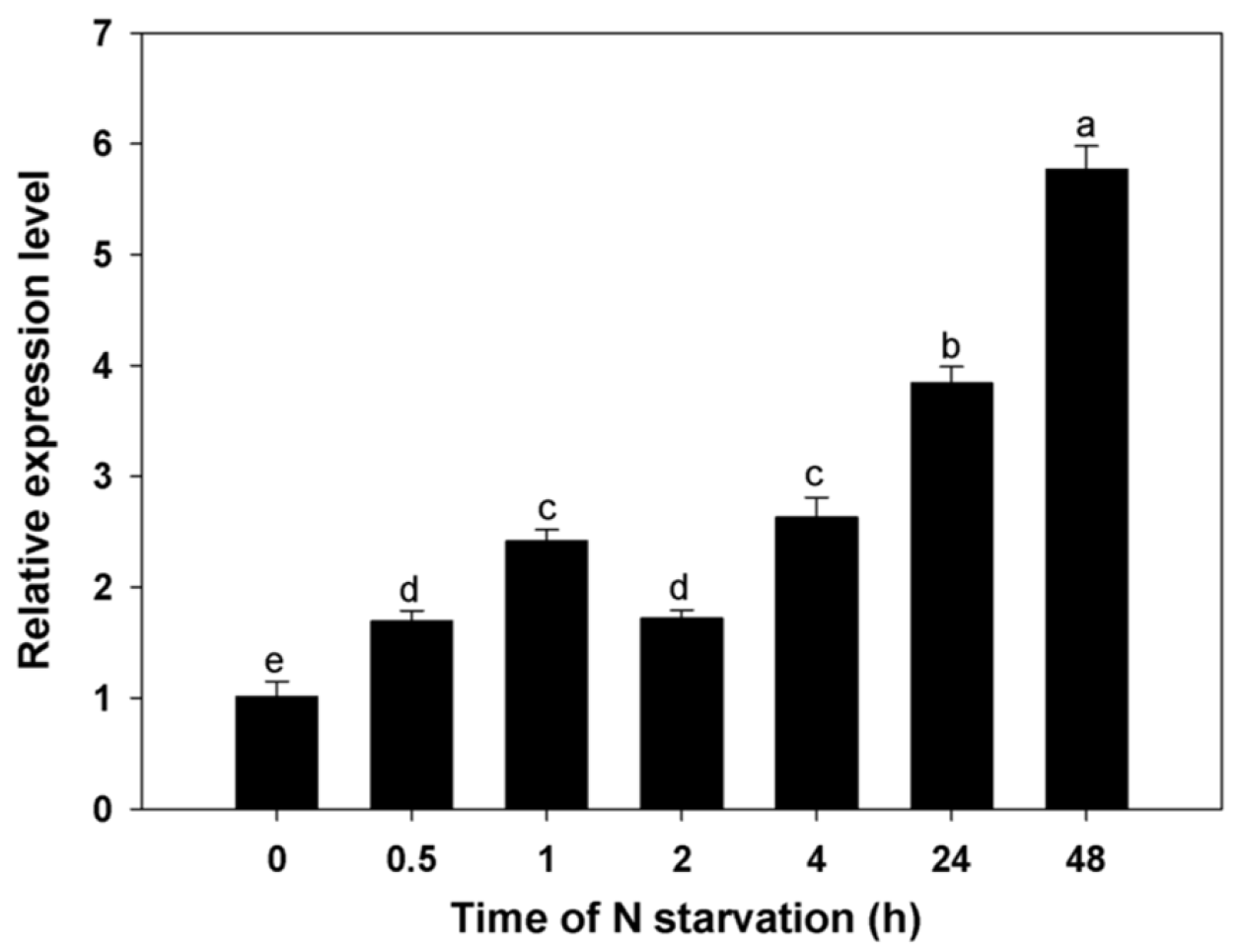
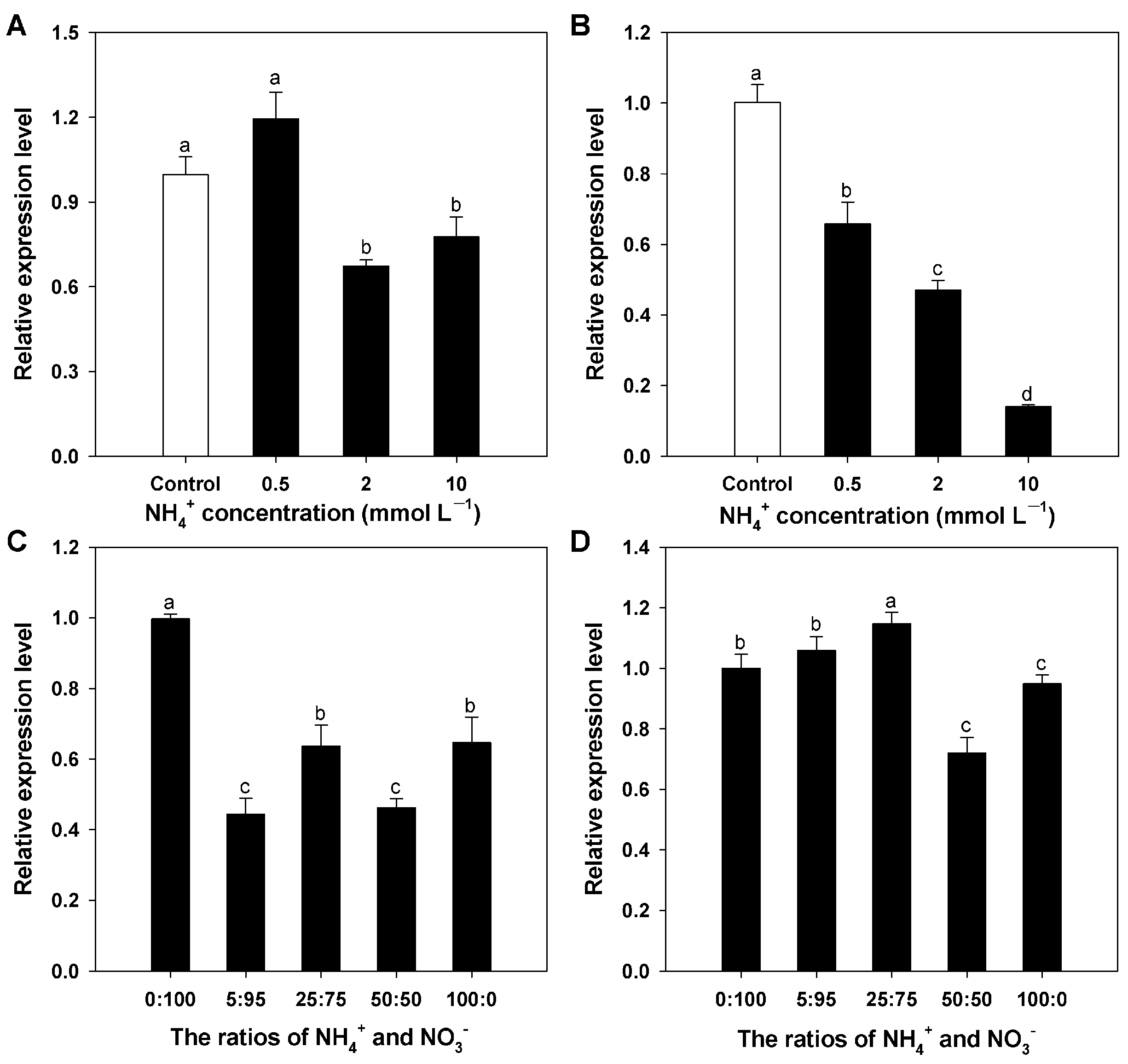
3.4. Expression Profiles for BcAMT1;5 Gene in Different N Regimes
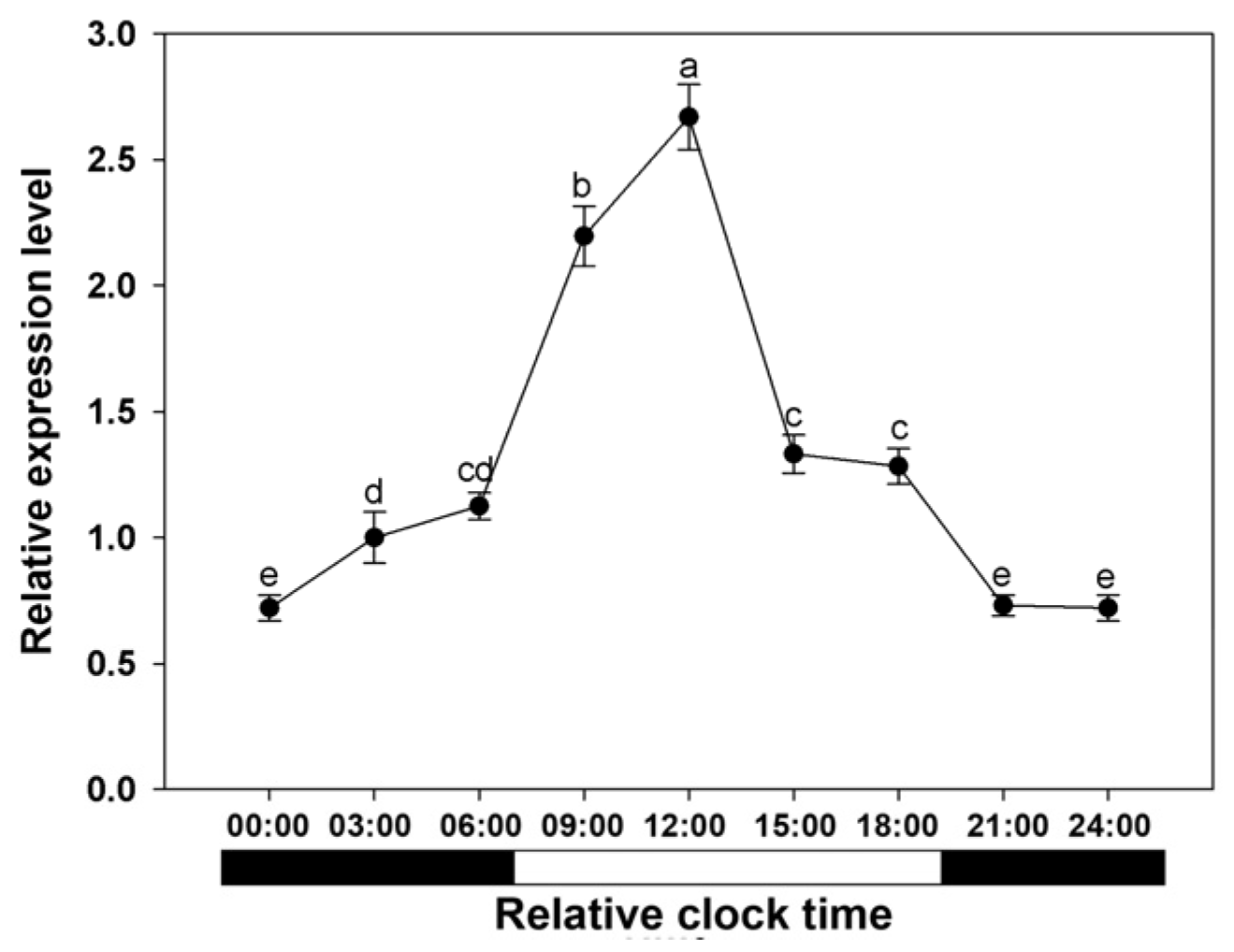
3.5. Expression of BcAMT1;5 Gene in Light Regimes
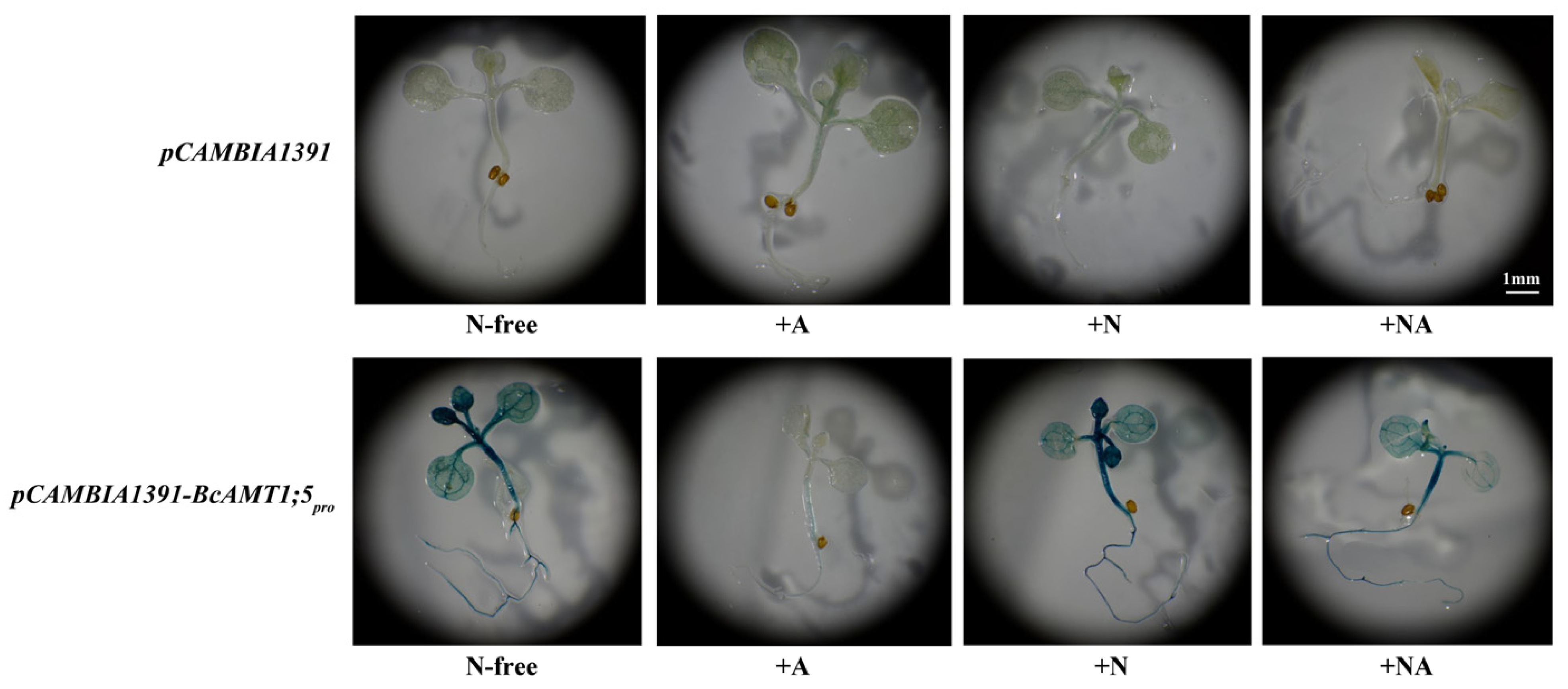
3.6. The GUS Activity of BcAMT1;5pro::GUS in Response to Different N Conditions
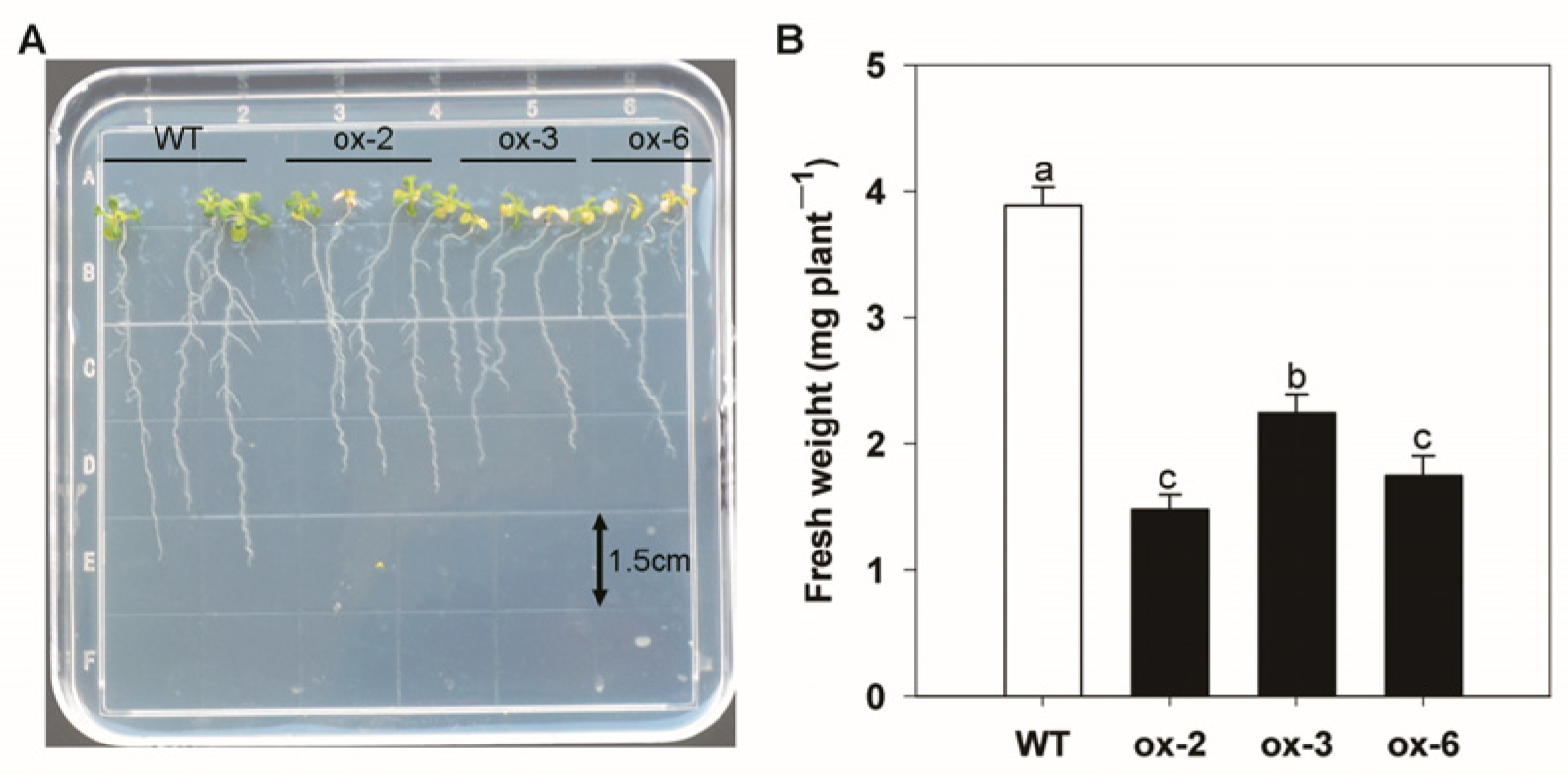
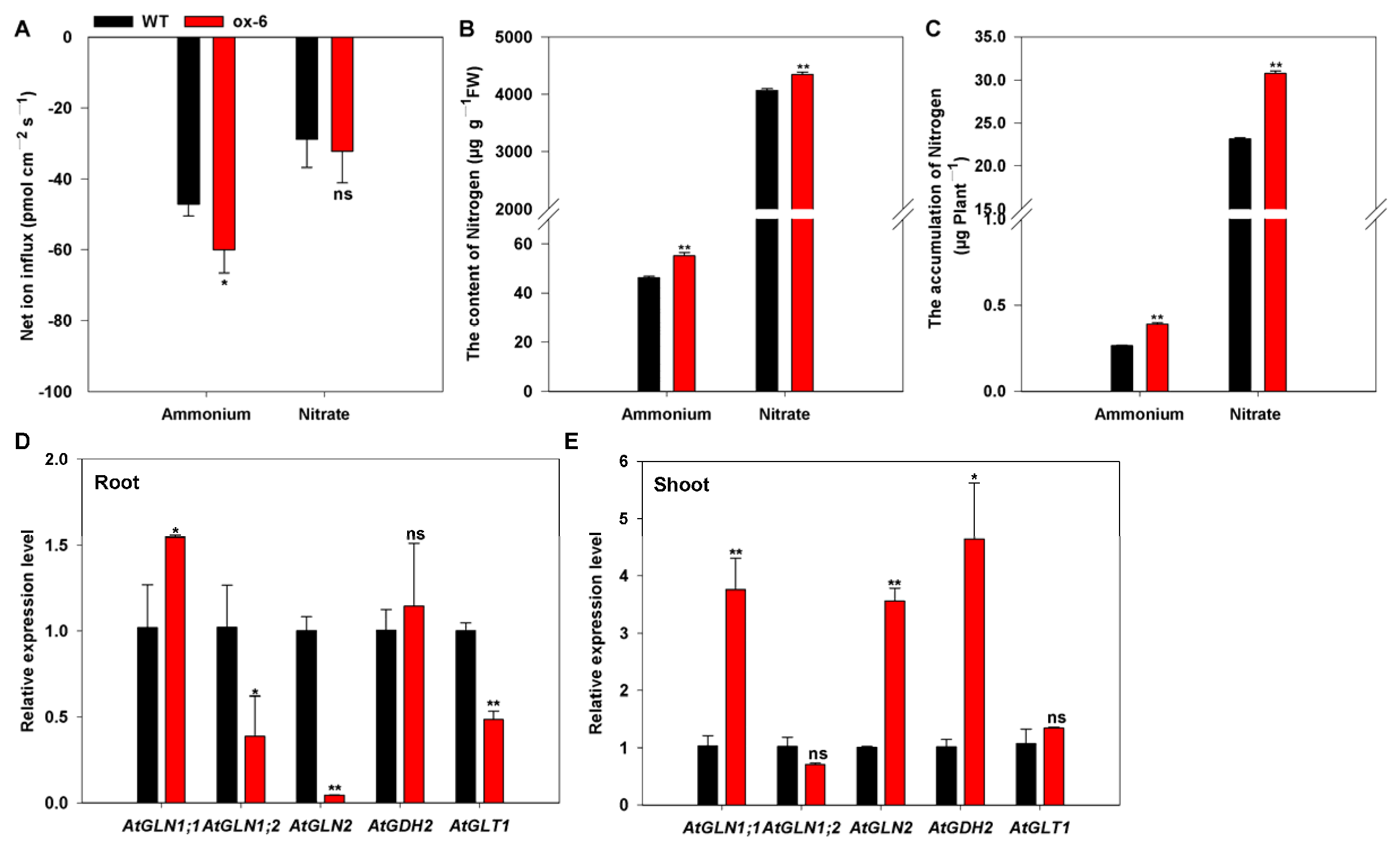
3.7. Heterologous Expression of BcAMT1;5 in Arabidopsis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gu, J.; Yang, J. Nitrogen (N) transformation in paddy rice field: Its effect on N uptake and relation to improved N management. Crop Environ. 2022, 1, 7–14. [Google Scholar] [CrossRef]
- Bloom, A.J.; Burger, M.; Asensio, J.S.R.; Cousins, A.B. Carbon dioxide enrichment inhibits nitrate assimilation in wheat and Arabidopsis. Science 2010, 328, 899–903. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhou, W.; Wu, J.; Xie, K.; Li, X. LjAMT2;2 Promotes ammonium nitrogen transport during arbuscular mycorrhizal fungi symbiosis in Lotus japonicus. Int. J. Mol. Sci. 2022, 23, 9522. [Google Scholar] [CrossRef] [PubMed]
- Gazzarrini, S.; Lejay, L.; Gojon, A.; Ninnemann, O.; Frommer, W.B.; von Wiren, N. Three functional transporters for constitutive, diurnally regulated, and starvation-induced uptake of ammonium into Arabidopsis roots. Plant Cell 1999, 11, 937–948. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.; Yang, S.; Huang, Y.; Su, Y. Identification of structural elements involved in fine-tuning of the transport activity of the rice ammonium transporter OsAMT1;3. Plant Physiol. Biochem. 2016, 108, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Hao, D.; Zhou, J.; Yang, S.; Qi, W.; Yang, K.; Su, Y. Function and regulation of ammonium transporters in plants. Int. J. Mol. Sci. 2020, 21, 3557. [Google Scholar] [CrossRef] [PubMed]
- McDonald, T.R.; Ward, J.M. Evolution of electrogenic ammonium transporters (AMTs). Front. Plant Sci. 2016, 7, 352. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Loqué, D.; Kojima, S.; Rauch, S.; Ishiyama, K.; Inoue, E.; Takahashi, H.; von Wirén, N. The organization of high-affinity ammonium uptake in Arabidopsis roots depends on the spatial arrangement and biochemical properties of AMT1-Type transporters. Plant Cell 2007, 19, 2636–2652. [Google Scholar] [CrossRef]
- Yuan, L.; Graff, L.; Loqué, D.; Kojima, S.; Tsuchiya, Y.N.; Takahashi, H.; von Wirén, N. AtAMT1;4, a pollen-specific high-affinity ammonium transporter of the plasma membrane in Arabidopsis. Plant Cell Physiol. 2009, 50, 13–25. [Google Scholar] [CrossRef]
- Nacry, P.; Bouguyon, E.; Gojon, A. Nitrogen acquisition by roots: Physiological and developmental mechanisms ensuring plant adaptation to a fluctuating resource. Plant Soil 2013, 370, 1–29. [Google Scholar] [CrossRef]
- Ijato, T.; Porras Murillo, R.; Ganz, P.; Ludewig, U.; Neuhäuser, B. Concentration-dependent physiological and transcriptional adaptations of wheat seedlings to ammonium. Physiol. Plant. 2021, 171, 328–342. [Google Scholar] [CrossRef] [PubMed]
- Konishi, N.; Ma, J.F. Three polarly localized ammonium transporter 1 members are cooperatively responsible for ammonium uptake in rice under low ammonium condition. New Phytol. 2021, 232, 1778–1792. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Li, J.; Zhang, B.; Hao, Y.; Ma, F. Genome-wide identification and expression analysis of AMT gene family in apple (Malus domestica Borkh.). Horticulturae 2022, 8, 457. [Google Scholar] [CrossRef]
- Zhu, Y.; Huang, X.; Hao, Y.; Su, W.; Liu, H.; Sun, G.; Chen, R.; Song, S. Ammonium transporter (BcAMT1.2) mediates the interaction of ammonium and nitrate in Brassica campestris. Front. Plant Sci. 2020, 10, 1776. [Google Scholar] [CrossRef] [PubMed]
- Zhong, L.; Huang, X.; Zhu, Y.; Kou, E.; Liu, H.; Sun, G.; Chen, R.; Song, S. Characterization and expression analysis of BcAMT1;4, an ammonium transporter gene in flowering Chinese cabbage. Hortic. Environ. Biotechnol. 2019, 60, 563–572. [Google Scholar] [CrossRef]
- Ludewig, U. Ion transport versus gas conduction: Function of AMT/Rh-type proteins. Transfus. Clin. Biol. 2006, 13, 111–116. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Marini, A.; Springael, J.; Frommer, W.B.; André, B. Cross-talk between ammonium transporters in yeast and interference by the soybean SAT1 protein. Mol. Microbiol. 2000, 35, 378–385. [Google Scholar] [CrossRef]
- Wiktorek-Smagur, A.; Hnatuszko-Konka, K.; Kononowicz, A.K. Flower bud dipping or vacuum infiltration-two methods of Arabidopsis thaliana transformation. Russ. J. Plant Physl. 2009, 56, 560–568. [Google Scholar] [CrossRef]
- Ivančič, I.; Degobbis, D. An optimal manual procedure for ammonia analysis in natural waters by the indophenol blue method. Water Res. 1984, 18, 1143–1147. [Google Scholar] [CrossRef]
- Couturier, J.; Montanini, B.; Martin, F.; Brun, A.; Blaudez, D.; Chalot, M. The expanded family of ammonium transporters in the perennial poplar plant. New Phytol. 2007, 174, 137–150. [Google Scholar] [CrossRef] [PubMed]
- Song, S.; He, Z.; Huang, X.; Zhong, L.; Liu, H.; Sun, G.; Chen, R. cloning and characterization of the ammonium transporter genes BaAMT1;1 and BaAMT1;3 from Chinese kale. Hortic. Environ. Biotechnol. 2017, 58, 178–186. [Google Scholar] [CrossRef]
- Li, H.; Yang, Q.S.; Liu, W.; Lin, J.; Chang, Y.H. The AMT1 family genes from Malus robusta display differential transcription features and ammonium transport abilities. Mol. Biol. Rep. 2017, 44, 379–390. [Google Scholar] [CrossRef]
- Ninnemann, O.; Jauniaux, J.C.; Frommer, W.B. Identification of a high affinity NH4+ transporter from plants. EMBO J. 1994, 13, 3464–3471. [Google Scholar] [CrossRef] [PubMed]
- Hachiya, T.; Sakakibara, H. Interactions between nitrate and ammonium in their uptake, allocation, assimilation, and signaling in plants. J. Exp. Bot. 2017, 68, 2501–2512. [Google Scholar] [CrossRef] [PubMed]
- Jian, S.; Liao, Q.; Song, H.; Liu, Q.; Lepo, J.E.; Guan, C.; Zhang, J.; Ismail, A.M.; Zhang, Z.; Abdelbagi, M.; et al. NRT1.1-related NH4+ toxicity is associated with a disturbed balance between NH4+ uptake and assimilation. Plant Physiol. 2018, 178, 1473–1488. [Google Scholar] [CrossRef]
- Li, H.; Cong, Y.; Chang, Y.; Lin, J. Two AMT2-Type ammonium transporters from Pyrus betulaefolia demonstrate distinct expression characteristics. Plant Mol. Biol. Rep. 2016, 34, 707–719. [Google Scholar] [CrossRef]
- Hachiya, T.; Mizokami, Y.; Miyata, K.; Tholen, D.; Watanabe, C.K.; Noguchi, K. Evidence for a nitrate-independent function of the nitrate sensor NRT1.1 in Arabidopsis thaliana. J. Plant Res. 2011, 124, 425–430. [Google Scholar] [CrossRef]
- Camañes, G.; Cerezo, M.; Primo-Millo, E.; Gojon, A.; García-Agustín, P. Ammonium transport and CitAMT1 expression are regulated by N in Citrus plants. Planta 2009, 229, 331–342. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Hao, Y.; Liu, H.; Sun, G.; Chen, R.; Song, S. Identification and characterization of two ammonium transporter genes in flowering Chinese cabbage (Brassica campestris). Plant Biotechnol. 2018, 35, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Luo, L.; Zhu, M.; Jia, L.; Xie, Y.; Wang, Z.; Xuan, W. Ammonium transporters cooperatively regulate rice crown root formation responding to ammonium nitrogen. J. Exp. Bot. 2022, 73, 3671–3685. [Google Scholar] [CrossRef] [PubMed]
- Bittsánszky, A.; Pilinszky, K.; Gyulai, G.; Komives, T. Overcoming ammonium toxicity. Plant Sci. 2015, 231, 184–190. [Google Scholar] [CrossRef] [PubMed]
- The, S.V.; Snyder, R.; Tegeder, M. Targeting nitrogen metabolism and transport processes to improve plant nitrogen use efficiency. Front. Plant Sci. 2021, 11, 628336. [Google Scholar] [CrossRef]
- Liu, Y.; von Wirén, N. Ammonium as a signal for physiological and morphological responses in plants. J. Exp. Bot. 2017, 68, 2581–2592. [Google Scholar] [CrossRef]
- Guan, M.; Thomas, C.D.B.; Carsten, P.; Schjoerring, J.K. Cytosolic glutamine synthetase Gln1;2 is the main isozyme contributing to GS1 activity and can be up-regulated to relieve ammonium toxicity. Plant Physiol. 2016, 171, 1921–1933. [Google Scholar] [CrossRef]
- Pereira, E.G.; Sperandio, M.V.L.; Santos, L.A.; Bucher, C.A.; Coelho, C.P.; Fernandes, M.S. Rice varieties with contrasting nitrogen use efficiency present different expression of amino acid transporters and ammonium transporters. Arch. Agron. Soil Sci. 2022, 1–15. [Google Scholar] [CrossRef]
- Ranathunge, K.; El-kereamy, A.; Gidda, S.; Bi, Y.; Rothstein, S.J. AMT1;1 transgenic rice plants with enhanced NH4+ permeability show superior growth and higher yield under optimal and suboptimal NH4+ conditions. J. Exp. Bot. 2014, 65, 965–979. [Google Scholar] [CrossRef]
- Lee, S.; Marmagne, A.; Park, J.; Fabien, C.; Yim, Y.; Kim, S.J.; Kim, T.H.; Lim, P.O.; Masclaux Daubresse, C.; Nam, H.G. Concurrent activation of OsAMT1;2 and OsGOGAT1 in rice leads to enhanced nitrogen use efficiency under nitrogen limitation. Plant J. 2020, 103, 7–20. [Google Scholar] [CrossRef]
- Hui, J.; Liu, Z.; Duan, F.; Zhao, Y.; Li, X.; An, X.; Wu, X.; Yuan, L. Ammonium-dependent regulation of ammonium transporter ZmAMT1s expression conferred by glutamine levels in roots of maize. J. Integr. Agr. 2022, 21, 2413–2421. [Google Scholar] [CrossRef]
- Hachiya, T.; Inaba, J.; Wakazaki, M.; Sato, M.; Toyooka, K.; Miyagi, A.; Kawai-Yamada, M.; Sugiura, D.; Nakagawa, T.; Kiba, T.; et al. Excessive ammonium assimilation by plastidic glutamine synthetase causes ammonium toxicity in Arabidopsis thaliana. Nat. Commun. 2021, 12, 4944. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, Y.; Zhong, L.; Huang, X.; Su, W.; Liu, H.; Sun, G.; Song, S.; Chen, R. BcAMT1;5 Mediates Nitrogen Uptake and Assimilation in Flowering Chinese Cabbage and Improves Plant Growth When Overexpressed in Arabidopsis. Horticulturae 2023, 9, 43. https://doi.org/10.3390/horticulturae9010043
Zhu Y, Zhong L, Huang X, Su W, Liu H, Sun G, Song S, Chen R. BcAMT1;5 Mediates Nitrogen Uptake and Assimilation in Flowering Chinese Cabbage and Improves Plant Growth When Overexpressed in Arabidopsis. Horticulturae. 2023; 9(1):43. https://doi.org/10.3390/horticulturae9010043
Chicago/Turabian StyleZhu, Yunna, Lihua Zhong, Xinmin Huang, Wei Su, Houcheng Liu, Guangwen Sun, Shiwei Song, and Riyuan Chen. 2023. "BcAMT1;5 Mediates Nitrogen Uptake and Assimilation in Flowering Chinese Cabbage and Improves Plant Growth When Overexpressed in Arabidopsis" Horticulturae 9, no. 1: 43. https://doi.org/10.3390/horticulturae9010043
APA StyleZhu, Y., Zhong, L., Huang, X., Su, W., Liu, H., Sun, G., Song, S., & Chen, R. (2023). BcAMT1;5 Mediates Nitrogen Uptake and Assimilation in Flowering Chinese Cabbage and Improves Plant Growth When Overexpressed in Arabidopsis. Horticulturae, 9(1), 43. https://doi.org/10.3390/horticulturae9010043





