The Inheritance Pattern of Key Desirable Agronomic and Fruit Quality Traits in Elite Red Papaya Genotypes
Abstract
1. Introduction
2. Methods
2.1. Germplasm and Trial Sites
2.2. Seedling Cultivation and Sex Determination
2.3. Statistical Analyses
2.3.1. Linear Mixed Model
2.3.2. Histogram Analysis
2.3.3. Variance and Covariance Components and Broad-Sense Heritability
2.3.4. Genetic Advance (Percentages)
2.3.5. Trait Gain Percentages
3. Results
3.1. Mixed Models for Exploring G × E Interaction of Measured Traits in F5 RIL
3.2. Heritability and Genetic Advance of Key Agronomic and Fruit Quality Traits
3.3. Breeding Value or Genetic Worth of F5 RIL for Measured Traits
3.4. F5 RIL Trait Gain Percentage (Increase or Decrease) over RB1
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Horticulture Innovation. Australian Horticulture Statistics Handbook; Horticulture Innovation: Sydney, Australia, 2021; Available online: https://www.horticulture.com.au/growers/papaya-fund/ (accessed on 31 April 2022).
- United States Department of Agriculture. Statistics on Papayas, State of Hawaii, 2016–2020; United States Department of Agriculture: Honolulu, HI, USA, 2021. Available online: hdoa.hawaii.gov (accessed on 28 July 2022).
- Nantawan, M.; Kanchana-udomkan, C.; Bar, I.; Ford, R. Linkage mapping and quantitative trait loci analysis of sweetness and other fruit quality traits in papaya. BMC Plant Biol. 2019, 19, 449. [Google Scholar] [CrossRef]
- Acquaah, G. Breeding rice. In Principles of Plant Genetics and Breeding, 2nd ed.; John Wiley & Sons Ltd.: Chichester, UK, 2012; pp. 606–610. [Google Scholar]
- Acquaah, G. Introduction to quantitative genetics. In Principles of Plant Genetics and Breeding, 2nd ed.; John Wiley & Sons Ltd.: Chichester, UK, 2012; pp. 63–94. [Google Scholar]
- Celp, J.; Hola, D.; Stejskal, J.; Korecky, J.; Kocova, M.; Lhotakova, Z.; Tomaskova, I.; Palovska, M.; Rothova, O.; Whetten, R.W.; et al. Genetic variability and heritability of chlorophyll fluorescence parameters in Scots pine (Pinus sylvestris L.). Tree Phys. 2016, 36, 883–895. [Google Scholar]
- Hofmeyr, J.D.J. Determination of sex in Carica papaya. S. Afr. J. Sci. 1938, 13, 332. [Google Scholar]
- Ali, F.; Ahsan, M.; Ali, Q.; Kanwal, N. Phenotypic Stability of Zea mays Grain Yield and Its Attributing Traits under Drought Stress. Front. Plant Sci. 2017, 8, 1397. [Google Scholar] [CrossRef]
- Griffiths, J.F.; Griffiths, A.J.; Wessler, S.R.; Lewontin, R.C.; Gelbart, W.M.; Suzuki, D.T.; Miller, J.H. Quantifying heritability. In An Introduction to Genetic Analysis, 7th ed; W.H. Freeman: New York, NY, USA, 2000. Available online: https://www.ncbi.nlm.nih.gov/books/NBK21866/ (accessed on 26 July 2022).
- Piepho, H.P.; Möhring, J. Computing heritability and selection response from unbalanced plant breeding trials. Genetics 2007, 177, 1881–1888. [Google Scholar] [CrossRef]
- Kumar, S.; Chagne, D.; Bink, M.C.A.M.; Volz, R.K.; Whitworth, C.; Carlisle, C. Genomic Selection for Fruit Quality Traits in Apple (Malus × domestica Borkh.). PLoS ONE 2012, 7, e36674. [Google Scholar] [CrossRef]
- Imai, A.; Kuniga, T.; Yoshioka, T.; Nonaka, K.; Mitani, N.; Fukamachi, H.; Hiehata, N.; Yamamoto, M.; Hayashi, T. Predicting segregation of multiple fruit-quality traits by using accumulated phenotypic records in citrus breeding. PLoS ONE 2018, 16, e0202341. [Google Scholar] [CrossRef]
- Canas-Gutivrrez, G.P.; Sepulveda-Ortega, S.; Lpez-Hernndez, L.N.; Alejandro, A.; Corts Andrs, J. Inheritance of Yield Components and Morphological Traits in Avocado cv. Hass from “Criollo” “Elite Trees” via Half-Sib Seedling Rootstocks. Front. Plant Sci. 2022, 13, 843099. [Google Scholar] [CrossRef]
- Silva, F.F.; Pereira, M.G.; Ramos, H.C.C.; Damasceno Júnior, P.C.; Pereira, T.N.S.; Gabriel, A.P.C.; Viana, A.P.; Ferreguetti, G.A. Selection and Estimation of the Genetic Gain in Segregating Generations of Papaya (Carica papaya L.). Crop Breed. Appl. Biotechnol. 2008, 8, 1–8. [Google Scholar] [CrossRef]
- Cortes, D.F.M.; Santa-Catarina, R.; Vettorazzi, J.C.F.; Ramos, H.C.C.R.; Viana, A.P.; Pereira, M.G. Development of superior lines of papaya from the Formosa group using the pedigree method and REML/Blup procedure. Bragantia 2019, 78, 350–360. [Google Scholar] [CrossRef]
- Oliveira, E.J.; Fraife, F.G.A.; Freitas, J.P.X.; Dantas, J.L.L.; Resende, M.D.V. Plant selection in F2 segregating populations of papaya from commercial hybrids. Crop Breed. Appl. Biotechnol. 2012, 12, 191–198. [Google Scholar] [CrossRef]
- Kanchana-udomkan, C.; Nantawan, U.; Ford, R. New Genetic Targets to Improve Quality in Papaya (PP15000); Horticulture Innovation: Sydney, NSW, Australia, 2018; p. 53. [Google Scholar]
- Marler, T.E.; Clemente, H.S. Papaya seedling growth response to wind and water deficit is additive. Hortic. Sci. 2006, 41, 96–98. [Google Scholar] [CrossRef]
- Available online: www.bom.gov.au (accessed on 1 August 2022).
- Kanchana-udomkan, C.; Nantawan, U.; Drew, R.; Ford, R. Molecular marker-assisted papaya sex determination for improved grower efficiency. Acta Hortic. 2018, 1205, 697–704. [Google Scholar] [CrossRef]
- Kanchana-udomkan, C.; Ford, R. Papaya Evaluation Handbook, 2nd ed.; (PP18000); Horticulture Innovation: Sydney, NSW, Australia, 2021; pp. 1–30. [Google Scholar]
- Jahufer, M.Z.Z.; Luo, D. “DeltaGen”—A comprehensive decision support tool for plant breeders. Crop Sci. 2018, 58, 1118–1131. [Google Scholar] [CrossRef]
- Falconer, D.S. Introduction to Qualitative Genetics, 2nd ed.; Longman: London, UK, 1989; p. 340. [Google Scholar]
- Singh, R.K.; Chaudhary, B.D. Biometrical Methods in Quantitative Genetics Analysis; Kalyani Publishers: New Delhi, India, 1999; pp. 1–318. [Google Scholar]
- Chavez-Pesqueira, M.; Suarez-Montes, P.; Castillo, G.; Nunez-Farfan, J. Habitat fragmentation threatens wild populations of Carica papaya (Caricaceae) in a lowland rainforest. Am. J. Bot. 2014, 101, 1092–1101. [Google Scholar] [CrossRef]
- Johnson, H.W.; Robinson, H.F.; Comstock, R. Estimates of Genetic and Environmental Variability in Soybeans. Agron. J. 1955, 47, 314–318. [Google Scholar] [CrossRef]
- Burton, G.W. Quantitative inheritance in grasses. In Proceedings of the 6th International Grassland Congress, State College, PA, USA, 17–23 August 1952; Volume 1, pp. 277–283. [Google Scholar]
- Lush, J.L. Heritability of quantitative characters in farm animals. In Proceedings of the 8th International Congress of Genetics; Berlingska Boktrycheriet: Lund, Sweden, 1949; pp. 356–357. [Google Scholar]
- Zhou, Z.; Bar, I.; Ford, R.; Smyth, H.; Kanchana-udomkan, C. Biochemical, Sensory, and Molecular Evaluation of Flavour and Consumer Acceptability in Australian Papaya (Carica papaya L.) Varieties. Int. J. Mol. Sci. 2022, 23, 6313. [Google Scholar] [CrossRef]
- Lush, J.L. Intrasine correlation and regression of offspring on dams as a method of estimating heritability of character. Proc. Am. Soc. Anim. Prod. 1940, 293–301. [Google Scholar]
- Quaggio, J.A.; Mattos, D., Jr.; Cantarella, H.; Almeida, E.L.E.; Cardoso, S.A.B. Lemon yield and fruit quality affected by NPK fertilization. Sci. Hortic. 2002, 96, 151–162. [Google Scholar] [CrossRef]
- Quaggio, J.A.; Mattos, D., Jr.; Cantarella, H. Fruit yield and quality of sweet oranges affected by nitrogen, phosphorus and potassium fertilization in tropical soils. Fruits 2006, 61, 293–302. [Google Scholar] [CrossRef]
- Adachi, S.; Yoshikawaw, K.; Yamanouchi, U.; Tanabata, T.; Sun, J.; Ookawa, T.; Yamamoto, T.; Sage, R.F.; Hirasawa, T.; Yonemaru, J. Fine mapping of carbon assimilation rate 8, a quantitative trait locus for flag leaf nitrogen content, stomatal conductance and photosynthesis in rice. Front. Plant Sci. 2017, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Bonomelli, C.; Gil, P.M.; Schaffer, B. Effect of Soil Type on Calcium Absorption and Partitioning in Young Avocado (Persea americana Mill.) Trees. Agronomy 2019, 9, 837. [Google Scholar] [CrossRef]
- Lawson, T.; Blatt, M. Stomatal size, speed, and responsiveness impact on photosynthesis and water use efficiency. Plant Phys. 2014, 164, 1556–1570. [Google Scholar] [CrossRef] [PubMed]
- Long, S.P.; Marshall-Colon, A.; Zhu, X.G. Meeting the global food demand of the future by engineering crop photosynthesis and yield potential. Cell 2015, 161, 56–66. [Google Scholar] [CrossRef]
- Yoon, D.K.; Ishiyama, K.; Suganami, M.; Tazoe, Y.; Watanabe, M.; Imaruoka, S.; Ogura, M.; Ishida, H.; Suzuki, Y.; Obara Mae, T.; et al. Transgenic rice overproducing Rubisco exhibits high yields with high nitrogen use efficiency in a paddy field. Nat. Food 2020, 1, 134–139. [Google Scholar] [CrossRef]
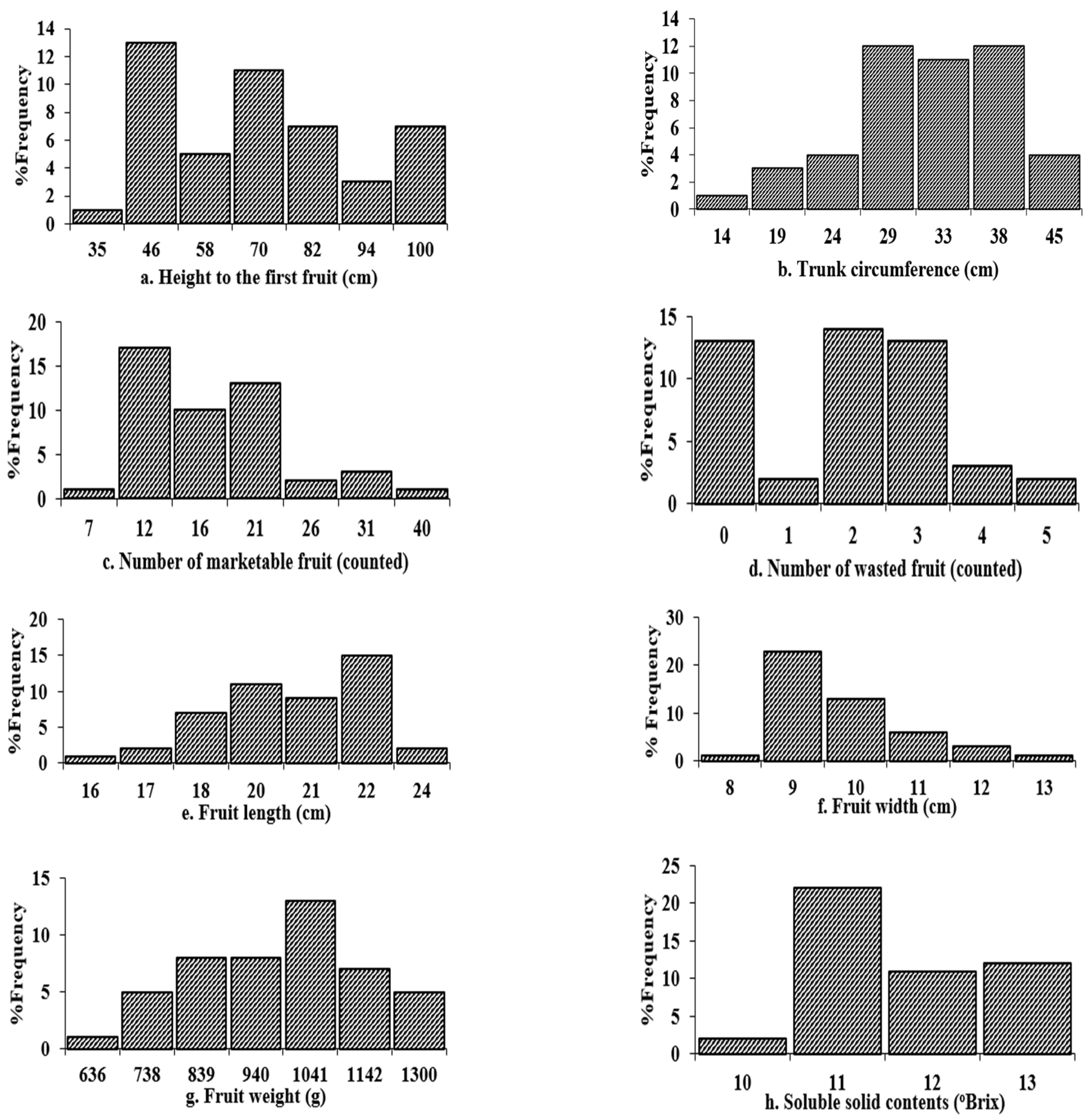
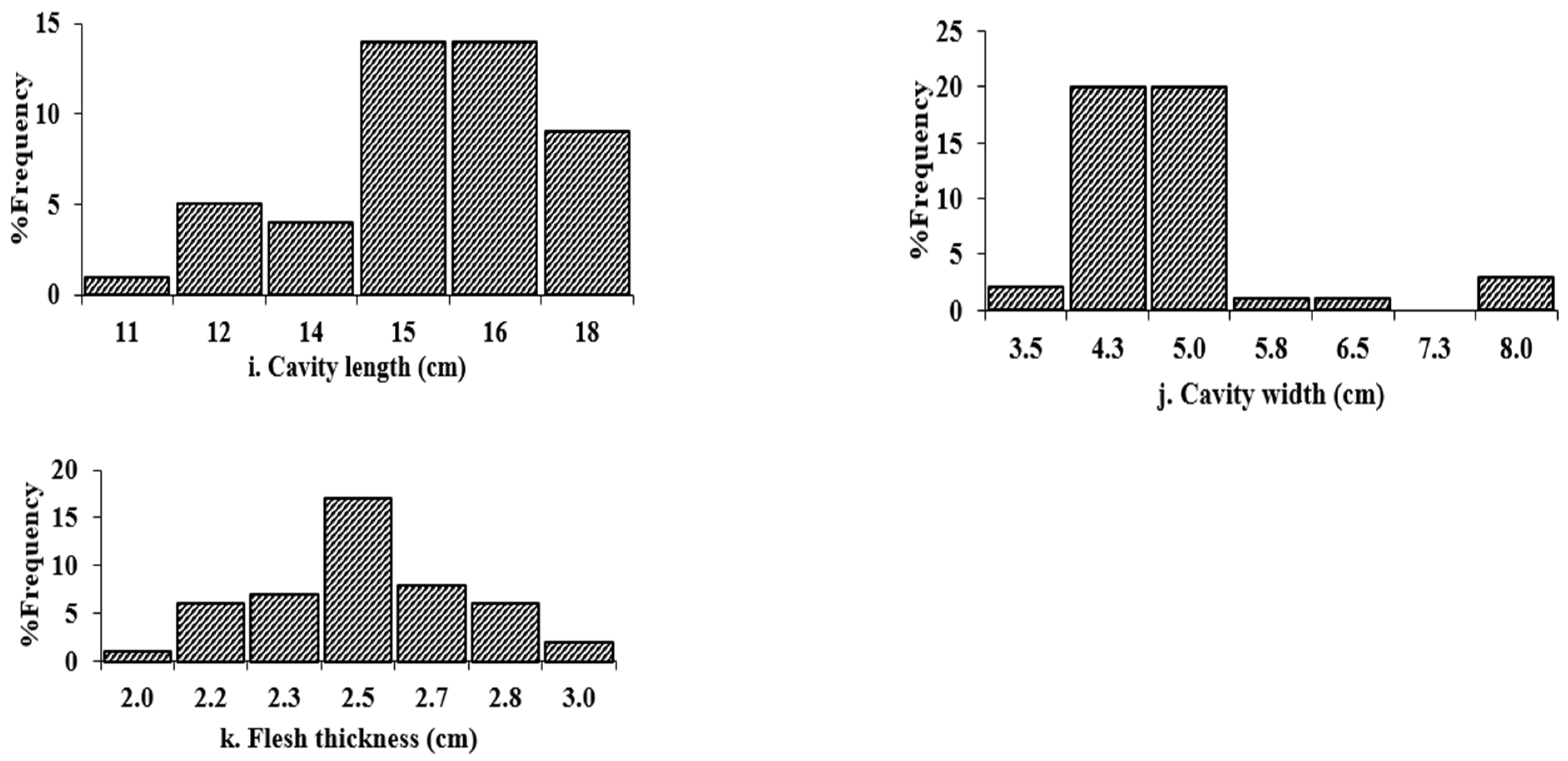
| Random Factor | Traits | Among Two Field Sites within the Tablelands (T1 + T2) Environment | Among Two Field Sites within the Coastal (C1 + C2) Environment | Across Two Distinct Agro-Geographical Climates (Across Four Trial Sites): Tablelands (T1 and T2) and Coastal (C1 and C2) | |||
|---|---|---|---|---|---|---|---|
| Variance | p-Value | Variance | p-Value | Variance | p-Value | ||
| Genotype among two field sites within the Coastal & Tablelands environment × Environment | Height to the first fruit (cm) | 35.49 | 0.21 | 45.91 | 0.14 | 89.10 | 0.000 |
| Trunk circumference (cm) | 7.14 | 0.17 | 3.21 | 0.12 | 20.01 | 0.000 | |
| Number of marketable fruit (counted) | 7.15 | 0.13 | 2.12 | 0.24 | 10.05 | 0.01 | |
| Number of wasted fruit (counted) | 0.41 | 0.22 | 0.39 | 0.25 | 0.63 | 0.001 | |
| Fruit weight (g) | 3814 | 0.12 | 2125 | 0.22 | 18,424 | 0.001 | |
| Fruit width (cm) | 0.17 | 0.23 | 0.11 | 0.19 | 0.36 | 0.24 | |
| Fruit length (cm) | 0 | 0.24 | 0 | 0.17 | 0 | 0.13 | |
| Flesh thickness (cm) | 0.04 | 0.11 | 0.03 | 0.26 | 0.42 | 0.005 | |
| Cavity width (cm) | 0.08 | 0.09 | 0.02 | 0.21 | 0.47 | 0.03 | |
| Cavity length (cm) | 0.05 | 0.13 | 0.12 | 0.20 | 0.21 | 0.14 | |
| Soluble solid contents (°Brix) | 0.32 | 0.25 | 0.24 | 0.17 | 0.59 | 0.001 | |
| Traits | h2b.s at T1 | h2b.s at T2 | h2b.s at C1 | h2b.s at C2 |
|---|---|---|---|---|
| Height to the first fruit (cm) | 0.62 | 0.81 | 0.89 | 0.16 |
| Trunk circumference (cm) | 0.12 | 0.80 | 0.88 | 0.44 |
| Number of marketable fruit (counted) | 0.71 | 0.74 | 0.88 | 0.65 |
| Number of wasted fruit (counted) | 0.86 | 0.10 | 0.13 | 0.82 |
| Fruit weight (g) | 0.89 | 0.18 | 0.48 | 0.00 |
| Fruit width (cm) | 0.90 | 0.73 | 0.44 | 0.15 |
| Fruit length (cm) | 0.90 | 0.11 | 0.16 | 0.29 |
| Flesh thickness (cm) | 0.84 | 0.90 | 0.73 | 0.12 |
| Cavity width (cm) | 0.63 | 0.10 | 0.79 | 0.11 |
| Cavity length (cm) | 0.90 | 0.83 | 0.42 | 0.12 |
| Soluble solid contents (°Brix) | 0.90 | 0.67 | 0.68 | 0.83 |
| Traits | GA at T1 (%) | GA at T2 (%) | GA at C1 (%) | GA at C2 (%) |
|---|---|---|---|---|
| Height to the first fruit (cm) | 16.83 | 17.00 | 5.37 | 14.51 |
| Trunk circumference (cm) | 10.06 | 4.38 | 0.58 | 9.41 |
| Number of marketable fruit (counted) | 0.15 | 0.11 | 0.32 | 0.31 |
| Number of wasted fruit (counted) | 1.15 | 2.31 | 0.15 | 3.18 |
| Fruit weight (g) | 13.38 | 14.76 | 16.94 | 17.77 |
| Fruit width (cm) | 0.72 | 0.91 | 1.09 | 0.76 |
| Fruit length (cm) | 0.58 | 0.13 | 0.09 | 0.58 |
| Flesh thickness (cm) | 0.15 | 0.12 | 0.15 | 0.11 |
| Cavity width (cm) | 0.12 | 0.13 | 3.38 | 0.22 |
| Cavity length (cm) | 0.66 | 0.02 | 0.30 | 0.16 |
| Soluble solid contents (°Brix) | 0.26 | 0.58 | 0.39 | 0.57 |
| Traits | Line | Location | BLUP | Standard Error (±) |
|---|---|---|---|---|
| Height to the first fruit (cm) | T2-5-5.27 | T2 | 60.55 | 3.36 |
| T2-5-3.12 | T1 | 59.06 | 3.32 | |
| T2-5-3.12 | T2 | 56.37 | 3.36 | |
| T3-5-6.10 | T1 | 55.96 | 3.36 | |
| T1-5-2.3 | T1 | 55.84 | 3.43 | |
| T1-5-5.9 | T1 | 55.44 | 3.32 | |
| T3-5-6.10 | T2 | 54.87 | 3.60 | |
| T2-5-5.27 | T1 | 54.16 | 3.32 | |
| T1-5-5.9 | T2 | 53.90 | 3.36 | |
| RB1 | T1 | 71.1 | 11.15 | |
| RB1 | T2 | 72.6 | 10.12 | |
| Trunk circumference (cm) | T3-5-6.10 | T1 | 31.92 | 1.65 |
| T3-5-6.10 | T2 | 30.66 | 1.93 | |
| T1-5-2.3 | T1 | 31.96 | 1.65 | |
| T1-5-5.9 | T1 | 30.56 | 1.65 | |
| T1-5-5.9 | T2 | 34.66 | 1.69 | |
| T2-5-5.27 | T1 | 32.15 | 1.65 | |
| T2-5-5.27 | T2 | 29.62 | 1.69 | |
| T2-5-3.12 | T1 | 32.57 | 1.65 | |
| T2-5-3.12 | T2 | 33.19 | 1.69 | |
| RB1 | T1 | 32.11 | 1.03 | |
| RB1 | T2 | 31.55 | 1.12 | |
| Number of marketable fruit (counted) | T3-5-6.10 | T1 | 13.44 | 1.33 |
| T3-5-6.10 | T2 | 11.77 | 1.55 | |
| T1-5-2.3 | T1 | 12.28 | 1.34 | |
| T1-5-5.9 | T1 | 13.64 | 1.32 | |
| T1-5-5.9 | T2 | 13.73 | 1.32 | |
| T2-5-5.27 | T1 | 13.84 | 1.32 | |
| T2-5-5.27 | T2 | 13.89 | 1.32 | |
| T2-5-3.12 | T1 | 10.08 | 1.32 | |
| T2-5-3.12 | T2 | 13.55 | 1.32 | |
| RB1 | T1 | 14.88 | 1.62 | |
| RB1 | T2 | 14.95 | 1.51 | |
| Number of Wasted fruit (counted) | T3-5-6.10 | T1 | 2.64 | 0.44 |
| T3-5-6.10 | T2 | 1.72 | 0.59 | |
| T1-5-2.3 | T1 | 1.04 | 0.44 | |
| T1-5-5.9 | T1 | 1.38 | 0.44 | |
| T1-5-5.9 | T2 | 1.53 | 0.45 | |
| T2-5-5.27 | T1 | 1.56 | 0.44 | |
| T2-5-5.27 | T2 | 1.53 | 0.45 | |
| T2-5-3.12 | T1 | 1.52 | 0.44 | |
| T2-5-3.12 | T2 | 1.79 | 0.45 | |
| RB1 | T1 | 1.98 | 0.01 | |
| RB1 | T2 | 2.11 | 0.05 | |
| Fruit weight (g) | T3-5-6.10 | T1 | 958.13 | 0.73 |
| T3-5-6.10 | T2 | 957.67 | 0.11 | |
| T1-5-2.3 | T1 | 916.95 | 0.18 | |
| T1-5-5.9 | T1 | 1016.39 | 0.14 | |
| T1-5-5.9 | T2 | 937.04 | 0.91 | |
| T2-5-5.27 | T1 | 879.92 | 1.11 | |
| T2-5-5.27 | T2 | 989.99 | 0.88 | |
| T2-5-3.12 | T1 | 935.07 | 0.51 | |
| T2-5-3.12 | T2 | 982.06 | 0.42 | |
| RB1 | T1 | 1026.67 | 75.51 | |
| RB1 | T2 | 1028.11 | 72.23 | |
| Fruit width (cm) | T3-5-6.10 | T1 | 9.87 | 0.34 |
| T3-5-6.10 | T2 | 9.95 | 0.38 | |
| T1-5-2.3 | T1 | 9.73 | 0.36 | |
| T1-5-5.9 | T1 | 10.38 | 0.34 | |
| T1-5-5.9 | T2 | 9.79 | 0.34 | |
| T2-5-5.27 | T1 | 9.54 | 0.34 | |
| T2-5-5.27 | T2 | 9.73 | 0.34 | |
| T2-5-3.12 | T1 | 9.66 | 0.34 | |
| T2-5-3.12 | T2 | 10.49 | 0.34 | |
| RB1 | T1 | 11.36 | 1.45 | |
| RB1 | T2 | 12.11 | 1.56 | |
| Flesh thickness (cm) | T3-5-6.10 | T1 | 2.446 | 0.01 |
| T3-5-6.10 | T2 | 2.446 | 0.01 | |
| T1-5-2.3 | T1 | 2.446 | 0.01 | |
| T1-5-5.9 | T1 | 2.446 | 0.01 | |
| T1-5-5.9 | T2 | 2.446 | 0.01 | |
| T2-5-5.27 | T1 | 2.446 | 0.01 | |
| T2-5-5.27 | T2 | 2.446 | 0.01 | |
| T2-5-3.12 | T1 | 2.446 | 0.01 | |
| T2-5-3.12 | T2 | 2.446 | 0.01 | |
| RB1 | T1 | 2.92 | 0.41 | |
| RB1 | T2 | 2.94 | 0.45 | |
| Cavity width (cm) | T3-5-6.10 | T1 | 4.55 | 0.15 |
| T3-5-6.10 | T2 | 4.69 | 0.18 | |
| T1-5-2.3 | T1 | 4.56 | 0.15 | |
| T1-5-5.9 | T1 | 4.72 | 0.15 | |
| T1-5-5.9 | T2 | 4.40 | 0.15 | |
| T2-5-5.27 | T1 | 4.34 | 0.15 | |
| T2-5-5.27 | T2 | 4.67 | 0.15 | |
| T2-5-3.12 | T1 | 4.55 | 0.01 | |
| T2-5-3.12 | T2 | 4.73 | 0.01 | |
| RB1 | T1 | 5.81 | 1.11 | |
| RB1 | T2 | 5.98 | 1.52 | |
| Fruit length (cm) | T3-5-6.10 | T1 | 19.78 | 0.01 |
| T3-5-6.10 | T2 | 19.78 | 0.01 | |
| T1-5-2.3 | T1 | 19.78 | 0.01 | |
| T1-5-5.9 | T1 | 19.78 | 0.01 | |
| T1-5-5.9 | T2 | 19.78 | 0.01 | |
| T2-5-5.27 | T1 | 19.78 | 0.01 | |
| T2-5-5.27 | T2 | 19.78 | 0.01 | |
| T2-5-3.12 | T1 | 19.78 | 0.01 | |
| T2-5-3.12 | T1 | 19.78 | 0.01 | |
| RB1 | T1 | 21.32 | 1.51 | |
| RB1 | T2 | 21.55 | 1.52 | |
| Cavity length (cm) | T3-5-6.10 | T1 | 14.94 | 0.46 |
| T3-5-6.10 | T2 | 15.43 | 0.47 | |
| T1-5-2.3 | T1 | 14.96 | 0.49 | |
| T1-5-5.9 | T1 | 15.07 | 0.45 | |
| T1-5-5.9 | T2 | 15.39 | 0.45 | |
| T2-5-5.27 | T1 | 15.33 | 0.45 | |
| T2-5-5.27 | T2 | 14.90 | 0.45 | |
| T2-5-3.12 | T1 | 15.37 | 0.45 | |
| T2-5-3.12 | T2 | 15.04 | 0.45 | |
| RB1 | T1 | 16.45 | 1.31 | |
| RB1 | T2 | 16.99 | 1.22 | |
| Soluble solid contents (°Brix) | T3-5-6.10 | T1 | 11.95 | 0.47 |
| T3-5-6.10 | T2 | 11.06 | 0.51 | |
| T1-5-2.3 | T1 | 11.84 | 0.51 | |
| T1-5-5.9 | T1 | 10.85 | 0.47 | |
| T1-5-5.9 | T2 | 11.10 | 0.47 | |
| T2-5-5.27 | T1 | 11.97 | 0.47 | |
| T2-5-5.27 | T2 | 11.88 | 0.47 | |
| T2-5-3.12 | T1 | 11.89 | 0.47 | |
| T2-5-3.12 | T2 | 11.14 | 0.47 | |
| RB1 | T1 | 10.41 | 0.93 | |
| RB1 | T2 | 10.44 | 0.95 |
| Traits | Line | Location | BLUP | Standard Error (±) |
|---|---|---|---|---|
| Height to the first fruit (cm) | C1-5-4.1 | C1 | 91.15 | 6.68 |
| C1-5-4.1 | C2 | 75.84 | 7.11 | |
| C1-5-4.2 | C1 | 89.92 | 6.76 | |
| C1-5-4.3 | C1 | 82.70 | 6.76 | |
| C3-3-5.24 | C1 | 63.69 | 6.68 | |
| C3-3-5.24 | C2 | 64.94 | 7.11 | |
| C2-5-5 | C2 | 81.81 | 7.18 | |
| RB1 | C1 | 100.92 | 7.89 | |
| RB1 | C2 | 101.23 | 8.12 | |
| Trunk circumference (cm) | C1-5-4.1 | C1 | 31.59 | 2.68 |
| C1-5-4.1 | C2 | 27.68 | 2.88 | |
| C1-5-4.2 | C1 | 33.05 | 2.69 | |
| C1-5-4.3 | C1 | 28.89 | 2.69 | |
| C3-3-5.24 | C1 | 20.92 | 2.68 | |
| C3-3-5.24 | C2 | 31.82 | 2.88 | |
| C2-5-5 | C2 | 31.61 | 2.89 | |
| RB1 | C1 | 31.10 | 1.85 | |
| RB1 | C2 | 31.25 | 1.92 | |
| Number of marketable fruit (counted) | C1-5-4.1 | C1 | 19.06 | 1.62 |
| C1-5-4.1 | C2 | 20.83 | 1.62 | |
| C1-5-4.2 | C1 | 20.05 | 1.66 | |
| C1-5-4.3 | C1 | 19.62 | 1.66 | |
| C3-3-5.24 | C1 | 19.03 | 1.62 | |
| C3-3-5.24 | C2 | 19.35 | 1.62 | |
| C2-5-5 | C1 | 19.06 | 1.62 | |
| RB1 | C1 | 14.21 | 2.34 | |
| RB1 | C2 | 15.23 | 2.45 | |
| Number of Wasted fruit (counted) | C1-5-4.1 | C1 | 0.84 | 0.44 |
| C1-5-4.1 | C2 | 1.81 | 0.45 | |
| C1-5-4.2 | C1 | 0.85 | 0.46 | |
| C1-5-4.3 | C1 | 0.85 | 0.46 | |
| C3-3-5.24 | C1 | 1.01 | 0.44 | |
| C3-3-5.24 | C2 | 0.55 | 0.45 | |
| C2-5-5 | C2 | 0.67 | 0.47 | |
| RB1 | C1 | 1.51 | 0.43 | |
| RB1 | C2 | 1.55 | 0.49 | |
| Fruit weight (g) | C1-5-4.1 | C1 | 959.46 | 0.01 |
| C1-5-4.1 | C2 | 959.46 | 0.01 | |
| C1-5-4.2 | C1 | 959.46 | 0.01 | |
| C1-5-4.3 | C1 | 959.46 | 0.01 | |
| C3-3-5.24 | C1 | 959.46 | 0.01 | |
| C3-3-5.24 | C2 | 959.46 | 0.01 | |
| C2-5-5 | C2 | 959.46 | 0.01 | |
| RB1 | C1 | 1056.45 | 84.42 | |
| RB1 | C2 | 1102.56 | 79.23 | |
| Fruit width (cm) | C1-5-4.1 | C1 | 9.54 | 0.01 |
| C1-5-4.1 | C2 | 9.54 | 0.01 | |
| C1-5-4.2 | C1 | 9.54 | 0.01 | |
| C1-5-4.3 | C1 | 9.54 | 0.01 | |
| C3-3-5.24 | C1 | 9.54 | 0.01 | |
| C3-3-5.24 | C2 | 9.54 | 0.01 | |
| C2-5-5 | C2 | 9.54 | 0.01 | |
| RB1 | C1 | 9.56 | 0.45 | |
| RB1 | C2 | 9.61 | 0.61 | |
| Flesh thickness (cm) | C1-5-4.1 | C1 | 2.40 | 0.11 |
| C1-5-4.1 | C2 | 2.46 | 0.11 | |
| C1-5-4.2 | C1 | 2.35 | 0.11 | |
| C1-5-4.3 | C1 | 2.32 | 0.11 | |
| C3-3-5.24 | C1 | 2.74 | 0.11 | |
| C3-3-5.24 | C2 | 2.36 | 0.11 | |
| C2-5-5 | C2 | 2.35 | 0.11 | |
| RB1 | C1 | 2.33 | 0.26 | |
| RB1 | C2 | 2.31 | 0.23 | |
| Cavity width (cm) | C1-5-4.1 | C1 | 4.38 | 0.56 |
| C1-5-4.1 | C2 | 4.67 | 0.58 | |
| C1-5-4.2 | C1 | 3.91 | 0.56 | |
| C1-5-4.3 | C1 | 5.61 | 0.56 | |
| C3-3-5.24 | C1 | 6.39 | 0.56 | |
| C3-3-5.24 | C2 | 4.67 | 0.58 | |
| C2-5-5 | C2 | 4.41 | 0.58 | |
| RB1 | C1 | 6.86 | 0.49 | |
| RB1 | C2 | 6.89 | 4.51 | |
| Fruit length (cm) | C1-5-4.1 | C1 | 19.78 | 0.43 |
| C1-5-4.1 | C2 | 19.54 | 0.43 | |
| C1-5-4.2 | C1 | 19.50 | 0.44 | |
| C1-5-4.3 | C1 | 19.63 | 0.44 | |
| C3-3-5.24 | C1 | 19.18 | 0.43 | |
| C3-3-5.24 | C2 | 19.81 | 0.43 | |
| C2-5-5 | C2 | 19.15 | 0.44 | |
| RB1 | C1 | 19.41 | 0.31 | |
| RB1 | C2 | 19.52 | 0.30 | |
| Cavity length (cm) | C1-5-4.1 | C1 | 15.13 | 0.51 |
| C1-5-4.1 | C2 | 14.61 | 0.52 | |
| C1-5-4.2 | C1 | 14.39 | 0.53 | |
| C1-5-4.3 | C1 | 14.99 | 0.53 | |
| C3-3-5.24 | C1 | 13.65 | 0.51 | |
| C3-3-5.24 | C2 | 15.28 | 0.52 | |
| C2-5-5 | C2 | 14.16 | 0.53 | |
| RB1 | C1 | 16.96 | 0.44 | |
| RB1 | C2 | 16.45 | 0.41 | |
| Soluble solid contents (°Brix) | C1-5-4.1 | C1 | 10.92 | 0.29 |
| C1-5-4.1 | C2 | 11.68 | 0.30 | |
| C1-5-4.2 | C1 | 11.81 | 0.29 | |
| C1-5-4.3 | C1 | 11.05 | 0.29 | |
| C3-3-5.24 | C1 | 11.70 | 0.29 | |
| C3-3-5.24 | C2 | 10.87 | 0.30 | |
| C2-5-5 | C2 | 10.96 | 0.30 | |
| RB1 | C1 | 9.49 | 0.25 | |
| RB1 | C2 | 9.52 | 0.31 |
| Traits | T3-5-6.10 | T1-5-2.3 | T1-5-5.9 | T2-5-5.27 | T2-5-3.12 | C1-5-4.1 | C1-5-4.1 | C1-5-4.3 | C3-3-5.24 | C2-5-5 |
|---|---|---|---|---|---|---|---|---|---|---|
| Height to the first fruit (cm) | −49.2 | −47.6 | −53.0 | −54.6 | −12.7 | −0.48 | −1.13 | −9.81 | −32.96 | −30.71 |
| Trunk circumference (cm) | −29.7 | −29.4 | 36.5 | 28.5 | 7.0 | −38.06 | −33.96 | −44.78 | 29.93 | 22.39 |
| Number of Marketable fruit (counted) | 18.4 | −0.2 | −13.9 | 21.6 | 14.8 | 53.52 | 52.20 | 40.85 | 50.01 | 53.52 |
| Number of Wasted fruit (counted) | −100 | −33.3 | −100 | −100 | 11.1 | 100.00 | −100.00 | −100.00 | −100.00 | −100.00 |
| Fruit weight (g) | −13.7 | −33.2 | −6.4 | −39.4 | −9.3 | 12.56 | −16.18 | 9.79 | −7.18 | −3.20 |
| Fruit width (cm) | −16.9 | −19.8 | −8.4 | −24.4 | −3.3 | 3.16 | −4.91 | 18.25 | 8.77 | −11.40 |
| Fruit length (cm) | 3.5 | −6.7 | 13.5 | −5.5 | 1.8 | 8.70 | 5.80 | 4.35 | 27.54 | −3.62 |
| Flesh thickness (cm) | −28.6 | −28.1 | −22.6 | −35.7 | −20.9 | −6.21 | −18.62 | 26.21 | 46.90 | −17.24 |
| Cavity width (cm) | 1.8 | −6.6 | 6.3 | −6.2 | 7.4 | 6.55 | 0.69 | 3.28 | −4.31 | −6.47 |
| Cavity length (cm) | −2.6 | −21.6 | 6.0 | −7.8 | 15.0 | 17.22 | 7.59 | 15.44 | 0.25 | 5.70 |
| Soluble solid contents (°Brix) | 1.17 | 7.8 | 20.56 | 21.64 | 4.9 | 18.15 | 9.89 | 9.93 | 21.47 | 13.70 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ali, F.; Kanchana-udomkan, C.; Ford, R. The Inheritance Pattern of Key Desirable Agronomic and Fruit Quality Traits in Elite Red Papaya Genotypes. Horticulturae 2022, 8, 845. https://doi.org/10.3390/horticulturae8090845
Ali F, Kanchana-udomkan C, Ford R. The Inheritance Pattern of Key Desirable Agronomic and Fruit Quality Traits in Elite Red Papaya Genotypes. Horticulturae. 2022; 8(9):845. https://doi.org/10.3390/horticulturae8090845
Chicago/Turabian StyleAli, Fawad, Chutchamas Kanchana-udomkan, and Rebecca Ford. 2022. "The Inheritance Pattern of Key Desirable Agronomic and Fruit Quality Traits in Elite Red Papaya Genotypes" Horticulturae 8, no. 9: 845. https://doi.org/10.3390/horticulturae8090845
APA StyleAli, F., Kanchana-udomkan, C., & Ford, R. (2022). The Inheritance Pattern of Key Desirable Agronomic and Fruit Quality Traits in Elite Red Papaya Genotypes. Horticulturae, 8(9), 845. https://doi.org/10.3390/horticulturae8090845








