Abstract
Previous studies identified that strigolactones (SLs) and gibberellins (GAs) interacted when controlling branching in plant shoots, but the underlying mechanism remains unknown. qRT-PCR analysis suggested that the SL receptor gene CsDAD2 was significantly upregulated in the leaves, stems, and nodes of cucumber after treatment with 50 mg/L of GA3. Furthermore, the CsDAD2 gene was cloned and introduced into wild-type Arabidopsis plants via Agrobacterium-mediated transformation. For the CsDAD2-OE lines, the endogenous content of GA3 was subsequently higher at the seedling stage, with the number of primary cauline branches also significantly increased at the maturity stage compared with WT. Additionally, GA-related genes were up-regulated in the first inter-nodes and the third nodes of the CsDAD2-OE lines, thus indicating that GA was metabolically active in these tissues. The expression of the branch inhibitor gene AtBRC1 decreased at the seedling stage as well as at the maturity stage of the CsDAD2-OE lines. These findings suggest that CsDAD2 might have important functions in the interactions between GAs and SLs as it can promote the accumulation of GAs in plant nodes and suppress the expression of BRC1, hence increasing primary cauline branching.
1. Introduction
Plant architecture is a crucial agronomic characteristic that affects crop yield and biomass. In addition to being influenced by shoot branching, it also represents a defense strategy for higher plants to adapt to their surroundings and to avoid injury [,]. Within plants, axillary meristem and axillary bud development depend on the coordinated regulation of numerous signal pathways, triggered by environmental stimuli (temperature, light, nutrition, decapitation) as well as endogenous factors (plant hormones, sucrose) [,], with plant hormones playing an essential role for this purpose. For instance, strigolactones (SLs), which are carotenoid-derived metabolites generated in shoots as well as roots, play crucial roles in plant development [,,,,,], and mutations in their biosynthesis or signaling genes have been shown to promote plant branching [,,,,,]. Similarly, gibberellins (GAs) have been known as growth regulators for almost a century. In fact, many faulty phenotypes, including germination suppression, male sterility, dwarfing and increased tillering buds, are known to occur as a result of mutations in GA biosynthesis or signaling genes [,]). Furthermore, GAs are generally considered to inhibit stem branching [], as confirmed in studies on peas (Pisum sativum L.), rice (Oryza sativa L.), and Arabidopsis (Arabidopsis thaliana L.) where they negatively regulated shoot branching [,,]. However, in perennial strawberry (Fragaria × ananassa Duch.) and some other perennial woody plants, such as Jatropha (Jatropha. curcas L.) and hybrid aspen (Populus tremula L. × P. tremuloides Michx.), this hormone could positively regulate the growth of axillary buds [,,]. Hence, the regulation of plant branching by GAs differs between species. Surprisingly, both GA and SL signal transduction systems use α/β-hydrolase-derived receptors which are involved in the E3 ubiquitin ligase-mediated protein degradation process, thereby revealing that these pathways could have a common evolutionary basis []. Further studies have shown that GA3 treatment could actually lower the expression of SL biosynthesis genes, thereby reducing the level of SLs in roots. Hence, GAs could dampen the biosynthesis of SLs [,,]. The down-regulation of GA biosynthesis genes coupled with the up-regulation of GA inactivation genes in d17 and d14 mutants has also resulted in lower bioactive GA1 content compared with wild types []. Altogether, the above studies suggest that both SLs and GAs play vital roles in plant branching, with potential crosstalk occurring between them during the process.
Cucumber (Cucumis sativus L.) is a major vegetable crop consumed fresh or processed into pickles []. However, it is important to physically trim unnecessary branches to improve both its quality and yield. Thus, knowledge of the regulatory mechanism for the development of lateral branches in cucumber can be essential to assist breeding programs that yield cucumbers of various branching types. Branching is a quantitative trait [] whose molecular mechanism is not fully understood. Recently, it was found that the cucumber gene CsBRC1 inhibited lateral branch outgrowth by directly suppressing the functions of the auxin efflux carrier CsPIN3, thus leading to auxin accumulation in the axillary buds []. However, only few reports are available on the interactions between SLs and GAs in cucumber. Therefore, it is important that such interactions are better studied in order to better understand branching development in cucumber.
In this study, an SL receptor gene CsDAD2 was cloned from cucumber, with subsequent expression analysis showing that transcripts of CsDAD2 were higher in the roots and stems of cucumber as well as in the plant’s nodes following GA3 treatment. In addition, overexpression of CsDAD2 in Arabidopsis increased the number of primary cauline branches. The expression of AtGA3ox2 also decreased significantly, although AtGA2ox6 was up-regulated in the nodes at the maturity stage. Furthermore, the CsDAD2-OE lines showed an increase in endogenous GA3, with GA3 treatment significantly down-regulating and upregulating AtGA3ox2 and AtGA2ox6, respectively, during the seedling stage. The altered expression of GA-related genes not only suggested a higher GA3 content in nodes but also that BRC1 was maintained at low levels, resulting in increased branching in mature plants.
2. Materials and Methods
2.1. Plant Materials and Growth Conditions
The typical commercial cucumber ‘Ba er te’ and the wild-type Arabidopsis ecotype Columbia were used in this study. Plant materials were grown in pots containing black charcoal soil and under a 16 h:8 h photoperiod at 25 °C in a glasshouse. To examine the expression of SL-related genes, five-leaf cucumber plants were randomly divided into three groups: a control group sprayed with water, an A1 group sprayed with 50 mg/L of GA3 solution, and an A2 group sprayed with 100 mg/L of GA3 solution. Each plant had its leaves evenly sprayed on the adaxial and abaxial sides until drops of water were visible. Leaves, roots, stems, and nodes were taken 12 h after treatment for real-time quantitative reverse transcription-polymerase chain reaction (qRT-PCR), with three biological replicates set for each treatment.
2.2. Gene Isolation and Bioinformatics Analysis of CsDAD2
The cDNA sequence of the CsDAD2 gene was first obtained from phytozome v12 (http://www.Phytozome.net/soybean.php (accessed on 12 December 2018) by using cDNA sequences of the Arabidopsis homologs as query sequences (TBLASTN) before designing primers to clone the coding sequence of the gene from the cDNA of cucumber (Table S1). The gene was then inserted into a pGM-Simple-T Fast vector (TIANGEN, Beijing, China), with the latter subsequently used to transform the E. coli DH5α competent cells. Plasmids from the recombinant cells were eventually extracted and sent to Sangon Biotech (Shanghai, China) for identification. Pfam (http://pfam.xfam.org/search#tabview=tab0 (accessed on 15 October 2019)) was then used to determine the functional domain structure of CsDAD2. This protein’s tertiary structure was obtained with the Phyre2 tool (http://www.sbg.bio.ic.ac.uk/phyre2 (accessed on 13 October 2019)) and edited with Pymol 2.4.0 software. CsDAD2 and its closest homologs from other species were selected and multiple sequence alignment was carried out using the DNAMAN software (Lynnon Biosoft, San Ramon, California, USA). Based on the results, a phylogenetic tree was finally generated in MEGA 6.0 software, using the neighbor-joining (NJ) method with 1000 bootstrap replications. The accession numbers of DAD2 selected sequences are listed in Table S2.
2.3. Vector Construction and Plant Transformation
Fragments resulting from the digestion of pGM-CsDAD2 by KpnI and PstI restriction enzymes were ligated to pCAMBIA2300 which was previously digested with the same restriction enzymes in order to generate an over-expression construct 35S-CsDAD2. This recombinant vector was then transformed into E. coli DH5α competent cells for amplification before being introduced into Agrobacterium tumefaciens GV3101. The latter was subsequently used to infect wild-type (WT) Arabidopsis thaliana by the inflorescence infection method to generate CsDAD2-overexpression lines (CsDAD2-OE lines). Successfully transformed plants were screened using 1/2 MS medium plates containing 25 mg/L kanamycin and identified by PCR.
2.4. Treatment of Transgenic Arabidopsis
To examine the expression of GA-related genes, Arabidopsis from the CsDAD2-OE lines and wild-type ones were cultured in two forms. In the first case, the transgenic Arabidopsis and WT were grown in plates containing 1/2 MS medium. When the plants grew to eight leaves, whole seedlings were taken for qRT-PCR and the GA3 content was analyzed by HPLC. The remaining plants were then treated as follows: (1) Control group sprayed with water. (2) C1 group sprayed with 50 mg/L of GA3 solution. (3) C2 group sprayed with 100 mg/L of GA3 solution. (4) C3 group sprayed with 50 mg/L of paclobutrazol (PAC) solution. (5) C4 group sprayed with 100 mg/L of PAC solution. Whole seedlings of these five groups were taken for qRT-PCR three hours after treatment. In addition, plant materials grown in pots containing black charcoal soil had their roots, leaves, first nodes, first internodes, and third nodes taken for qRT-PCR when the plants grew to maturity. The number of rosette branches and primary cauline branches was also recorded. Three biological replicates were set for each treatment.
2.5. Extraction and Quantification of GA3 in Arabidopsis
After mixing approximately 0.1 g of the Arabidopsis seedlings with 1 mL of precooled reagent (methanol:water:acetic acid = 80:20:1), the mixture was allowed to leach for 12 h at 4 °C. Centrifugation was then performed at 8000 rpm for 10 min and after collecting the supernatant, the remaining residue was mixed with 0.5 mL of precooled reagent (methanol:water:acetic acid = 80:20:1). The above process was again repeated and both supernatants were combined. This was followed by the removal of any organic phase by blowing nitrogen at 40 °C. Residues were subsequently mixed with 0.5 mL of petroleum ether three times for decolorization at 60–90 °C before discarding the organic phase. After adjusting the pH to 2.8 with saturated aqueous citric acid, ethyl acetate extraction was carried out three times, with the organic phase merged together. The latter was dried by blowing nitrogen gas and after adding methanol (0.5 mL) for eddy shock dissolution, the sample was filtered.
HPLC analysis of the extracted hormones was performed using a Rigol L3000 system (Beijing Puyuan Jingdian Technology, Beijing, China). Different GA3 standard solutions (0.1, 0.5, 1.0, 2.0, and 5.0 μg/mL) were prepared using methanol as the solvent, with two replicates for each standard concentration. The HPLC, equipped with a Kromasil C18 reversed-phase chromatographic column (250 mm × 4.6 mm, 5 µm), was then performed under the following conditions: ultraviolet wavelength of 210 nm, a column temperature of 30 °C, mobile phase (A:0.1% phosphoric acid, B:methanol, A:B = 6.5:3.5), an injection volume of 10 μL, a flow rate of 1 mL/min, and a retention time of 40 min. After the baseline had stabilized, the mobile phase was run through the column and samples were added.
2.6. RNA Isolation and qRT-PCR
Total RNA was first extracted using an RNA pure plant kit (Kangwei Century Company, Beijing, China) according to the manufacturer’s instructions before synthesizing cDNA using the First Strand cDNA Synthesis Kit (TOYOBO, Shanghai, China). The cDNA was then diluted for qRT-PCR in three biological repeats according to the UltraSYBR Mixture kit (Kangwei Century Company, Beijing, China). Selected primers for the qRT-PCR are shown in Tables S3–S4. The cycling conditions, as applied on a 7500 Real-Time PCR System (Applied Biosystems, Waltham, MA, USA), were as follows: 94 °C for 20 s, 56 °C for 20 s, and 72 °C for 30 s. In this case, actin was used as an internal reference for the gene expression analysis. The 2−ΔΔCt method was eventually applied to assess the relative expression level of each gene.
2.7. Statistical Analysis
Statistical analysis of the data was performed using Microsoft Office Excel 2016 and IBM SPSS Statistics 20. All results were presented as the mean ± standard deviation (SD) of three replicates (n = 3). Gene expression levels were statistically evaluated by analysis of test results. Other data were statistically evaluated by analysis of variance (ANOVA), and the means were compared by Duncan’s multiple comparisons. Significant differences between different treatments were determined at p < 0.05 and p < 0.01 significance levels. Charts were generated with Origin 2019, Microsoft PowerPoint, and R (version 4.2.2) software.
3. Results
3.1. CsDAD2 Cloning and Bioinformatics Analysis
The expression of the SL biosynthesis gene CsCCD7, receptor genes CsD14 and CsDAD2, and branching regulatory genes CsTCP2 and CsTCP18 (BRC1) were analyzed in cucumber which had been treated with different concentrations of GA3 in order to assess whether the GAs had any effects on the SLs and on the development of branches. The results showed that GA3 treatment influenced the expression of both the SL-related genes and the branching regulatory genes. In particular, increased expression of CsDAD2 was noted in leaves, stems, and nodes after two concentrations of GA3 treatment (Figure S1). Therefore, CsDAD2 was selected for subsequent experiments to explore the molecular mechanism of the interaction between SLs and GAs.
The CDS region of the cucumber CsDAD2 gene was obtained by PCR amplification and its sequence of 908 bp encoded a protein of 276 aa, with the 29–269 aa being the functional domain of abhydrolase-6. Thus, CsDAD2 belonged to the alpha/beta hydrolase family (Figure 1a). A total of 271 aa residues were modeled, with 100% confidence and using a single template with the highest score. The image of the tertiary structure of CsDAD2 was colored by a rainbow from the N to the C end (Figure 1b). After protein sequence alignment, it was found that the CsDAD2 protein sequence had a large number of conserved sites with homologous proteins from other species. In particular, the similarity with Benincasa hispida reached 87.73% (Figure 1c). A phylogenetic tree was then built using CsDAD2 and the homologous proteins from other species. In this case, it was found that this protein had a very close evolutionary relationship with Benincasa hispida, while being further from Arabidopsis thaliana (Figure 1d).
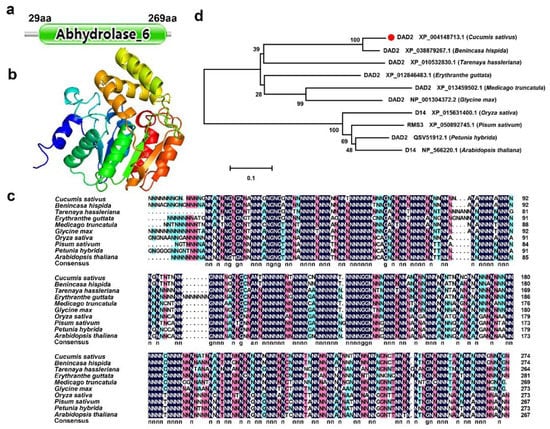
Figure 1.
Sequence analysis of CsDAD2. (a) The functional domain of CsDAD2; (b) the tertiary structure of CsDAD2. Using a single template with the highest score, a total of 271 amino acid residues were modeled with 100% confidence. This picture is colored by the end of the rainbow from N to C; (c) sequence alignment of CsDAD2 and its homologs. The black, pink, and blue backgrounds represent 100%, 75%, and 50% similarities; (d) the phylogenetic relationship between CsDAD2 and its orthologs in other plants. Supplementary Table S2 lists the accession number of DAD2.
3.2. Analysis of CsDAD2 Expression in Different Tissues
The expression level of CsDAD2 in different tissues of cucumber (Figure 2a) was analyzed using gene-specific primers. The results showed that transcripts of CsDAD2 were present in leaves, roots, stems, nodes and cotyledons, but the expression levels were higher in roots and stems, where they were 85.48- and 1.38-fold higher compared with those in leaves. On the other hand, the expression levels in nodes and cotyledons were 0.41- and 1.09-fold higher than those in leaves (Figure 2b). These results indicated that CsDAD2 was mainly expressed in the roots of cucumber.
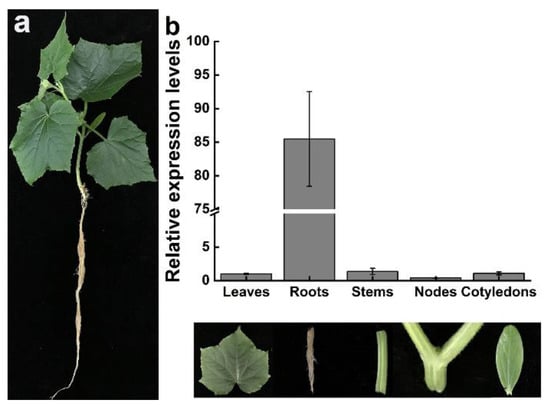
Figure 2.
Spatio-temporal expression of CsDAD2 in different tissues using qRT-PCR. (a) Cucumber plant morphology; (b) relative expression of CsDAD2 in leaves, roots, stems, nodes, and cotyledons.
3.3. Construction of the Plant Expression Vector, Generation of Transgenic Arabidopsis, and Branching in CsDAD2-OE Lines
To further characterize the functions of CsDAD2, two T3 homozygous transgenic Arabidopsis strains (#1 and #2) were generated to overexpress the CsDAD2 gene under the control of the strong constitutive CaMV35S promoter (Figure 3a,c).
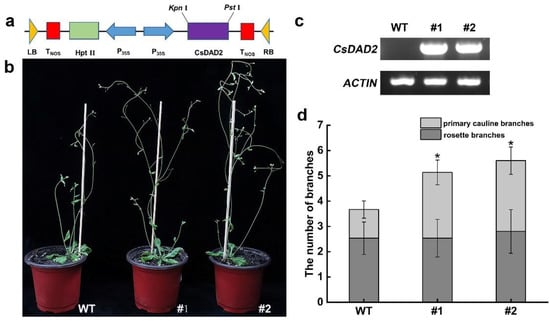
Figure 3.
Identification and phenotypic analysis of CsDAD2-overexpressed Arabidopsis. (a) Schematic representation of a construct used for Agrobacterium tumefaciens-mediated transformation with the CsDAD2 gene; (b) branching phenotypes of wild-type and representative individuals of two independent 35s::CsDAD2 lines; (c) expression levels of CsDAD2 in WT and two 35s::CsDAD2 lines as determined by PCR; (d) counts of the number of rosette branches and primary cauline branches, with 15 plants used per genotype to analyze the plants’ phenotypes (Student’s t test: * p < 0.05).
The number of primary cauline branches in the two transgenic lines increased significantly compared with WT; the increase in line #1 and #2 was 2.6- and 2.8-fold higher than in WT (Figure 3b,d). Line #2 was selected for subsequent experiments.
3.4. The Expression of GA-Related Genes and the GA3 Content in CsDAD2-OE Lines
The expression of GA-related and bud dormancy genes was determined in transgenic lines at the eight-leaf stage for examining the effects of the CsDAD2 gene on GA signal transduction. A subset of genes consisting of the GA biosynthesis genes AtGA20ox1, AtGA20ox2, and AtGA3ox2; the GA signaling genes AtGID1a, AtDELLA genes (AtRGA1, AtRGL), and AtSLY1; the GA degradation genes AtGA2ox2 and AtGA2ox6; and the bud dormancy gene AtBRC1 were studied. The results showed that the gene expression for GA biosynthesis was reduced, with expression levels of AtGA20ox1 and AtGA20ox2 being 0.36- and 0.08-fold that of WT (Figure 4a). Similarly, transcripts of the GA degradation genes AtGA2ox2 and AtGA2ox6 were reduced (Figure 4b). Overall, the expression of GA signaling genes was downregulated, leading to obvious decreases in AtGID1a and AtRGL1 expression (Figure 4c). In the case of the bud dormancy gene AtBRC1, the expression level was 0.47-fold that of WT (Figure 4b).
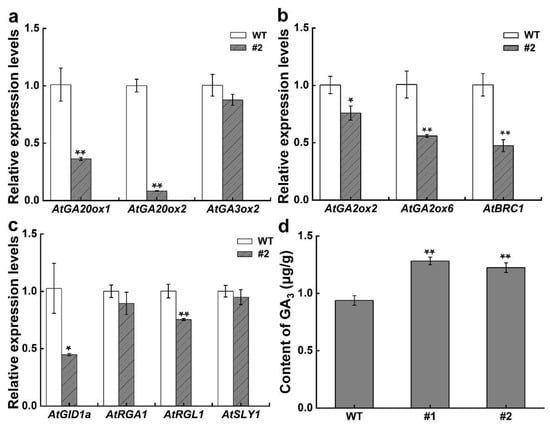
Figure 4.
Expression of GA-related genes and the GA3 content in WT as well as in CsDAD2-overexpressed Arabidopsis. Expression of GA-related genes in WT and #2 was analyzed using qRT-PCR while the GA3 content in WT, #1, and #2 was determined by HPLC at the eight-leaf stage. (a) GA biosynthesis genes; (b) GA degradation genes and bud dormancy gene AtBRC1; (c) GA signaling genes; (d) endogenous content of GA3. Data represent the mean ± standard deviation (SD) of three biological replicates (Student’s t test: * p < 0.05, ** p < 0.01).
Furthermore, GA3 content was significantly increased in the CsDAD2-OE line, with 0.31- and 0.37-fold increases in #1 and #2 compared with WT (Figure 4d).
3.5. Analysis of GA-Related Gene Expression in Different Tissues of CsDAD2-OE Lines
In different tissues of the CsDAD2-OE lines, the transcript levels of GA-related genes were analyzed to assess whether GA synthesis and signal transduction were regulated by CsDAD2 locally (Figure 5). It was found that the transcripts of GA-related genes generally decreased in the roots of transgenic Arabidopsis compared with WT. In the case of leaves, the expression of the genes also decreased, except for AtGA2ox6 and AtRGA1 for which increased expression was noted. In the first nodes, the expression of the GA degradation genes AtGA2ox2 and AtGA2ox6 and the signaling gene AtRGA1 increased, while the expression of all the GA biosynthesis genes as well as other signaling genes decreased. However, overall, the expression of the GA biosynthesis genes AtGA20ox1 and AtGA20ox2; the GA degradation ones, AtGA2ox2 and AtGA2ox6; and the signaling genes AtGID1a and AtRGA1 were up-regulated in the first inter-nodes as well as the third nodes of transgenic Arabidopsis compared with WT (Figure 5). The transcript level of the bud dormancy gene AtBRC1 decreased in the first and third nodes (Figure S2).
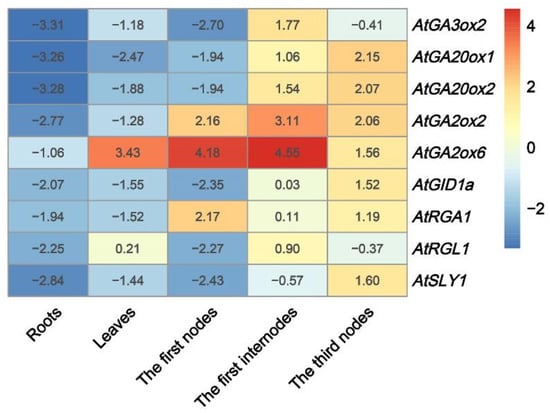
Figure 5.
Expression of GA-related genes after CsDAD2 overexpression in different tissues of Arabidopsis. The numbers represent the log2fold-change of gene expression values between #2 and WT. Positive and negative numbers respectively represent up-regulated and down-regulated gene expression in #2 relative to WT.
3.6. Analysis of GA-Related and SL-Related Gene Expression in CsDAD2-OE Lines after GA3 and PAC Treatments
To explore the molecular mechanism of interactions between CsDAD2 and GAs further, the transcript levels of GA-related genes and SL-related genes were assayed in CsDAD2-OE lines after GA3 and PAC (an inhibitor of GA3 biosynthesis) treatment. The expression of GA synthesis genes was significantly decreased in transgenic lines treated with different concentrations of GA3 compared with the control (CK/#2). Conversely, AtGA3ox2 was significantly more expressed after PAC treatment at two concentrations (Figure 6a). Similarly, the transcript levels of the GA degradation genes AtGA2ox2 and AtGA2ox6 were significantly increased with the latter showing a 6.71-fold increase compared with CK at the lower concentrations of GA3. However, the expression of AtGA2ox6 decreased significantly after PAC treatment (Figure 6b). Furthermore, the expression of AtRGA1 increased during treatment with lower concentrations of GA3, while that of other signal transduction genes were suppressed. The expression of AtGID1a increased 2.75-fold after treatment with lower concentrations of PAC, although AtRGA1 increased significantly after treatment with PAC; however, the gene expression for AtRGL1 and AtSLY1 decreased considerably (Figure 6c). These results suggested that exogenous GA and PAC may cause the feedback regulation mechanism response of the GA signal in CsDAD2-OE lines.
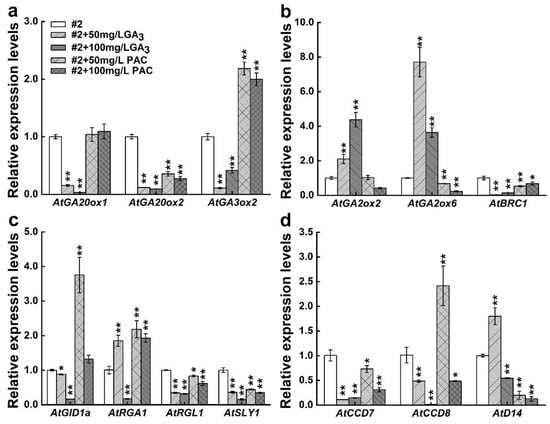
Figure 6.
Expression of GA-related genes and SL-related ones in CsDAD2-overexpressed Arabidopsis treated with GA3 and PAC. The expression of GA-related and SL-related genes was detected by qRT-PCR in CsDAD2-overexpressed Arabidopsis at the eight-leaf stage after three hours of GA3 and PAC treatment (0, 50, 100 mg/L GA3 and 50, 100 mg/L PAC). (a) GA biosynthesis genes; (b) GA degradation genes; and bud dormancy gene AtBRC1; (c) GA signaling genes; (d) SL related genes. Data represent the mean ± SD of three biological replicates (Student’s t test: * p < 0.05, ** p < 0.01).
The expression of the SL biosynthesis genes AtCCD7 and AtCCD8 significantly decreased in transgenic lines treated with the two concentrations of GA3 compared with CK. Moreover, the transgenic plants showed a significant increase in the expression of the SL signaling gene AtD14 after treatment with lower concentrations of GA3. However, a significant decrease was observed at the higher concentrations. The expression of AtCCD8 increased by 1.42-fold at lower concentrations of PAC treatment, while the expression of other SL-related genes significantly decreased (Figure 6d). The expression levels of bud dormancy gene AtBRC1 was significantly decreased after GA3 and PAC treatment (Figure 6b).
4. Discussion
A hormone network regulates shoot branching, and in this process strigolactones, auxin, and cytokinin were found to interact, thereby leading to the proposal of two models, namely, the auxin canalization model [,,] and the messenger model [], both of which can be used to explain the development of shoot branches. However, GAs also have substantial regulatory effects on branching, but studies on their relationships with other hormones are relatively scarce. It was found that the genes that encode the SL receptors, D14a and D14b, were strongly upregulated by GA3 and GA4 in hybrid aspen []. Based on the literature data, it was hypothesized that SL receptor genes could have a role in the interactions between GAs and SLs. In this study, the expression of the SL-related gene CsCCD7 and SL receptor genes (CsD14 and CsDAD2) were analyzed in cucumber after GA3 treatment. Surprisingly, two concentrations of GA3 significantly promoted the expression of CsDAD2 in cucumber stems and nodes (Figure S1). Therefore, this gene was subsequently studied due to its potential role in mediating the interactions between GAs and SLs. DAD2 is a homolog of D14 and belongs to the alpha/beta hydrolase fold superfamily (Figure 1a). It can recognize SLs for hydrolysis, thereby guiding the progress of the SL signaling [,,]. PhDAD2 expression was also higher in the leaves and axillary buds of wild petunia compared with other tissues []. It was further found that CsDAD2 expression was higher in roots compared with other tissues of cucumber (Figure 2b). Hence, the transcript levels of this gene in different tissues could be related to plant species or the developmental stage of plants. Previous reports have shown that Atd14 mutants in Arabidopsis could display an increase in rosette branches compared with wild-type ones [,], while hvd14 mutants in Hordeum vulgare produced a higher number of tillers in comparison with the wild-type parent cultivar []. However, in this study, it was found that the number of rosette branches remained unchanged, but the primary cauline branches actually increased in the CsDAD2-OE lines (Figure 3). The above results suggested that CsDAD2 overexpression may have potential side effects, which lead to an increase in the number of primary stem branches.
Interactions through the regulation of hormone biosynthesis has been reported in some studies, although direct evidence for this has been rarely presented [,,]. In this study, the expression of GA-related genes in CsDAD2-OE lines was reduced (Figure 4a–c), and the GA3 content increased. This might have been because GA metabolism was at a low level, but this was facilitated by the accumulation of endogenous GA3 during the seedling stage in the CsDAD2-OE lines. It is proposed that SLs act locally in axillary buds by upregulating the expression of the BRC1, which is well-established as a regulator of bud outgrowth. Indeed, brc1 mutants display increased SL-resistance to branching [,,]. It has been suggested that SLs promote the expression of PsBRC1 through a signal sensed by the receptor D14 []. This study found that the overexpression of CsDAD2 inhibited the expression of AtBRC1 in Arabidopsis seedlings (Figure 4b), and this might be related to increased endogenous GA3 content. After further analyzing the expression of GA-related genes in various tissues of the CsDAD2-OE lines, it was found that the expression levels of AtGA3ox2 involved in GA biosynthesis were low in the first and third nodes, but those of the GA degradation genes AtGA2ox2 and AtGA2ox6 increased (Figure 5), thereby indicating that CsDAD2 retained GA biosynthesis and promoted GA degradation in the nodes of the transgenic plants. It was shown that high expression of the AtGA20x gene resulted in low levels of endogenous active GAs in Arabidopsis and Paspalum notatum []. These results could lead to a lower GA content, but it is likely that higher content of endogenous GA could result in feedback regulation of GA production in the nodes of transgenic Arabidopsis. AtBRC1 expression was lower in the first and third nodes of CsDAD2-OE lines compared with WT at the maturity stage (Figure S2). This result was consistent with the decreased expression of AtBRC1 at the seedling stage, indicating that the transgenic Arabidopsis may also contain higher concentrations of GA3 at the maturity stage. Ultimately, decreased AtBRC1 expression led to the increase in primary cauline branches of the CsDAD2-OE lines (Figure 3c). Hence, the results suggested that the potential side effect of CsDAD2 overexpression led to reduced AtBRC1 expression.
A previous study showed that RGA would not be degraded by D14 in an SL-dependent manner. Yet, the interaction between D14 and DELLA was SL-dependent [,]. Based on this, it was speculated that CsDAD2 may interact with RGA. This result was confirmed in the CsDAD2-OE lines at the maturity stage where the overexpression of CsDAD2 promoted the expression of AtRGA1 in the first and third nodes (Figure 5). Therefore, it is proved that DAD2 positively regulates the expression of RGA1.
Analysis of the expression of GA-related genes in CsDAD2-OE line seedlings indicated that GA3 led to a significant decrease in the expression of the biosynthesis genes, while PAC promoted the expression of AtGA3ox2 (Figure 6a). In addition, the GA degradation genes AtGA2ox2 and AtGA2ox6 significantly increased in expression after GA3 treatment, and AtGA2ox6 significantly decreased after PAC treatment (Figure 6b). These results were similar to previous reports where GA3 strongly upregulated GA2ox genes [,]. The results, therefore, showed that exogenous GA3 and PAC could induce feedback regulation on the endogenous content of GAs, especially through AtGA3ox2 and AtGA2ox6, in transgenic plants. Changes in these two genes after GA3 treatment were consistent with those in transgenic Arabidopsis nodes (Figure 5), suggesting that high concentrations of GA3 might be present at this site. The application of GA3 decreased the expression of CsBRC1 in the CsDAD2-OE lines (Figure 6b). Reduced AtBRC1 could be the cause of GA3 content in the nodes of transgenic plants. However, GAs were negative regulators of rice tillering, and the expression of TB1 (BRC1) was increased with the use of GA3 in wild-type rice plants []. Therefore, the regulation of plant branching by GAs differs between species. Overexpression of CsDAD2 is thought to impact other regulatory factors such as cytokinin, thus reducing the expression of AtBRC1 [,,,]. These results suggested that a complex regulatory network could be formed to inhibit the expression of AtBRC1 including the interaction between DAD2 and GA, thereby improving the branching process.
In fact, GA3 treatment was shown to reduce the expression of SL synthesis genes such as OsD10/CCD8 and OsD17/CCD7, thereby reducing the level of SLs in rice roots [,]. It was also found that the expression of AtCCD7 and AtCCD8 decreased significantly in CsDAD2-OE lines after GA3 treatment, thus revealing that gibberellin has an effect on SL biosynthesis in CsDAD2-overexpressed plants. Simply lowering the concentration of PCA further promoted the expression of AtCCD8, unlike the GA3 treatment, whereas the expression of AtCCD7 still decreased (Figure 6d). Therefore, AtCCD8 and AtCCD7 had different effects on PAC, especially in terms of the concentrations. The expression of D14a and D14b was increased in hybrid aspen fed with GA3 at concentrations of 10 µm []. Similarly, AtD14 expression increased significantly in CsDAD2-OE lines after treatment with lower concentrations of GA3, although the expression significantly decreased after PAC treatment at both concentrations. However, it was significantly decreased after treatment with higher concentrations of GA3 (Figure 6d). Therefore, the expression of AtD14 in the CsDAD2-OE lines did display unsimilar responses at different concentrations of GA3.
5. Conclusions
In summary, this study showed that CsDAD2 could be involved in the interactions between GAs and SLs. These interactions were discovered using CsDAD2-overexpressed lines. CsDAD2 might promote GA metabolism at a low level and then facilitates the accumulation of endogenous GA3 during the seedling stage. Furthermore, CsDAD2 overexpression might promote the accumulation of GA3 content in plant nodes, and a surge of GA3 leads to the decreased expression of the GA biosynthesis gene AtGA3ox2 and the elevated expression of the GA degradation gene AtGA2ox6, before ultimately leading to a decrease in the expression of the branching inhibitor gene BRC1, thereby promoting primary cauline branches at the maturity stage.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae9010023/s1.
Author Contributions
Conceptualization, Y.C. and Y.D.; methodology, R.P.; software, Q.L. and R.Z.; validation, Y.C. and M.L.; formal analysis, C.C.; investigation, Y.C. and X.J.; resources, X.J. and C.C.; data curation, Y.C. and X.J.; writing—original draft preparation, Y.C. and Y.D., writing—review and editing, X.J. and C.C.; visualization, Y.C. and Q.L.; supervision, X.J. and C.C.; project administration, X.J.; funding acquisition, X.J. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Natural Science Foundation of Heilongjiang Province (LH2021C052) and the 2021 Heilongjiang University Student Innovation Training Program (202110231012).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Nicotra, A.B.; Atkin, O.K.; Bonser, S.P.; Davidson, A.M.; Finnegan, E.J.; Mathesius, U.; Poot, P.; Purugganan, M.D.; Richards, C.L.; Valladares, F.; et al. Plant phenotypic plasticity in a changing climate. Trends. Plant Sci. 2010, 15, 684–692. [Google Scholar] [CrossRef]
- Drummond, R.S.; Janssen, B.J.; Luo, Z.; Oplaat, C.; Ledger, S.E.; Wohlers, M.W.; Snowden, K.C. Environmental Control of Branching in Petunia. Plant Physiol. 2015, 168, 735–751. [Google Scholar] [CrossRef]
- Mcsteen, P.; Leyser, O. Shoot branching. Annu. Rev. Plant. Biol. 2005, 56, 353–374. [Google Scholar] [CrossRef]
- Puig, J.; Pauluzzi, G.; Guiderdoni, E.; Gantet, P. Regulation of shoot and root development through mutual signaling. Mol. Plant 2012, 5, 974–983. [Google Scholar] [CrossRef]
- Napoli, C. Highly Branched Phenotype of the Petunia dad1-1 Mutant Is Reversed by Grafting. Plant Physiol. 1996, 111, 27–37. [Google Scholar] [CrossRef]
- Beveridge, C.A.; Symons, G.M.; Murfet, I.C.; Ross, J.J.; Rameau, C. The rms1 mutant of pea has elevated indole-3-acetic acid levels and reduced root-sap zeatin riboside content but increased branching controlled by graft-transmissible signal(s). Plant Physiol. 1997, 115, 1251–1258. [Google Scholar] [CrossRef]
- Gomez-Roldan, V.; Fermas, S.; Brewer, P.B.; Puech-Pagès, V.; Dun, E.A.; Pillot, J.-P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.-C.; et al. Strigolactone inhibition of shoot branching. Nature 2008, 455, 189–194. [Google Scholar] [CrossRef]
- Umehara, M.; Hanada, A.; Yoshida, S.; Akiyama, K.; Arite, T.; Takeda-Kamiya, N.; Magome, H.; Kamiya, Y.; Shirasu, K.; Yoneyama, K.; et al. Inhibition of shoot branching by new terpenoid plant hormones. Nature 2008, 455, 195–200. [Google Scholar] [CrossRef]
- Ruyter-Spira, C.; Kohlen, W.; Charnikhova, T.; Zeijl, A.V.; Bezouwen, L.V.; Ruijter, N.D.; Cardoso, C.; Lopez-Raez, J.A.; Matusova, R.; Bours, R.; et al. Physiological effects of the synthetic strigolactone analog GR24 on root system architecture in Arabidopsis: Another belowground role for strigolactones? Plant Physiol. 2011, 155, 721–734. [Google Scholar] [CrossRef]
- Sun, H.; Tao, J.; Hou, M.; Huang, S.; Chen, S.; Liang, Z.; Xie, T.; Wei, Y.; Xie, X.; Yoneyama, K.; et al. A strigolactone signal is required for adventitious root formation in rice. Ann. Bot. 2015, 115, 1155–1162. [Google Scholar] [CrossRef]
- Arite, T.; Umehara, M.; Ishikawa, S.; Hanada, A.; Maekawa, M.; Yamaguchi, S.; Kyozuka, J. d14, a strigolactone-insensitive mutant of rice, shows an accelerated outgrowth of tillers. Plant Cell Physiol. 2009, 50, 1416–1424. [Google Scholar] [CrossRef]
- Drummond, R.S.M.; Martinez-Sanchez, N.M.; Janssen, B.J.; Templeton, K.R.; Simons, J.L.; Quinn, B.D.; Karunairetnam, S.; Snowden, K.C. Petunia hybrida CAROTENOID CLEAVAGE DIOXYGENASE7 is involved in the production of negative and positive branching signals in petunia. Plant Physiol. 2009, 151, 1867–1877. [Google Scholar] [CrossRef]
- Dun, E.A.; Brewer, P.B.; Beveridge, C.A. Strigolactones: Discovery of the elusive shoot branching hormone. Trends Plant Sci. 2009, 14, 364–372. [Google Scholar] [CrossRef]
- Beveridge, C.A.; Kyozuka, J. New genes in the strigolactone-related shoot branching pathway. Curr. Opin. Plant Biol. 2010, 13, 34–39. [Google Scholar] [CrossRef]
- Domagalska, M.A.; Leyser, O. Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 2011, 12, 211–221. [Google Scholar] [CrossRef]
- Waters, M.T.; Brewer, P.B.; Bussell, J.D.; Smith, S.M.; Beveridge, C.A. The Arabidopsis ortholog of rice DWARF27 acts upstream of MAX1 in the control of plant development by strigolactones. Plant Physiol. 2012, 159, 1073–1085. [Google Scholar] [CrossRef]
- Silverstone, A.L.; Chang, C.-W.; Krol, E.; Sun, T.-P. Developmental regulation of the gibberellin biosynthetic gene GA1 in Arabidopsis thaliana. Plant J. 1997, 12, 9–19. [Google Scholar] [CrossRef]
- Lo, S.-F.; Yang, S.-Y.; Chen, K.-T.; Hsing, Y.-I.; Zeevaart, J.A.; Chen, L.-J.; Yu, S.-M. A Novel Class of Gibberellin 2-Oxidases Control Semidwarfism, Tillering, and Root Development in Rice. Plant Cell 2008, 20, 2603–2618. [Google Scholar] [CrossRef]
- Yan, Y.; Zhao, N.; Tang, H.; Gong, B.; Shi, Q. Shoot branching regulation and signaling. Plant Growth Regul. 2020, 92, 131–140. [Google Scholar] [CrossRef]
- Mauriat, M.; Sandberg, L.G.; Moritz, T. Proper gibberellin localization in vascular tissue is required to control auxin-dependent leaf development and bud outgrowth in hybrid aspen. Plant J. 2011, 67, 805–816. [Google Scholar] [CrossRef]
- Zawaski, C.; Busov, V.B. Roles of gibberellin catabolism and signaling in growth and physiological response to drought and short-day photoperiods in populus trees. PLoS ONE 2014, 9, e86217. [Google Scholar] [CrossRef] [PubMed]
- Rameau, C.; Bertheloot, J.; Leduc, N.; Andrieu, B.; Foucher, F.; Sakr, S. Multiple pathways regulate shoot branching. Front. Plant Sci. 2015, 5, 741. [Google Scholar] [CrossRef] [PubMed]
- Rinne, P.L.H.; Welling, A.; Vahala, J.; Ripel, L.; Ruonala, R.; Kangasjärvi, J.; van der Schoot, C. Chilling of Dormant Buds Hyperinduces FLOWERING LOCUS T and Recruits GA-Inducible 1,3-β-Glucanases to Reopen Signal Conduits and Release Dormancy in Populus. Plant Cell 2011, 23, 130–146. [Google Scholar] [CrossRef] [PubMed]
- Ni, J.; Gao, C.; Chen, M.-S.; Pan, B.-Z.; Ye, K.; Xu, Z.-F. Gibberellin promotes shoot branching in the perennial woody plant Jatropha curcas. Plant Cell Physiol. 2015, 56, 1655–1666. [Google Scholar] [CrossRef] [PubMed]
- Tenreira, T.; Lange, M.J.P.; Lange, T.; Bres, C.; Labadie, M.; Monfort, A.; Hernould, M.; Rothan, C.; Denoyes, B. A Specific Gibberellin 20-Oxidase Dictates the Flowering-Runnering Decision in Diploid Strawberry. Plant Cell 2017, 29, 2168–2182. [Google Scholar] [CrossRef]
- Lantzouni, O.; Klermund, C.; Schwechheimer, C. Largely additive effects of gibberellin and strigolactone on gene expression in Arabidopsis thaliana seedlings. Plant J. 2017, 92, 924–938. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Muszynska, A. In silico analysis of the genes encoding proteins that are involved in the biosynthesis of the RMS/MAX/D pathway revealed new roles of strigolactones in plants. Int. J. Mol. Sci. 2015, 16, 6757–6782. [Google Scholar] [CrossRef]
- Ito, S.; Yamagami, D.; Umehara, M.; Hanada, A.; Yoshida, S.; Sasaki, Y.; Yajima, S.; Kyozuka, J.; Ueguchi-Tanaka, M.; Matsuoka, M.; et al. Regulation of strigolactone biosynthesis by gibberellin signaling. Plant Physiol. 2017, 174, 1250–1259. [Google Scholar] [CrossRef] [PubMed]
- Mori, N.; Nomura, T.; Akiyama, K. Identification of two oxygenase genes involved in the respective biosynthetic pathways of canonical and non-canonical strigolactones in Lotus japonicus. Planta 2020, 251, 40. [Google Scholar] [CrossRef] [PubMed]
- Zou, X.; Wang, Q.; Chen, P.; Yin, C.; Lin, Y. Strigolactones regulate shoot elongation by mediating gibberellin metabolism and signaling in rice (Oryza sativa L.). J. Plant Physiol. 2019, 237, 72–79. [Google Scholar] [CrossRef]
- You, Y.; Zheng, Y.; Wang, J.; Chen, G.; Li, S.; Shao, J.; Qi, G.; Xu, F.; Wang, G.; Chen, Z.-H.; et al. Molecular evolution and genome-wide analysis of the SBP-box family in cucumber (Cucumis sativas). Plant Growth Regul. 2020, 93, 175–187. [Google Scholar] [CrossRef]
- Li, B.; Gao, J.; Chen, J.; Wang, Z.; Shen, W.; Yi, B.; Wen, J.; Ma, C.; Shen, J.; Fu, T.; et al. Identification and fine mapping of a major locus controlling branching in Brassica napus. Theor. Appl. Genet. 2020, 133, 771–783. [Google Scholar] [CrossRef] [PubMed]
- Shen, J.; Zhang, Y.; Ge, D.; Wang, Z.; Song, W.; Gu, R.; Che, G.; Cheng, Z.; Liu, R.; Zhang, X. CsBRC1 inhibits axillary bud outgrowth by directly repressing the auxin efflux carrier CsPIN3 in cucumber. Proc. Natl. Acad. Sci. USA 2019, 116, 17105–17114. [Google Scholar] [CrossRef] [PubMed]
- Bennett, T.; Leyser, O. Something on the side: Axillary meristems and plant development. Plant Mol. Biol. 2006, 60, 843–854. [Google Scholar] [CrossRef]
- Leyser, O. The control of shoot branching: An example of plant information processing. Plant Cell Environ. 2009, 32, 694–703. [Google Scholar] [CrossRef]
- Ravazzolo, L.; Boutet-Mercey, S.; Perreau, F.; Forestan, C.; Varotto, S.; Ruperti, B.; Quaggiotti, S. Strigolactones and auxin coop-erate to regulate maize root development and response to nitrate. Plant Cell Physiol. 2021, 62, 610–623. [Google Scholar] [CrossRef]
- Brewer, P.B.; Dun, E.A.; Ferguson, B.J.; Rameau, C.; Beveridge, C.A. Strigolactone acts downstream of auxin to regulate bud outgrowth in pea and Arabidopsis. Plant Physiol. 2009, 150, 482–493. [Google Scholar] [CrossRef]
- Katyayini, N.U.; Rinne, P.; Tarkowska, D.; Strnad, M.; Schoot, C. Dual role of gibberellin in perennial shoot branching: Inhibition and activation. Front. Plant Sci. 2020, 11, 736. [Google Scholar] [CrossRef]
- Gaiji, N.; Cardinale, F.; Prandi, C.; Bonfante, P.; Ranghino, G. The computational-based structure of Dwarf14 provides evidence for its role as potential strigolactone receptor in plants. BMC Res. Notes 2012, 5, 307. [Google Scholar] [CrossRef]
- Yao, R.; Li, J.; Xie, D. Recent advances in molecular basis for strigolactone action. Sci. China Life Sci. 2017, 61, 277–284. [Google Scholar] [CrossRef]
- Lee, H.W.; Sharma, P.; Janssen, B.J.; Drummond, R.S.M.; Luo, Z.; Hamiaux, C.; Collier, T.; Allison, J.R.; Newcomb, R.D.; Snowden, K.C. Flexibility of the petunia strigolactone receptor DAD2 promotes its interaction with signaling partners. J. Biol. Chem. 2020, 295, 4181–4193. [Google Scholar] [CrossRef] [PubMed]
- Hamiaux, C.; Drummond, R.S.M.; Janssen, B.J.; Ledger, S.E.; Cooney, J.M.; Newcomb, R.D.; Snowden, K.C. DAD2 is an α/β hydrolase likely to be involved in the perception of the plant branching hormone, strigolactone. Curr. Biol. 2012, 22, 2032–2036. [Google Scholar] [CrossRef] [PubMed]
- Chevalier, F.; Nieminen, K.; Sánchez-Ferrero, J.C.; Rodríguez, M.L.; Chagoyen, M.; Hardtke, C.S.; Cubas, P. Strigolactone Promotes Degradation of DWARF14, an α/β Hydrolase Essential for Strigolactone Signaling in Arabidopsis. Plant Cell 2014, 26, 1134–1150. [Google Scholar] [CrossRef] [PubMed]
- Seto, Y.; Yasui, R.; Kameoka, H.; Tamiru, M.; Cao, M.; Terauchi, R.; Sakurada, A.; Hirano, R.; Kisugi, T.; Hanada, A.; et al. Strigolactone perception and deactivation by a hydrolase receptor DWARF14. Nat. Commun. 2019, 10, 1189–1190. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M.; Gruszka, D.; Tylec, P.; Szarejko, I. Identification and functional analysis of the HvD14 gene involved in strigolactone signaling in Hordeum vulgare. Physiol. Plant. 2016, 158, 341–355. [Google Scholar] [CrossRef]
- Hedden, P.; Thomas, S.G. Gibberellin biosynthesis and its regulation. Biochem. J. 2012, 444, 11–25. [Google Scholar] [CrossRef]
- Liu, J.; Mehdi, S.; Topping, J.; Friml, J.; Lindsey, K. Interaction of PLS and PIN and hormonal crosstalk in Arabidopsis root development. Front. Plant Sci. 2013, 4, 75. [Google Scholar] [CrossRef]
- Aerts, N.; Pereira Mendes, M.; Van Wees, S.C.M. Multiple levels of crosstalk in hormone networks regulating plant defense. Plant J. 2021, 105, 489–504. [Google Scholar] [CrossRef]
- Borghi, L.; Liu, G.-W.; Emonet, A.; Kretzschmar, T.; Martinoia, E. The importance of strigolactone transport regulation for symbiotic signaling and shoot branching. Planta 2016, 243, 1351–1360. [Google Scholar] [CrossRef]
- Aguilar-Martínez, J.A.; Poza-Carrión, C.; Cubas, P. Arabidopsis BRANCHED1 Acts as an Integrator of Branching Signals within Axillary Buds. Plant Cell 2007, 19, 458–472. [Google Scholar] [CrossRef]
- Confraria, A.; Muñoz-Gasca, A.; Ferreira, L.; Baena-González, E.; Cubas, P. Shoot branching phenotyping in Arabidopsis and tomato. In Environmental Responses in Plants; Methods in Molecular Biology; Duque, P., Szakonyi, D., Eds.; Humana: New York, NY, USA, 2022; Volume 2494, pp. 47–59. [Google Scholar] [CrossRef]
- Janssen, B.J.; Drummond, R.S.M.; Snowden, K.C. Regulation of axillary shoot development. Curr. Opin. Plant Biol. 2014, 17, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Agharkar, M.; Lomba, P.; Altpeter, F.; Zhang, H.; Kenworthy, K.; Lange, T. Stable expression of AtGA2ox1 in a low-input turfgrass (Paspalum notatum Flugge) reduces bioactive gibberellin levels and improves turf quality under field conditions. Plant Biotechnol. J. 2007, 5, 791–801. [Google Scholar] [CrossRef] [PubMed]
- Marzec, M. Strigolactones and gibberellins: A new couple in the phytohormone world? Trends Plant Sci. 2017, 22, 813–815. [Google Scholar] [CrossRef] [PubMed]
- Nagar, S.; Singh, V.P.; Arora, A.; Dhakar, R.; Singh, N.; Singh, G.P.; Meena, S.; Kumar, S.; Shiv Ramakrishnan, R. Understanding the role of gibberellic acid and paclobutrazol in terminal heat stress tolerance in wheat. Front. Plant Sci. 2021, 12, 692252. [Google Scholar] [CrossRef]
- Takeda, T.; Suwa, Y.; Suzuki, M.; Kitano, H.; Ueguchi-Tanaka, M.; Ashikari, M.; Matsuoka, M.; Ueguchi, C. The OsTB1 gene negatively regulates lateral branching in rice. Plant J. 2003, 33, 513–520. [Google Scholar] [CrossRef]
- Xu, J.; Zha, M.; Li, Y.; Ding, Y.; Chen, L.; Ding, C.; Wang, S. The interaction between nitrogen availability and auxin, cytokinin, and strigolactone in the control of shoot branching in rice (Oryza sativa L.). Plant Cell Rep. 2015, 34, 1647–1662. [Google Scholar] [CrossRef]
- Muhr, M.; Prüfer, N.; Paulat, M.; Teichmann, T. Knockdown of strigolactone biosynthesis genes in Populus affects BRANCHED1 expression and shoot architecture. New Phytol. 2016, 212, 613–626. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).