Epidemiological Role of Dictyophara europaea (Hemiptera: Dictyopharidae) in the Transmission of ‘Candidatus Phytoplasma solani’
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sites
2.2. Survey of D. europaea and Reservoir Plants
2.3. DNA Extraction from Insects, Herbaceous Hosts, and Experimental Plants
2.4. ‘Ca. P. solani’ Identification
2.5. Experimental Transmission of ‘Ca. P. solani’ with Field-Collected Dictyophara europaea
2.6. Multilocus Sequence Analysis (MLSA) of ‘Ca. P. solani’ Isolates
3. Results
3.1. ‘Ca. Phytoplasma solani’ Identification in Dictyophara europaea and Natural Reservoir Plants
3.2. ‘Ca. Phytoplasma solani’ Genotypes Associated with Dictyophara europaea and Reservoir Plants
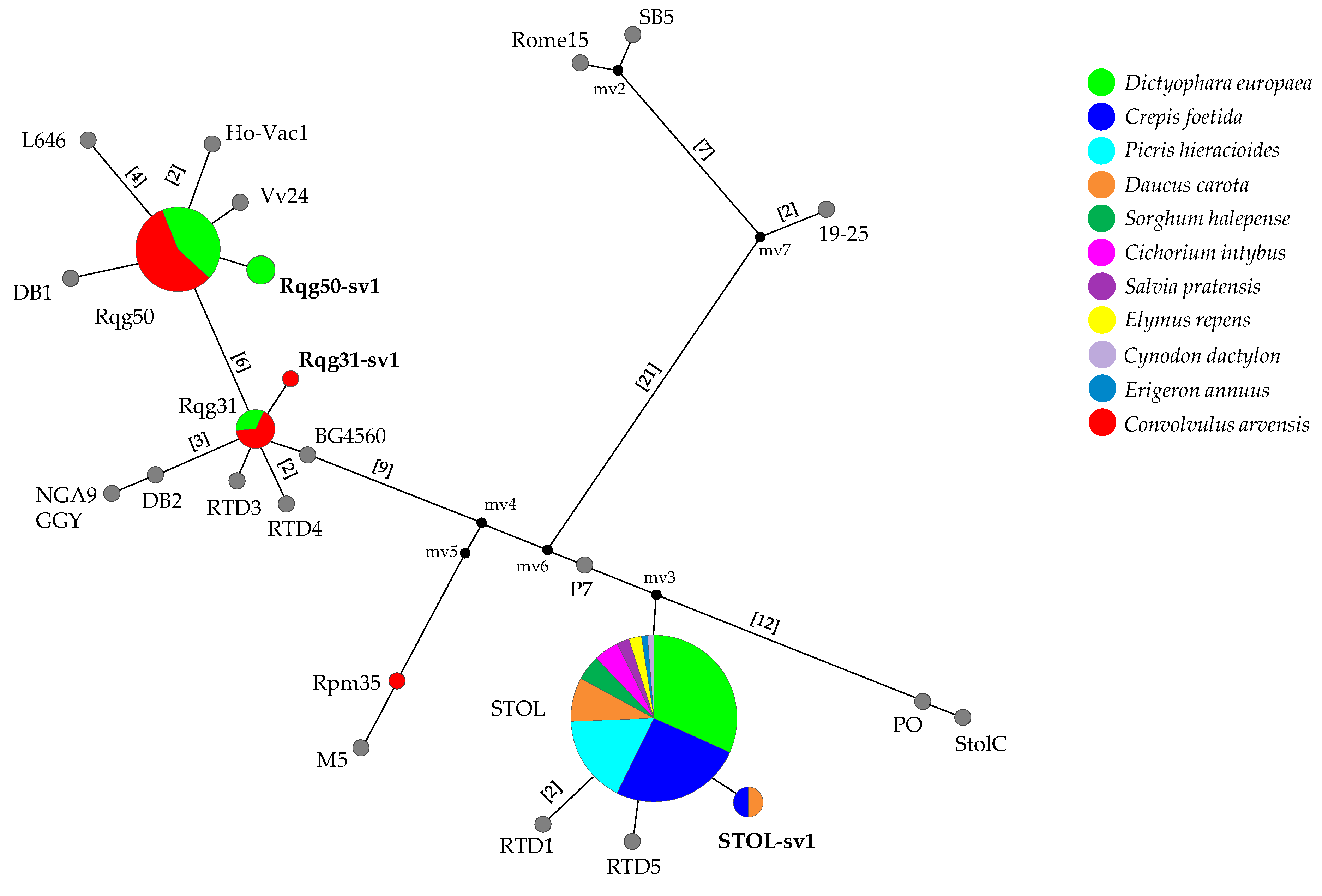
3.3. Multilocus Sequence Analysis
3.4. Transmission Trials
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Quaglino, F.; Zhao, Y.; Casati, P.; Bulgari, D.; Bianco, P.A.; Wei, W.; Davis, R.E. ‘Candidatus Phytoplasma solani’, a novel taxon associated with stolbur- and bois noir-related diseases of plants. Int. J. Syst. Evol. Microbiol. 2013, 63, 2879–2894. [Google Scholar] [CrossRef] [Green Version]
- Langer, M.; Maixner, M. Molecular characterisation of grapevine yellows associated phytoplasmas of the stolbur group based on RFLPanalysis of non-ribosomal DNA. Vitis 2004, 43, 191–199. [Google Scholar]
- Danet, J.L.; Foissac, X.; Zreik, L.; Salar, P.; Verdin, E.; Nourrisseau, J.G.; Garnier, M. “Candidatus Phlomobacter fragariae” is the prevalent agent of marginal chlorosis of strawberry in French production fields and is transmitted by the planthopper Cixius wagneri (China). Phytopathology 2003, 93, 644–649. [Google Scholar] [CrossRef] [Green Version]
- Jović, J.; Cvrković, T.; Mitrović, M.; Krnjanjić, S.; Petrović, A.; Redinbaugh, M.G.; Pratt, R.C.; Hogenhout, S.A.; Toševski, I. Stolbur phytoplasma transmission to maize by Reptalus panzeri and the disease cycle of maize redness in Serbia. Phytopathology 2009, 99, 1053–1061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jović, J.; Ember, I.; Mitrović, M.; Cvrković, T.; Krstić, O.; Krnjajić, S.B.; Ács, Z.; Kolber, M.; Toševski, I. Molecular detection of potato stolbur phytoplasma in Serbia. Bull. Insectol. 2011, 64, S83–S84. [Google Scholar]
- Murolo, S.; Marcone, C.; Prota, V.; Garau, R.; Foissac, X.; Romanazzi, G. Genetic variability of the stolbur phytoplasma vmp1 gene in grapevines, bindweeds and vegetables. J. Appl. Microbiol. 2010, 109, 2049–2059. [Google Scholar] [CrossRef] [PubMed]
- Sémétey, O.; Gaudin, J.; Danet, J.L.; Salar, P.; Theil, S.; Fontaine, M.; Krausz, M.; Chaisse, E.; Eveillard, S.; Verdin, E.; et al. Lavender decline in France is associated with chronic infection by lavender-specific strains of “Candidatus Phytoplasma solani”. Appl. Environ. Microbiol. 2018, 84, e01507-18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johannesen, J.; Lux, B.; Michel, K.; Seitz, A.; Maixner, M. Invasion biology and host specificity of the grapevine yellows disease vector Hyalesthes obsoletus in Europe. Entomol. Exp. Appl. 2008, 126, 217–227. [Google Scholar] [CrossRef]
- Ember, I.; Bodor, P.; Zsofi, Z.; Palfi, Z.; Ladanyi, M.; Pasti, G.; Deak, T.; Nyitraine, D.S.; Balo, B.; Szekeres, A.; et al. Bois noir affects the yield and wine quality of Vitis vinifera L. cv. ‘Chardonnay’. Eur. J. Plant. Pathol. 2018, 152, 185–197. [Google Scholar] [CrossRef] [Green Version]
- Johannesen, J.; Foissac, X.; Kehrli, P.; Maixner, M. Impact of vector dispersal and host-plant fidelity on the dissemination of an emerging plant pathogen. PLoS ONE 2012, 7, e51809. [Google Scholar] [CrossRef] [Green Version]
- Cvrković, T.; Jović, J.; Mitrović, M.; Krstić, O.; Toševski, I. Experimental and molecular evidence of Reptalus panzeri as a natural vector of bois noir. Plant Pathol. 2014, 63, 42–53. [Google Scholar] [CrossRef] [Green Version]
- Kosovac, A.; Jakovljević, M.; Krstić, O.; Cvrković, T.; Mitrović, M.; Toševski, I.; Jović, J. Role of plant-specialized Hyalesthes obsoletus associated with Convolvulus arvensis and Crepis foetida in the transmission of ‘Candidatus Phytoplasma solani’-inflicted bois noir disease of grapevine in Serbia. Eur. J. Plant Pathol. 2019, 153, 183–195. [Google Scholar] [CrossRef]
- Jakovljević, M.; Jović, J.; Krstić, O.; Mitrović, M.; Marinković, S.; Toševski, I.; Cvrković, T. Diversity of phytoplasmas identified in the polyphagous leafhopper Euscelis incisus (Cicadellidae, Deltocephalinae) in Serbia: Pathogen inventory, epidemiological significance and vectoring potential. Eur. J. Plant Pathol. 2020, 156, 201–221. [Google Scholar] [CrossRef]
- Cimerman, A.; Pacifico, D.; Salar, P.; Marzachì, C.; Foissac, X. Striking diversity of vmp1, a variable gene encoding a putative membrane protein of the stolbur phytoplasma. Appl. Environ. Microbiol. 2009, 75, 2951–2957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabre, A.; Danet, J.L.; Foissac, X. The stolbur phytoplasma antigenic membrane protein gene stamp is submitted to diversifying positive selection. Gene 2011, 472, 37–41. [Google Scholar] [CrossRef] [PubMed]
- Atanasova, B.; Jakovljević, M.; Spasov, D.; Jović, J.; Mitrović, M.; Toševski, I.; Cvrković, T. The molecular epidemiology of bois noir grapevine yellows caused by ‘Candidatus Phytoplasma solani’ in the Republic of Macedonia. Eur. J. Plant Pathol. 2015, 142, 759–770. [Google Scholar] [CrossRef] [Green Version]
- Aryan, A.; Brader, G.; Mörtel, J.; Pastar, M.; Riedle-Bauer, M. An abundant ‘Candidatus Phytoplasma solani’ tuf b strain is associated with grapevine, stinging nettle and Hyalesthes obsoletus. Eur. J. Plant Pathol. 2014, 140, 213–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kosovac, A.; Johannesen, J.; Krstić, O.; Mitrović, M.; Cvrković, T.; Toševski, I.; Jović, J. Widespread plant specialization in the polyphagous planthopper Hyalesthes obsoletus (Cixiidae), a major vector of stolbur phytoplasma: Evidence of cryptic speciation. PLoS ONE 2018, 13, e0196969. [Google Scholar] [CrossRef]
- Maixner, M. Transmission of German grapevine yellows (Vergilbungskrankheit) by the planthopper Hyalesthes obsoletus (Auchenorrhyncha: Cixiidae). Vitis 1994, 33, 103–104. [Google Scholar]
- Kosovac, A.; Radonjić, S.; Hrnčić, S.; Krstić, O.; Toševski, I.; Jović, J. Molecular tracing of the transmission routes of bois noir in Mediterranean vineyards of Montenegro and experimental evidence for the epidemiological role of Vitex agnus-castus (Lamiaceae) and associated Hyalesthes obsoletus (Cixiidae). Plant Pathol. 2016, 65, 285–298. [Google Scholar] [CrossRef] [Green Version]
- Biedermann, R.; Niedringhaus, R. Die ZikadenDeutschlands—BestimmungstafelnfüralleArten; WABV Fründ: Scheessel, Germany, 2004; pp. 1–409. [Google Scholar]
- Krstić, O.; Cvrković, T.; Mitrović, M.; Toševski, I.; Jović, J. Dictyophara europaea (Hemiptera: Fulgoromorpha: Dictyopharidae): Description of immatures, biologyandhost plant associations. Bull. Entomol. Res. 2016, 106, 395–405. [Google Scholar] [CrossRef] [PubMed]
- Filippin, L.; Jović, J.; Cvrković, T.; Forte, V.; Clair, D.; Toševski, I.; Boudon-Padieu, E.; Borgo, M.; Angelini, E. Molecular characteristics of phytoplasmas associated with Flavescence dorée in clematis and grapevine and preliminary results on the role of Dictyophara europaea as a vector. Plant Pathol. 2009, 58, 826–837. [Google Scholar] [CrossRef] [Green Version]
- Cvrković, T.; Jović, J.; Mitrović, M.; Krstić, O.; Krnjajić, S.; Toševski, I. Potential new hemipteran vectors of stolbur phytoplasma in Serbian vineyards. Bull. Insectology 2010, 64, 129–130. [Google Scholar]
- Mitrović, M.; Jović, J.; Cvrković, T.; Krstić, O.; Trkulja, N.; Toševski, I. Characterisation of a 16SrII phytoplasma strain associated with bushy stunt of hawkweed oxtongue (Picris hieracioides) in south-eastern Serbia and the role of the leafhopper Neoaliturus fenestratus (Deltocephalinae) as a natural vector. Eur. J. Plant Pathol. 2012, 134, 647–660. [Google Scholar] [CrossRef]
- Krstić, O.; Cvrković, T.; Mitrović, M.; Radonjić, S.; Hrnčić, S.; Toševski, I.; Jović, J. Wolbachia infection in natural populations of Dictyophara europaea, an alternative vector of grapevine Flavescence dorée phytoplasma: Effects and interactions. Ann. Appl. Biol. 2018, 172, 47–64. [Google Scholar] [CrossRef]
- Quaglino, F.; Sanna, F.; Moussa, A.; Faccincani, M.; Passera, A.; Casati, P.; Bianco, P.A.; Mori, N. Identification and ecology of alternative insect vectors of ‘Candidatus Phytoplasma solani’ to grapevine. Sci. Rep. 2019, 9, 19522. [Google Scholar] [CrossRef] [PubMed]
- Balakishiyeva, G.; Bayramova, J.; Mammadov, A.; Salar, P.; Danet, J.L.; Ember, I.; Verdin, E.; Foissac, X.; Huseynova, I. Important genetic diversity of “Candidatus Phytoplasma solani” related strains associated with Bois noir grapevine yellows and planthoppers in Azerbaijan. Eur. J. Plant. Pathol. 2018, 151, 937–946. [Google Scholar] [CrossRef]
- Holzinger, W.E.; Kammerlander, I.; Nickel, H. The Auchenorrhyncha of Central Europe, Fulgoromorpha, Cicadomorpha Excl. Cicadellidae; Brill Academic Publishers: Leiden, The Netherlands, 2003; pp. 1–673. [Google Scholar]
- Angelini, E.; Clair, D.; Borgo, M.; Bertaccini, A.; Boudon-Padieu, E. Flavescence dorée in France and Italy—occurrence of closely related phytoplasma isolates and their near relationships to Palatinate grapevine yellows and an alder yellows phytoplasma. Vitis 2001, 40, 79–86. [Google Scholar]
- Daire, X.; Clair, D.; Reinert, W.; Boudon-Padieu, E. Detection and differentiation of grapevine yellows phytoplasmas belonging to the elm yellows group and to the stolbur subgroup by PCR amplification of non-ribosomal DNA. Eur. J. Plant Pathol. 1997, 103, 507–514. [Google Scholar] [CrossRef]
- Clair, D.; Larrue, J.; Aubert, G.; Gillet, J.; Cloquemin, G.; Boudon-Padieu, E. A multiplex nested-PCR assay for sensitive and simultaneous detection and direct identification of phytoplasma in the Elm yellows group and Stolbur group and its use in survey of grapevine yellows in France. Vitis 2003, 42, 151–157. [Google Scholar]
- Radonjić, S.; Hrnčić, S.; Jović, J.; Cvrković, T.; Krstić, O.; Krnjajić, S.; Toševski, I. Occurrence and distribution of grapevine yellows caused by stolbur phytoplasma in Montenegro. J. Phytopathol. 2009, 157, 682–685. [Google Scholar] [CrossRef]
- Fialová, R.; Válová, P.; Balakishiyeva, G.; Danet, J.L.; Šafárová, D.; Foissac, X.; Navrátil, M. Genetic variability of stolbur phytoplasma in annual crop and wild plant species in south Moravia. J. Plant Pathol. 2009, 91, 411–416. [Google Scholar]
- Delić, D.; Balech, B.; Radulović, M.; Lolić, B.; Karačić, A.; Vukosavljević, V.; Đurić, G.; Cvetković, T.J. Vmp1 and stamp genes variability of ‘Candidatus phytoplasma solani’ in Bosnian and Herzegovinian grapevine. Eur. J. Plant Pathol. 2016, 145, 221–225. [Google Scholar] [CrossRef]
- Mitrović, M.; Jakovljević, M.; Jović, J.; Krstić, O.; Kosovac, A.; Trivellone, V.; Jermini, M.; Toševski, I.; Cvrković, T. ‘Candidatus Phytoplasma solani’ genotypes associated with potato stolbur in Serbia and the role of Hyalesthes obsoletus and Reptalus panzeri (Hemiptera, Cixiidae) as natural vectors. Eur. J. Plant Pathol. 2016, 144, 619–630. [Google Scholar] [CrossRef]
- Plavec, J.; Križanac, I.; Budinšćak, Ž.; Škorić, D.; Šeruga Musić, M. A case study of FD and BN phytoplasma variability in Croatia: Multigene sequence analysis approach. Eur. J. Plant Pathol. 2015, 142, 591–601. [Google Scholar] [CrossRef]
- Ćurčić, Ž.; Kosovac, A.; Stepanović, J.; Rekanović, E.; Kube, M.; Duduk, B. Multilocus Genotyping of ‘Candidatus Phytoplasma solani’Associated with Rubbery Taproot Disease of Sugar Beet in the Pannonian Plain. Microorganisms 2021, 9, 1950. [Google Scholar] [CrossRef]
- Bandelt, H.J.; Forster, P.; Röhl, A. Median-joining networks for inferring intraspecific phylogenies. Mol. Biol. Evol. 1999, 16, 37–48. [Google Scholar] [CrossRef]
- Jović, J.; Marinković, S.; Jakovljević, M.; Krstić, O.; Cvrković, T.; Mitrović, M.; Toševski, I. 2021. Symptomatology, (Co) occurrence and Differential Diagnostic PCR Identification of ‘Ca. Phytoplasma solani’ and ‘Ca. Phytoplasma convolvuli’ in Field Bindweed. Pathogens 2021, 10, 160. [Google Scholar] [CrossRef]
- Song, Z.S.; Liang, A.P. The Palaearctic planthopper genus Dictyophara Germar, 1833 (Hemiptera: Fulgoroidea: Dictyopharidae) in China. Ann. Zool. 2008, 58, 537–549. [Google Scholar] [CrossRef]
- Jović, J.; Cvrković, T.; Mitrović, M.; Krnjajić, S.; Redinbaugh, M.G.; Pratt, R.C.; Gingery, R.E.; Hogenhout, S.A.; Toševski, I. Roles of stolbur phytoplasma and Reptalus panzeri (Cixiinae, Auchenorrhyncha) in the epidemiology of Maize redness in Serbia. Eur. J. Plant Pathol. 2007, 118, 85–89. [Google Scholar] [CrossRef]
- Maixner, M. Phytoplasma Epidemiological Systems with Multiple Plant Hosts. In Phytoplasmas: Genomes, Plant Hosts and Vectors; Weintraub, P.G., Jones, P., Eds.; CABI Publishing: Wallingford, UK, 2010; pp. 213–232. [Google Scholar]
- Pierro, R.; Panattoni, A.; Passera, A.; Materazzi, A.; Luvisi, A.; Loni, A.; Ginanni, M.; Lucchi, A.; Bianco, P.A.; Quaglino, F. Proposal of a new Bois noir epidemiological pattern related to ‘Candidatus Phytoplasma solani’strains characterized by a possible moderate virulence in Tuscany. Pathogens 2020, 9, 268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabre, A.; Balakishiyeva, G.; Ember, I.; Omar, A.; Acs, Z.; Kölber, M.; Kauzner, L.; Della Bartola, M.; Danet, J.L.; Foissac, X. StAMP encoding the antigenic membrane protein of stolburphytoplasma is useful for molecular epidemiology. Bull. Insectol. 2011, 64, S21–S22. [Google Scholar]
- Salehi, M.; Izadpanah, K.; Siampour, M. Characterization of a phytoplasma associated with cabbage yellows in Iran. Plant Dis. 2007, 91, 625–630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, R.; Wang, G.; Wang, S.; Zhang, D.; Wei, L.; Chen, H.; Li, O.; Hu, X. Molecular identification and diversity of ‘Candidatus Phytoplasma solani’associated with red-leaf disease of Salvia miltiorrhiza in China. J. Phytopathol. 2016, 164, 882–889. [Google Scholar] [CrossRef]
- Chuche, J.; Danet, J.L.; Rivoal, J.B.; Arricau-Bouvery, N.; Thiéry, D. Minor cultures as hosts for vectors of extensive crop diseases: Does Salvia sclarea act as a pathogen and vector reservoir for lavender decline? J. Pest Sci. 2018, 91, 145–155. [Google Scholar] [CrossRef]
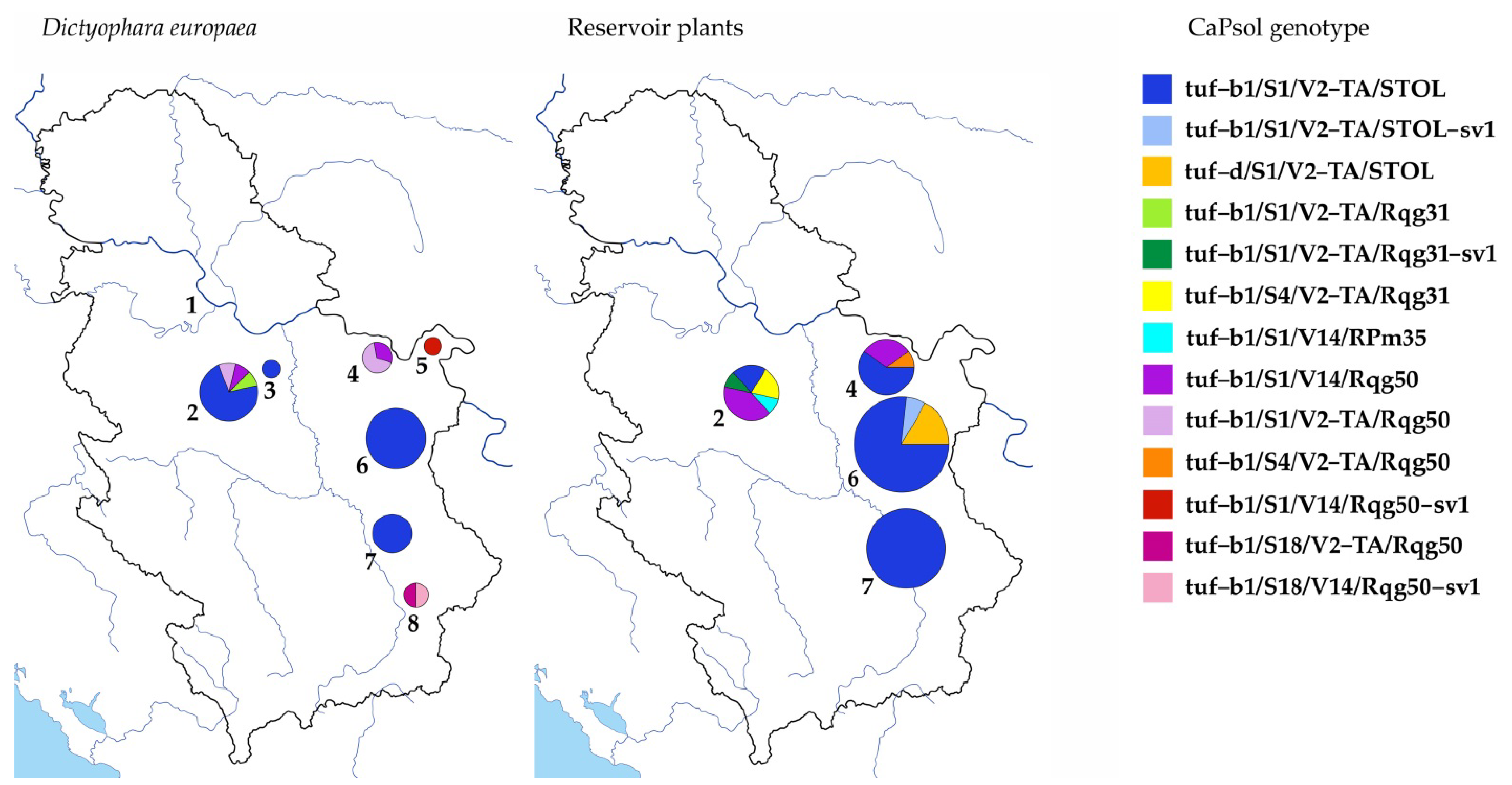
| Region | Location (GPS) 1 | Habitat Type | Date of Collection | No. of Analyzed/CPs-Positive Specimens (%) | ‘Ca. P. solani’ MLSA Genotype 2 | |||
|---|---|---|---|---|---|---|---|---|
| tuf Sequence | secY Sequence | vmp1 Profile | stamp Sequence | |||||
| Northern Serbia | 1. Dobanovci (44°50.890′ N 20°10.089′ E) | Semi-mesic meadow | August 2017 | 15/0 | - | - | - | - |
| Central Serbia | 2. Topola (44°13.705′ N 20°41.137′ E) | Vineyards region | July 2016 | 27/3 (11%) | tuf-b1 | S1 | V2-TA | STOL (2) |
| tuf-b1 | S1 | V2-TA | Rqg31 | |||||
| August 2016 | 14/1 (7%) | tuf-b1 | S1 | V2-TA | STOL | |||
| August 2017 | 75/7 (9%) | tuf-b1 | S1 | V2-TA | STOL (5) | |||
| tuf-b1 | S1 | V2-TA | Rqg50 | |||||
| tuf-b1 | S1 | V14 | Rqg50 | |||||
| 3. Krnjevo (44°25.787′ N 21°02.764′ E) | Vineyards region | September 2018 | 10/1 (10%) | tuf-b1 | S1 | V2-TA | STOL | |
| Eastern Serbia | 4. Donji Milanovac (44°31.501′ N 22°02.500′ E) | Abandoned pasture | July 2017 | 26/3 (12%) | tuf-b1 | S1 | V2-TA | Rqg50 |
| tuf-b1 | S1 | V14 | Rqg50 (2) | |||||
| 5. Brza Palanka (44°29.201′ N 22°27.381′ E) | Moderate hillside meadow | July 2018 | 12/1 (8%) | tuf-b1 | S1 | V14 | Rqg50-sv1 | |
| 6. Rogljevo (44°07.513′ N 22°34.078′ E) | Vineyards region | August 2016 | 48/5 (10%) | tuf-b1 | S1 | V2-TA | STOL (5) | |
| August 2017 | 23/3 (13%) | tuf-b1 | S1 | V2-TA | STOL (3) | |||
| August 2018 | 34/4 (12%) | tuf-b1 | S1 | V2-TA | STOL (4) | |||
| Southern Serbia | 7. Jasenovik (43°21.909′ N 22°01.341′ E) | Stony meadow | August 2017 | 48/5 (10%) | tuf-b1 | S1 | V2-TA | STOL (5) |
| 8. Vlasotince (42°56.283′ N 22°05.543′ E) | Xerothermic ruderal slope | July 2017 | 25/2 (8%) | tuf-b1 | S18 | V14 | Rqg50-sv1 | |
| tuf-b1 | S18 | V2-TA | Rqg50 | |||||
| Total | 357/35 (10%) | |||||||
| Location (GPS) 1 | Plant Species | No. of Analyzed/CPs-Positive Specimens (%) | ‘Ca. P. solani’ MLSA Genotype 2 | |||
|---|---|---|---|---|---|---|
| tuf Sequence | secY Sequence | vmp1 Profile | stamp Sequence | |||
| 2. Topola | Salvia pratensis | 20/2 (10%) | tuf-b1 | S1 | V2-TA | STOL (2) |
| Convolvulus arvensis | 20/8 (40%) | tuf-b1 | S1 | V14 | Rqg50 (4) | |
| tuf-b1 | S4 | V2-TA | Rqg31 (2) | |||
| tuf-b1 | S1 | V2-TA | Rqg31-sv1 | |||
| tuf-b1 | S1 | V14 | Rpm35 | |||
| Clematis vitalba | 6/0 | - | - | - | - | |
| 4. Donji Milanovac | Convolvulus arvensis | 12/4 (33%) | tuf-b1 | S1 | V14 | Rqg50 (3) |
| tuf-b1 | S4 | V2-TA | Rqg50 | |||
| Cichorium intybus | 6/2 (33%) | tuf-b1 | S1 | V2-TA | STOL (2) | |
| Daucus carota | 6/1 (17%) | tuf-b1 | S1 | V2-TA | STOL | |
| Sorghum halepense | 6/2 (33%) | tuf-b1 | S1 | V2-TA | STOL (2) | |
| Elymus repens | 6/1 (17%) | tuf-b1 | S1 | V2-TA | STOL | |
| 6. Rogljevo | Crepis foetida | 20/12 (60%) | tuf-b1 | S1 | V2-TA | STOL (8) |
| tuf-b1 | S1 | V2-TA | STOL-sv1 | |||
| tuf-d | S1 | V2-TA | STOL (3) | |||
| Picris hieracioides | 16/6 (38%) | tuf-b1 | S1 | V2-TA | STOL (6) | |
| Cichorium intybus | 6/2 (33%) | tuf-b1 | S1 | V2-TA | STOL (2) | |
| Daucus carota | 16/7 (44%) | tuf-b1 | S1 | V2-TA | STOL (4) | |
| tuf-b1 | S1 | V2-TA | STOL-sv1 | |||
| tuf-d | S1 | V2-TA | STOL (2) | |||
| Sorghum halepense | 6/2 (33%) | tuf-b1 | S1 | V2-TA | STOL (2) | |
| Elymus repens | 6/1 (17%) | tuf-b1 | S1 | V2-TA | STOL | |
| Achillea millefolium | 6/0 | - | - | - | - | |
| Falcaria vulgaris | 6/0 | - | - | - | - | |
| Clematis vitalba | 6/0 | - | - | - | - | |
| 7. Jasenovik | Crepis foetida | 18/10 (56%) | tuf-b1 | S1 | V2-TA | STOL (10) |
| Picris hieracioides | 20/8 (40%) | tuf-b1 | S1 | V2-TA | STOL (8) | |
| Erigeron annuus | 6/1 (17%) | tuf-b1 | S1 | V2-TA | STOL | |
| Daucus carota | 6/1 (17%) | tuf-b1 | S1 | V2-TA | STOL | |
| Cynodon dactylon | 8/1 (13%) | tuf-b1 | S1 | V2-TA | STOL | |
| Carlina vulgaris | 6/0 | - | - | - | - | |
| Senecio erucifolius | 6/0 | - | - | - | - | |
| Total | 246/71 (29%) | |||||
| Location of D. europaea Collection | Experimental Plant | No. of D. europaea Specimens per Plant | No. of CPs-Transmitted/Replicate Plants | tuf/secY/vmp1/stamp Genotype of Transmitted Isolates (Number of Isolates) |
|---|---|---|---|---|
| Rogljevo, Eastern Serbia | Vitis vinifera cv. Chardonnay | 12 | 3/12 | tuf-b1/S1/V2-TA/STOL (3) |
| Catharantus roseus | 12 | 4/12 | tuf-b1/S1/V2-TA/STOL (4) | |
| Topola, Central Serbia | Vitis vinifera cv. Chardonnay | 12 | 2/12 | tuf-b1/S1/V2-TA/STOL (2) |
| tuf-b1/S1/V2-TA/Rqg31 (1) | ||||
| Catharantus roseus | 12 | 3/12 | tuf-b1/S1/V2-TA/STOL (2) | |
| tuf-b1/S1/V2-TA/Rqg50 (1) | ||||
| Total | 12/48 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cvrković, T.; Jović, J.; Krstić, O.; Marinković, S.; Jakovljević, M.; Mitrović, M.; Toševski, I. Epidemiological Role of Dictyophara europaea (Hemiptera: Dictyopharidae) in the Transmission of ‘Candidatus Phytoplasma solani’. Horticulturae 2022, 8, 654. https://doi.org/10.3390/horticulturae8070654
Cvrković T, Jović J, Krstić O, Marinković S, Jakovljević M, Mitrović M, Toševski I. Epidemiological Role of Dictyophara europaea (Hemiptera: Dictyopharidae) in the Transmission of ‘Candidatus Phytoplasma solani’. Horticulturae. 2022; 8(7):654. https://doi.org/10.3390/horticulturae8070654
Chicago/Turabian StyleCvrković, Tatjana, Jelena Jović, Oliver Krstić, Slavica Marinković, Miljana Jakovljević, Milana Mitrović, and Ivo Toševski. 2022. "Epidemiological Role of Dictyophara europaea (Hemiptera: Dictyopharidae) in the Transmission of ‘Candidatus Phytoplasma solani’" Horticulturae 8, no. 7: 654. https://doi.org/10.3390/horticulturae8070654
APA StyleCvrković, T., Jović, J., Krstić, O., Marinković, S., Jakovljević, M., Mitrović, M., & Toševski, I. (2022). Epidemiological Role of Dictyophara europaea (Hemiptera: Dictyopharidae) in the Transmission of ‘Candidatus Phytoplasma solani’. Horticulturae, 8(7), 654. https://doi.org/10.3390/horticulturae8070654








