Abstract
Exploring the genetic diversity among plant accessions is important for conserving and managing plant genetic resources. In the current study, a collection of forty-six tomato accessions from Jordan were evaluated based on their performance and their morpho-physiological, in addition to molecularly characterizing to detect genetic diversity. Tomato accessions seedlings were exposed to drought stress with 70% field capacity and 40% field capacity under field conditions in Jordan. Drought stress had significantly negatively influenced the dry root weight, fresh root weight, root growth rate, fresh shoot weight, dry shoot weight, and shoot growth rate. Moreover, proline content showed a highly significant increase of 304.2% in response to drought stress. The analysis of twenty morphological characters revealed a wide range of variations among tomato accessions. Accessions were screened with fourteen SSR primers; six primers were informative to explain the genetic diversity. Based on resolving power, primers LEct004 and LEat018 were most significant with all 46 accessions. Interestingly, polymorphic information content (PIC) values ranged from 0.00 (Asr2 marker) to 0.499 (LEct004), which confirms that the SSR markers are highly informative. Our findings provide new insights into using informative molecular markers to elucidate such wide genetic variation discovered in our collections from Afraa and Abeel (the southern part of Jordan). Interestingly, the SSR markers were associated with genes, e.g., LEat018 with ACTIN_RELATED PROTEIN gene, the LEct004 with the HOMEOBOX PROTEIN TRANSCRIPTION FACTORS gene, and Asr2 with ABA/WDS. Moreover, the AUXIN RESPONSE FACTOR8 gene was associated with the LEta014 SSR marker and the LEta020 with the THIOREDOXIN FAMILY TRP26 gene. Therefore, the genetic diversity analysis and functional annotations of the genes associated with SSR information obtained in this study provide valuable information about the most suitable genotype that can be implemented in plant breeding programs and future molecular analysis. Furthermore, evaluating the performance of the collection under different water regimes is essential to produce new tomato varieties coping with drought stress conditions.
1. Introduction
Tomato (Solanum lycopersicum L.) is an annual herbaceous plant that belongs to the Solanaceae Family [1,2,3]. Tomato is the second most economically important vegetable grown worldwide, it forms a significant part of the agricultural industry and is also the second most consumed vegetable. It is known that tomato production is increased considerably worldwide [2,3,4,5]. In Jordan, a wide range of tomato cultivars and accessions were gathered from farmers and stored in the seed bank at the National Agricultural Research Center (NARC) [6,7,8]. In Jordan, the production of tomatoes increases year by year with high ability of export. Accession tomatoes are highly adapted to the Jordanian conditions with large hereditary types used in breeding for tomato productivity and adaptation improvement. Therefore, they have been used for some time in breeding to make a character of adaptation to abiotic stresses [6,7]. Accessions are an excellent source for improving the tomato crop for drought and have existed in Jordan for several years. The collections of accession frequently exist in the thousands, which can usually be subject to pre-screening under different stress conditions to identify the most promising genotype [9]. Tomato is known to be sensitive to environmental stresses, including drought stress which impacts seed germination and plant development and performance [10]. Cattivelli et al. [11] reported the effect of drought on stages of plant development to understand the drought tolerance from germination to reproduction through conventional and molecular approaches. Drought stress during the vegetative or early reproductive phase usually reduces yield. It often induces physiological and molecular changes in plant water relation parameters such as cell turgor pressure, osmotic pressure, and water potential [12,13]. Drought stress at different developmental stages causes several morphological and biochemical alternations in various plant species. Water deficit at the early seedling stage might lead to higher dry root weights, longer roots, coleoptiles, and higher root-to-shoot ratios [14,15,16], all of these changes are parameters of interest and have been widely used as reliable markers toward drought stress tolerance for evaluating various crop plants. Reduction in water potential induces stomatal closure resulting in a decrease in photosynthesis, leaf expansion and orientation, stomatal behavior, photosynthesis, respiration rate, solute translocation, and ultimately yields [13,17]. Toxic substances generated during stress, such as reactive oxygen species (ROS), cause oxidative damage to the cellular organization. Tomato plants have developed an antioxidant system that scavenges toxic elements and accumulates osmoprotectants, including proline, glycine betaine, and other osmoprotectants to keep osmotic balance [13,17]. Drought stress tolerance was evaluated according to different tolerance indices to characterize tomatoes’ physiological and genetic basis, including plant development, fruit set, fruit weight, shoot and root morphology, water use efficiency (WUE), and other physiological parameters [18]. Researchers have evaluated drought-tolerant tomato breeding cultivars in response to drought conditions drought stress [18]. Pakmore VF and the breeding line L03306 showed better performance in several deficit irrigation regimes. These genotypes are considered a resource for the drought tolerance breeding program.
In recent years, crop physiology and agronomics have led to new insights into drought tolerance. These insights have provided the breeders with new knowledge and tools for plant improvement and the ability to detect the variation between species, varieties, and accessions; for example, using several types of DNA molecular markers can be used such as random amplified polymorphic DNA (RAPD), restriction fragment length polymorphism (RFLP), amplified fragment length polymorphism (AFLP), minisatellites, variable number of tandem repeats (VNTRs), and simple sequence repeats (SSRs) [11,19,20,21]. SSRs are sections of DNA consisting of (1–6) or (2–7) base pair units tandemly repeated throughout the genome [20,22,23,24], to detect the variation degree [9,21,24] or phylogenetic [25,26,27]. The hypervariability character of SSR markers, based on microsatellite DNA loci, enables this method to be the main major in studying plant population genetics [8,20,28,29,30,31]. SSRs can be used in gene mapping studies, for example, using 65 SSR primer pairs by Liu et al. [32] in the cotton genome mapping [33]. Additionally, Shiri [34] used 38 maize hybrids and 12 SSR pairs to investigate the genetic diversity and then identify the informative SSRs for drought tolerance. The amplified bands were 40, the number of alleles ranged from 2 to 6, and the Polymorphism Information Content (PIC) ranged from 0.23 to 0.79. Tam et al. [35] detected the genetic diversity within the 34 lines of tomato and 35 lines of pepper using 29 SSR primers (16 for tomato and 13 for pepper). Our study noticed that the genetic variations within the tomato and pepper collections were similar because all the bands for both collections were polymorphic and the polymorphism percentages were equal (100%).
The specific goals of this research were to evaluate the genetic diversity of the tomato accessions grown in Jordan and to determine (1) the extent of morphological variations among tomato accessions, (2) the allele distribution in relation to the gene pool origins and probable drought tolerance based on geographic origin, (3) evaluate the effect of drought stress on tomato accessions at the seedling stage, (4) predict the function of candidate genes that are associated with our SSR primers in tomato, (5) determine the expression pattern of our target genes in plant tissues-specific, and (6) by combining phenotypes, SSR marker genotypes, and putative expression pattern, we can understand the functional roles of our genes which are related to the local adaptation to drought stress.
2. Materials and Methods
2.1. Plant Materials and Effect of Drought on Tomato Accessions at Germination Stage
Forty-six tomato accessions originating from different geographical regions in Jordan were ordered from the Genebank of the National Center for Agricultural Research and Extension (NCARE), Amman, Jordan, and were used in this study (Table 1). The seeds were grown in growth chambers at the plant production department laboratories as well as glasshouse of Jordan University of Science and Technology (JUST).At seedling stage, the leaf sampleswere collected for DNA analysis at the Princess Haya Biotechnology Center (PHBC) at the King Abdullah University Hospital (KAUH) and the University of Jordan.

Table 1.
Tomato accession collection regions used in this study.
2.2. Field Trial
Ten seeds of each tomato accession were sown at 8 cm depth (in 8 L pots containing a mixture of soil: sand: peat moss in a volume ratio of 2:1:1) under greenhouse conditions. Field capacity (FC) was determined by saturating the soil with water and recording the weight of the soil after drainage had stopped. Soil moisture content was measured gravimetrically by weighting soil samples before and after oven-drying at 105 °C for 24 h divided by the weight of the dry soil. Di-ammonium phosphate (DAP) fertilizer was added to the pots, the seeds were sown, and then the pots were covered with plastic to reduce evaporation during development. When plumule started to emerge, small holes were made carefully in the covers to enable the plants to grow, and 50 mL NPK (30:10:10) was added per pot (60 g per 20 L) to avoid the appearance of mineral deficiency in plants. Plants were exposed to drought stress at the beginning of the early seedling stage with 70% field capacity and 40% field capacity under field conditions, and three randomly selected accessions were used as a reference. Before irrigation, three reference pots were weighed and watered to adjust the corresponding FC. The experiment was carried out using a completely randomized design with three replications (ten seeds per replicate).
2.3. Vegetative Traits
After 60 days of growing, tomato plants were harvested. Three plants from each replicate of each treatment were randomly selected to be used for further analysis. Shoots and roots were separated manually to measure the fresh and dry weights after drying them at (65 °C for 72 h) in the oven. The relative shoot or root weight was calculated as follows:
Relative of shoot or root weight = [shoot or root weight at drought treatment/shoot or root weight at control] ×100.
The growth rate of shoot or root = shoot or root fresh weight/45 days.
Relative shoot or root growth rate = [shoot or root growth rate at drought treatment/shoot or root fresh growth rate at control] ×100.
2.4. Determination of Proline Content
The free content of proline was estimated and extracted according to the protocol of Bates et al. [36] _ENREF_32, and more measurement steps are discussed in the protocol by Senthilkumar et al. [37].
2.5. Morphological Characteristics
The following 20 morphological characters related to the tomato accessions (Table S4) during the seedling, immature, mature, and ripening stages were measured by a Tomato descriptor (IPGRI, 1999). Additionally, fruit shape-related perimeter traits and fruit shape index for the external shape and other characters were measured by a Tomato Analyzer (TA) software program version 3 (Rodríguez et al., 2010).
2.6. Statistical Analysis
2.6.1. Drought Data Analysis
The phenotypic data were statistically analyzed by the SPSS software (version 17). Analysis of variance (ANOVA) and means separation at LSD (0.05) in addition to T-Test were also calculated using SPSS software to compare the treatments.
2.6.2. Morphological Data Analysis
Data were statistically analyzed by the SPSS software (version 17). Means, range, maximum value, minimum value, standard deviation, standard error of the mean, and sum were measured. Hierarchical cluster analysis was used to calculate morphological similarity values between accessions using the Euclidean distance interval option and then classified them by dendrogram using average linkage (within groups).
2.7. DNA Extraction and Simple Sequence Repeat (SSR) Assays
Total genomic DNA was extracted from young leaves for five different plants per line using a DNA Plant Kit (Qiagen). DNA quantification was performed with an ND-1000 spectrophotometer (Nanodrop Technologies, Wilmington, DE, USA). The DNA quality was assessed using the absorbance ratio at 260 to that at 280 nm wavelengths (A260/A280). DNA quantity was calculated as DNA (µg/µL) = A260 × 50, where A260 is the absorbance at 260 nm. Thus, the concentration of DNA in µg/mL was calculated as DNA (µg/mL) = [A260 × 50] × DF where DF is the dilution factor. Fourteen SSR primers designed for tomato DNA fingerprinting were used. These SSR primer sequences were obtained from [38]. Six primers were selected for the next analysis to determine genetic diversity in tomato collection (LEat018, LEct004, LEta014, LEta020, CT114, and Asr2) based on the screening of fourteen SSR primers. Information about these primer sequences and their information are presented in Table 2.

Table 2.
Primers and their sequence, expected fragment, and melting temperature.
2.8. PCR Amplification and Product Electrophoresis
PCR amplification was performed for all the SSR markers and the best performing conditions were identified. During the primer testing, a fraction of the total number of plants was used for the polymerase chain reaction. PCR reactions were performed in 96-well plates using either the Perkin Elmer GeneAmp PCR system 9600 (PE Biosystems) or the TECHNE Genius thermal cycler (Techne Ltd., Cambridge, UK) with the same amplification program. Six SSR primer pairs were used for amplification reaction (Table 2). The DNA from the 46 tomato samples was amplified using SSR markers following the PCR amplification protocol by Promega (Madison, WI, USA). The PCR amplification conditions were set up for one cycle of denaturation for 2 min at 94 °C, followed by 33-cycle amplification with a 25 s denaturing at 94 °C, a 25 s annealing at the Tm (Tm varies for the individual primers), a 25 s extension at 68 °C, and a final extension cycle at 68 °C for 5 min.
The PCR products of SSRs were separated using 3% Metaphor Agarose gel (FMCBioProducts) that was recommended to separate small-sized bands from SSRs [39] and electrophoretic apparatus (MS Major Science, UK) and BIO-RAD (Criterion TM cassettes) 100 V for 1 h was applied. DNA loading dye of 1 X was added to the PCR products for visual capture of DNA migration during electrophoresis. Five μL of 1000 bp and 100 bp DNA ladder were used as a reference to estimate the size of specific DNA bands. Finally, Gel Works ID advanced software analyzed the amplified DNA banding patterns. The size of the allele fragments that SSR amplified was measured. Polymorphism information content (PIC) was calculated as PIC = 1 − Spi2, where pi is the allele frequency [40].
2.9. Data Scoring
The gel for each primer was analyzed separately by scoring the bands and coded by (0) and (1) for the absent and present amplification bands for all test markers, respectively, using SAGA 6 software). Genetic similarity values between accessions were calculated using the Dice coefficient according to Dice [41]:
2 × |X ∩ Y|/(|X| + |Y|)
From the NTSYS-pc, version 2.0 software [42]. The genetic similarity matrix was used to generate a dendrogram using the Unweighted Pair Group Method of Arithmetic Averages (UPGMA).
2.10. Functional Assignments for Gene-Associated SSRs in Tomato
The sequence of SSR markers was used as a query to search against the Solanum lycopersicum genomics that we already downloaded from NCBI genomics (https://www.ncbi.nlm.nih.gov/genome/?term=tomato, accessed on 12 April 2022). Then, the alignment sequence was compared using various databases, such as National Center for Biotechnology Information (NCBI) gene bank, Phytozome, InterPro, and KEGG databases to predict the candidate genes associated with our SSR primers in tomatoes. For the potential functions of these genes, Phytozome v13 was used to obtain the annotations by KOG (Eukaryotic Orthologous Groups), KEGG (Kyoto Encyclopedia of Genes and Genomes), ENZYME, Pathway, and the InterPro family of protein analysis (Classification of protein families) tools. In Phytozome, we made the blast sequence against five tomato genomics such as Solanum lycopersicum ITAG2.4, Solanum lycopersicum ITAG3.2, Solanum lycopersicum ITAG4.0, Solanum tuberosum v4.03, and Solanum tuberosum v6.1.
2.11. Putative Tissue Expression Pattern, Subcellular Localization, Root Cell Types and Tissues of Our Target Genes
Putative tissue-specific expression profiles of Solyc11g005330, Solyc02g089940, Solyc03g031970, Solyc01g097450, Solyc10g011690, and Solyc04g071580 genes that are associated with our SSR markers were extracted based on Solanum lycopersicum transcript expression database from nineteen tissues and organs including flowers, leaves, roots, and fruit from different developmental stages. Expression profiles were built using the tomato plant Electronic Fluorescent Pictograph Browsers (Tomato eFP browsers) (http://bar.utoronto.ca/eplant_tomato/) accessed on 12 April 2022 [43]. Moreover, the putative subcellular localizations of our previous gene from Solanum lycopersicum were examined based on tomato protein localization of fourteen different cell organs to recognize possible synthesis sites using the tomato Cell eFP browsers (Tomato eFP browsers) (http://bar.utoronto.ca/eplant_tomato/) accessed on 12 April 2022. Furthermore, the putative root cell types and tissues specific to our genes were examined using different root cell types and tissues under various promoter toolboxes, such as AtWER, SIPEP, AtPEP, SICO2, SISCR, SISHR, AtS32, AtS18, SIWOX5, SIRPL11C, and 35S promoters to determine the putative function of our genes at specific root cell types http://bar.utoronto.ca/eplant_tomato/ accessed on 12 April 2022.
3. Results
3.1. The Effects of Drought on Tomato Accessions at the Seedling Stage
Several parameters such as fresh root weight, dry root weight, root growth rate, fresh shoot weight, dry shoot weight, and shoot growth rate were measured on the 46 tomato accessions. Seedlings were grown under three levels of water stress (control, 70% FC, 40% FC). However, all the parameters showed significant differences at 40% FC compared to the control treatment, which was also significant at 70%FC (Table S1). As expected, all morphological traits had a lower mean performance under drought stress (70% FC and 40% FC) than under normal conditions. On average, all parameters had a reduction due to drought stress at 70% FC and 40% FC compared with the control treatment (Table S3).
Drought stress is significantly affected by the proline concentrations in the leaf tissue for 46 tomato accessions (Table S1). A highly significant increment in proline content was detected at 40% FC compared to the control treatment (100% FC) by 304.2%.
3.2. Morphological Characterization among Tomato Accessions
In this study, Tomato Analyzer (TA) was used to assess the fruit shape variation in the accessions by measuring the morphological characterizations rapidly and accurately and quantifying traits that are impossible to quantify manually. The analysis of variance for all the morphological characters indicates a wide range of variability among tomato accessions, including fruit shape-related perimeter traits and fruit shape index for the external shape fruit shape index internal (Table S4).
3.3. Genetic Variation among Tomato Accessions Revealed by SSRs
Our investigation tested 14 SSR primers; of these, six yielding polymorphic amplification products were used, and the remaining 8 SSR primers either yielded no amplification product or no polymorphic. The banding patterns of SSRs are shown in Supplementary Figures S1–S6. Of the 46 tomato accessions, the genetic relationship among thirty-six tomato accessions was analyzed using six SSR primer pairs. Two hundred and forty-seven amplified bands were produced for 12 loci; of them, (11) loci were polymorphic and (1) loci were monomorphic, shown in Table 3, indicating that there is high allelic variation. The molecular weights ranged from 128 to 1170 bp. The number of alleles per locus varied from 1 for (CT114 and Asr2) markers to 3 for (LEat018 and LEat020) SSR markers. The percentage of polymorphic was 91.67% with a range between (zero to 0.49) (Table 4). The highest values of the effective number of alleles (Ne*) [44], the gene diversity (h*) [45], and the Shannon Index (I*) [46] were recorded for the LEct004 primer (350 bp loci) with values of 1.99, 0.4996, and 0.6927, respectively. While the lowest value (zero) of the effective number of alleles (Ne*), the gene diversity (h*), and the Shannon Index (I*) was shown by Asr2 primer (536 bp loci) (Table 3 and Table 4).

Table 3.
SSR names, the total number of bands/primers, loci, monomorphic, polymorphic loci, and percentage of polymorphism.

Table 4.
Diversity parameters of tomato accessions obtained from the analysis of SSR alleles.
3.4. UPGMA Dendrogram and Similarity
Genetic variation among tomatoes was evaluated based on bands obtained from SSR profiling using Nie genetic distance and Unweighted Pair Group with Arithmetic Averages (UPGMA). The coefficient of genetic similarity ranged from 0.30 between JOR956 and JOR966 accessions to 1 between JOR950 and JOR951, JOR964 and JOR965, JOR955 and JOR979, JOR955 and JOR980, JOR979 and JOR980, JOR955 and JOR988, JOR979 and JOR988, and JOR980 and JOR988. The most similar tomato accessions reported above are from Kharja, Rhaba, and Ain Al-Biada, whereas the most different are from Qasfa and Rhaba shown in Table 1.
Moreover, similarity values for all tomato accessions using the UPGMA dendrogram are shown in Figure 1. At a genetic similarity value of 0.62, the dendrogram is divided into two groups except for 25 (JOR978) accessions collected from Rhaba. The first group consists of three sub-groups: 1 (JOR111), 5 (JOR953), and 30 (JOR984) accessions with 80% similarity, 6 (JOR954), 7 (JOR955), 26 (JOR979), 27 (JOR980), 34 (JOR988), 22 (JOR972), 19 (JOR968), 28 (JOR981), 18 (JOR967), 23 (JOR973), 20 (JOR970), 33 (JOR987), 35 (JOR989), and 36 (JOR990) accessions with 76% similarity, and 17 (JOR966) and 21 (JOR971) accessions with 88% similarity. The tomato accessions reported in the first group were from kharja, Al-al, Rhaba, Afra, Abel, and Ain Al-Baida. The second group consists of three sub-groups: 2 (JOR950), 3 (JOR951), 8 (JOR956), and 12 (JOR961) accessions with 76.5% similarity, 4 (JOR952), 11 (JOR959), 10 (JOR958), 15 (JOR964), 16 (JOR965), 9 (JOR957), 13 (JOR962), and 32 (JOR986) accessions with 85% similarity, and 14 (JOR963), 29 (JOR982), and 31 (JOR985) accessions with 77% similarity. The tomato accessions reported in the second group were from kharja, Qasfa, Shtafina, Al-al, Sakib, Hebras, Ain-Jannah, Abel, Afra, and Rhaba. The number of alleles and the PIC value for each SSR marker are presented in Table 4.
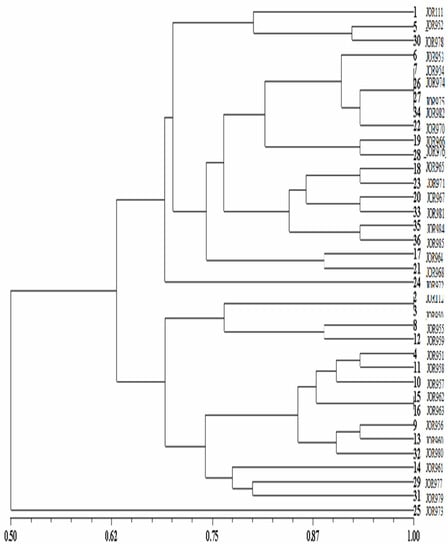
Figure 1.
Dendrogram of tomato accessions generated by UPGMA cluster analysis of the dissimilarity values based on [45] coefficient.
Polymorphic information content (PIC) values ranged from 0.00 to 0.499 (mean 0.33), confirming that the SSR markers are highly informative. The SSR marker had the highest PIC value (LEct004), followed by LEat018 (0.488), while the (Asr2) marker had the lowest PIC value. In this study, we found no relationship between the number of nucleotides per repeat and PIC shown in Table 4. For example, LEat018 with the lower PIC (0.34) has 29 repeats compared to LEta020, which has PIC (0.36) with 11 repeats.
3.5. The Functional Analysis of the Associated Genes with SSRs in Tomato
S. lycopersicum genomic sequence was used as a template for searching the sequence of SSR primers to predict the potential functions of our six genes. Then, databases such as Phytozome, NCBI, InterPro, and KEGG predicted more function annotations for these genes. In context, these genes were related to a wide range of functions, indicating that these gene-associated SSRs were potentially associated with essential biological functions, such as the LEat018 SSR marker associated with ACTIN_RELATED PROTEIN gene (Solyc11g005330) (Table 5). Additionally, the LEct004 SSR marker was associated with the HOMEOBOX PROTEIN TRANSCRIPTION FACTORS gene (Solyc02g089940). Moreover, the LEta014 SSR marker had an associated gene annotated as AUXIN RESPONSE FACTOR 8 gene (Solyc03g031970), and the LEta020 SSR marker was found to be associated with the gene THIOREDOXIN FAMILY TRP26 gene (Solyc01g097450). Furthermore, the CT114 marker was associated with the protein suppressor of the PHYA-105 1 (SPA1) gene (Solyc10g011690). In addition, the ABA/WDS-induced protein (ABA_WDS) gene (Solyc04g071580) was associated with the Asr2 SSR marker (Table 5).

Table 5.
BLAST corresponding Solyc (Solanum lycopersicum) gene sequences and annotation for SSR marker sequences.
3.6. Putative Tissue Expression Pattern of Genes S. lycopersicum Transcript Expression
The expression profile of the genes based on S. lycopersicum transcript expression was analyzed to understand their potential functions in different tissues (Figure 2). The results showed that the gene Solyc11g005330 which is related to the ACTIN_RELATED PROTEIN was highly expressed in all tomato tissues, especially in Mature Green Fruit, Breaker Fruit, 3 cm Fruit, Root, and Breaker Fruit + 10 (Figure 2 and Table 5), while the highest expression levels for Solyc02g089940 gene concerning HOMEOBOX PROTEIN TRANSCRIPTION FACTORS were observed in Fully Opened Flower, Leaves, Pimpinellifolium Leaf, Unopened Flower Bud, and Root (Figure 2 and Table 5). Additionally, the highest expression levels for the Solyc03g031970 gene encoding to AUXIN RESPONSE FACTOR 8 were recorded at 1 cm Fruit, 3 cm Fruit, 2 cm Fruit, Unopened Flower Bud, Mature Green Fruit, and Leaves. The Solyc01g097450 gene, which is related to THIOREDOXIN FAMILY TRP26, showed no clear expression level in any tomato tissue. On the other side, the highest expression levels for the Solyc10g011690 gene concerning the protein suppressor of PHYA-105 1 (SPA1) were observed in Root, 3 cm Fruit, Pimpinellifolium Immature Green Fruit, Pimpinellifolium Breaker Fruit, Mature Green Fruit, and Breaker Fruit. In addition, the Solyc04g071580 gene related to ABA/WDS-induced protein (ABA_WDS) was a high expression in Breaker Fruit + 10, Breaker Fruit, Mature Green Fruit, Pimpinellifolium Immature Green Fruit, 3 cm Fruit, and Root.
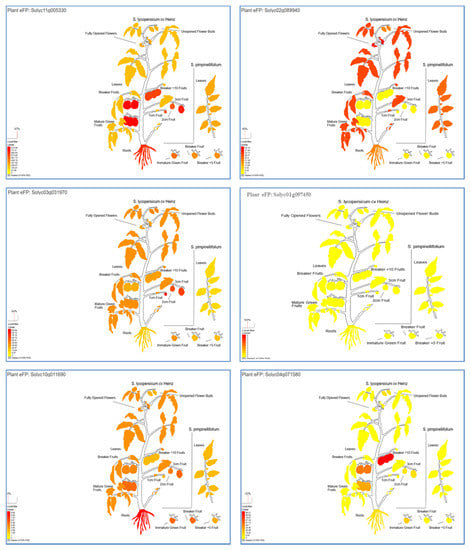
Figure 2.
The putative “plant electronic fluorescent pictograph” tissue expression of Solyc11g005330, Solyc02g089940, Solyc03g031970, Solyc01g097450, Solyc10g011690, and Solyc04g071580 genes of different tissues and developmental stages. The more intense the red color of the expression bar, the more gene expression detected [43].
3.7. Putative Subcellular Localizations of the Genes Based on S. lycopersicum Transcript Expression
Cell Electronic Fluorescent Pictograph tools were used to predict the putative subcellular localizations of the genes according to the protein localization of different cell organelles in the tomatoes. The subcellular localization profiles showed that Solyc11g005330 and Solyc01g097450 genes were highly expressed and presented in the cytosol (Figure 3), while the Solyc02g089940, Solyc03g031970, Solyc10g011690, and Solyc04g071580 genes were highly expressed and presented in the nucleus (Figure 3).
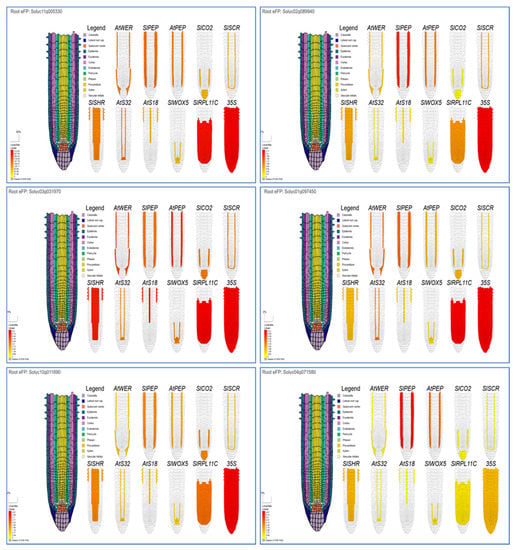
Figure 3.
The putative expression of Solyc11g005330, Solyc02g089940, Solyc03g031970, Solyc01g097450, Solyc10g011690, and Solyc04g071580 genes with different root cell types and/or tissues that related tovarious promoters. The more intense the red color of the expression bar, the more gene expression detected [43].
3.8. Putative Root Cell Types and Tissues Specific to the Genes Based on S. lycopersicum Transcript Expression
Root analysis using eplant_tomato tools observed the highest expression levels of the Solyc11g005330 gene in all root cell types under 35Spro, followed by phloem under AtS32pro, endodermis, and cortex under SIPEPpro (Figure 4). Additionally, the Solyc02g089940 gene was highly expressed in all root cell types under 35Spro, then endodermis and cortex under SIPEPpro, and cortex under AtPEPpro. Moreover, the highest expression levels of the Solyc03g031970 gene were reported for all root cell types under 35Spro, followed by epidermis and procambium under AtS18. In addition, a highly expressed Solyc01g097450 gene was observed for all root cell types under 35Spro, followed by endodermis and cortex under SIPEPpro, epidermis, and lateral root cap under AtWER. Furthermore, the highest expression levels of the Solyc10g011690 gene were observed for all root cell types under 35Spro, columella, and cortex under SICO2pro, then exodermis and cortex under SIPEP. At the same time, the Solyc04g071580 gene was highly expressed in exodermis and cortex under SIPEP, the cortex under AtPEPpro (Figure 4).
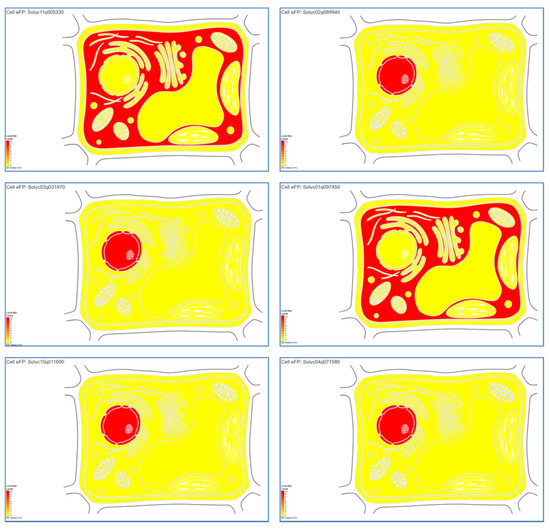
Figure 4.
Putative subcellular localizations of the genes and/or’ proteins for different cell organs using Cell eFP browsers in Tomato plant. The more intense the red color of the expression bar, the more gene expression detected [43].
4. Discussion
Natural variation in morphological parameters among genotypes is an essential analysis for understanding the genetic diversity that can be used to improve cultivars [47]. The current study revealed a wide range of diversity in tomatoes for most of the studied traits.
4.1. Drought Tolerance
Drought stress at different developmental stages causes various morpho-physiological changes in the plant. Water stress at the seedling stage might lead to higher dry root weights, longer roots, coleoptiles, and higher root/shoot ratios [14,15,16]. All of these changes are parameters of interest and have been widely used as reliable morph-physiological markers toward drought tolerance for various crop plants. Several parameters such as fresh root weight, dry root weight, root growth rate, fresh shoot weight, dry shoot weight, and shoot growth rate were measured for all tomato accessions. As expected, all morphological traits had a lower mean performance under drought stress (70% FC and 40% FC) than under normal conditions. This agreed with Vurayai et al. [48] who observed that the shoot: root ratio was significantly reduced by water stress imposed during the vegetative, flowering, and pod filling stages compared to the non-stressed control plant. Additionally, Yücel et al. [49] observed that the fresh root weight, fresh shoot weight, dry root weight, and dry shoot weight decreased by screening nine chickpea genotypes under water limited conditions. Drought stress is significantly affected by the proline concentrations in the leaf tissue for tomato accessions [8]. A highly significant increment in proline content was detected at 40%FC compared to the control treatment (100% FC). Differences between accessions and treatments showed a significantly positive relationship, which agreed with Vasquez-Robinet et al.’s study on the drought of potatoes [50]. The accumulation of osmolytes was also investigated during drought stress in durum wheat, e.g., proline was strongly upregulated by drought conditions that increased about twenty times in plants stressed at 12.5% SWC [51]. Understanding the physiological mechanisms in tomato accessions under drought stress conditions can help improve their performance and adaptation to harsh stress conditions, improving yield potential which is the ultimate target of crop breeding programs.
4.2. Morphological Characteristics
Several researchers were interested in the natural phenotypic variations which positively affect natural morphological diversity and genetic variations in a diverse population [3,27]. In this study, the analysis of variance for all the morphological characters showed a wide range of variation among tomato accessions, including fruit shape-related perimeter, fruit shape index, curved fruit shape index, and fruit shape index internal. These results are in line with Ranc et al. [52], which found a powerful link between the phenotypic variability present in the tomato germplasm and molecular polymorphisms using simple sequence repeat (SSR) markers. Additionally, Brdar-Jokanović et al. [53] studied the relationship between drought tolerance, growth type, and fruit size of different tomato accessions. They found that accessions with sizeable fruit sizes had comparatively higher water requirements.
4.3. Molecular Level
The genetic diversity analysis of crops is important for crop breeding. The selected six SSR loci that have been previously reported to be highly informative in distinguishing tomato genotypes [54,55] were used to detect the genetic diversities among the tomato accessions in our study. Additionally, Abd El-Hady et al. [56] stated that the SSR markers showed more polymorphic than RAPD markers. The average PIC value was higher than the result reported by Benor et al. [57] after testing 39 inbred lines of tomatoes using 35 polymorphic SSR loci (PIC = 0.31) and lower than the result reported by He et al. [54] after using 65 polymorphic SSR loci for testing 19 varieties of tomato (PIC = 0.37), indicating that SSR markers are of great utility for genetic diversity studies of tomatoes. This study revealed an exchange of genetic resources between farmers, particularly tomato accessions collected from Afra and Abel (southern part of Jordan). On the other hand, Rhaba accessions have special characteristics and agronomic traits such as sour taste and irregular shapes. Moreover, the results were similar to those reported by He et al. [54] _ENREF_57, but these results disagree with Smulders et al. [58], who found a positive relationship between the number of repeats and PIC in tomatoes. Taken together, we demonstrated the importance of these molecular markers and their allele detection and explained the vast genetic variation in collections from Afraa and Abeel (the southern part of Jordan).
Interestingly, our investigation detected that the plants’ most frequent type of SSRs was the TA/AT. This observation was subsequently confirmed with additional studies such as those by Rajput et al. [59]. Additionally, comparing the area of tomato fruit for all accessions, the number of alleles at each locus and the heterozygosity found that accession numbers JOR111, JOR958, JOR981, JOR986, and JOR989 had the highest fruit area. In contrast, the accession numbers JOR973, JOR961, JOR950, JOR951, and JOR955 had the lowest fruit area, and these accessions had more than one allele at the most heterozygous loci (LEct018, LEta020). The highest frequency was detected for the CC genotype (at LEct018) and AA genotype (at LEta020). It has been demonstrated that the SSR markers are recommended to distinguish closely related genotypes because of their high degree of variability and, therefore, become favored in population studies [60,61,62]. Consequently, it assessed the genetic variability among tomato accessions using the SSR method to identify the most suitable genotype for future use in plant breeding programs.
4.4. Putative Tissue Expression Analyses of Our Target Gene-Associated SSR Markers
BAR database tools were used to generate expression pattern profiles of our candidate genes with different tissues, cell organs, and root cell types. In this context, a putative expression and recognized synthesis sites of these genes provide an excellent way to understand the epistatic relationship between our gene’s synthesis site and a putative function. For example, the LEat018 SSR marker that associated with actin-related protein gene (ARP; Solyc11g005330), and this gene has the actin family domain (IPR004000). This domain is involved in the formation of filaments in the cytoskeletal system and plays important roles in various cellular functions in the cytoplasm and the nucleus [63]. Additionally, plants have many isoforms from actin protein which are probably involved in multiple functions such as graviperception, cell shape determination, tip growth, cytoplasmic streaming, cell wall deposition, etc. [64]. In addition, Nie et al. [65] reported that the AtARP4 gene from Arabidopsis thaliana is vital for plant growth and is related to hormone response such as salicylic acid, while any mutation in this gene can cause altered transcription response in hundreds of genes that affect plant development and lead to early flowering. Moreover, from our results, we found this gene was highly expressed in different tissues, presented in the cytoplasm, and observed in all root cell types, phloem, endodermis, and cortex under 35Spro, AtS32pro, and SIPEPpro promoters which can drive expression in various root cell types and tissues throughout the root, including the elongation zone and meristematic zone (Figure 2, Figure 3 and Figure 4). Previous studies reported that many actin-related protein (ARP) genes have distinct transcript expression patterns in different tissues (such as roots, seedlings, xylem precursor cells, pollen, flowers, leaves, and siliques) and cell organs such as cytoplasm and nucleus [66,67].
Furthermore, the LEct004 SSR marker was associated with the HOMEOBOX PROTEIN TRANSCRIPTION FACTORS gene (Solyc02g089940). This gene contains five domains, such as IPR009057 (homeodomain-like), PR006563 (POX domain), IPR008422 (homeobox KN domain), IPR016039 (thiolase-like), and IPR001356 (homeobox domain), that are reported to be key regulators for plant development and growth [68,69]. Additionally, many homeobox proteins were involved in transcriptional regulation and various metabolic pathways, such as OsHOX22 and OsHOX24 genes from rice which have a negative regulator role in abiotic stress response [70]. Moreover, this gene was found to be highly expressed in different tissues, presented in the nucleus, and observed in all root cell types, endodermis and cortex under 35Spro, SIPEPpro, and AtPEP promoters which can drive expression in various root cell types and tissues throughout the root, including the elongation zone and meristematic zone (Figure 2, Figure 3 and Figure 4). Sakamoto et al. [71] reported that these domains could play a role in suppressing target gene expression through their function as a nuclear localization signal.
In addition, AUXIN RESPONSE FACTOR 8 gene (Solyc03g031970) was associated with the LEta014 SSR marker, which has four domains, such as IPR003340 (B3 DNA binding domain), IPR010525 (auxin response factor), IPR003311 (AUX/IAA protein), and IPR015300 (DNA-binding pseudo barrel domain). Recent evidence suggests that these previous domains are key regulators of auxin-modulated gene expression, such as regulating diverse cellular and developmental responses in plants, including cell expansion, division, differentiation, light responses, patterning of embryo responses, and embryonic and post-embryonic development in some plants [72,73]. Thus, we found that this gene was highly expressed in different tissues, presented in the nucleus, and observed in all root cell types and Epidermis and Procambium under 35Spro and AtS18 prompters (Figure 2, Figure 3 and Figure 4). These results are in line with Kang et al. [74], who found that ARF was expressed and localized in the nucleus. Moreover, auxin response factors (ARFs) have played important roles in the process of plant growth and development as they increase the contents of carotenoids and enhance the tolerance to salt and drought in transgenic Arabidopsis [74]. Additionally, Bouzroud et al. [75] showed that many of ARFs genes were differentially expressed in tomato leaves and roots under salt, drought, and flooding stress conditions. Chen et al. [76] reported that the ARFs genes could play essential roles in various plant physiological processes by participating in ABA signaling pathways and regulating the expression of some genes such as SlABI5/ABF and SCL3, which influence stomatal morphology and vascular bundle development and ultimately improve tomato plant resistance to water deficit. Additionally, Salehin et al. [77] found the aliphatic Glucosinolate (GLSs) levels are regulated by the auxin-sensitive Aux/IAA repressors IAA5, IAA6, and IAA19, and any loss of these gene expressions results in reduced GLS levels and decreased drought tolerance in the Arabidopsis plant.
In addition, the LEta020 SSR marker was related to the THIOREDOXIN FAMILY TRP26 gene (Solyc01g097450), and this gene contains two domains IPR008979 (galactose-binding domain-like) and IPR010400 (PITH domain). Thioredoxin is a relatively small and very stable redox protein known to be present in many plants such as arabidopsis thaliana, Brassica napus, Zea may, Oryza sativa, Nicotiana tabacum, Spinacia olerace, and Pisum sativum. The higher plants have at least two types of thioredoxin, f-type, which is the only one related to activating fructose-1,6-bisphosphatase efficiently, and m-type, which can trigger the NADP-malate dehydrogenase [78]. In this context, the expression and activity of the Fructose-1,6 bisphosphatase gene are regulated in cytosolic by environmental factors such as light and drought conditions [79]. Moreover, thioredoxin plays a role in various critical biological processes, including anti-oxidative stress, cell cycle control, regulation of receptors/transcription factors, structural functions/protein folding, signal transduction (cell to cell), vacuolar inheritance, redox regulation of chloroplast enzymes, control of chloroplastic translation, the structure of the photosynthetic apparatus/folding, and other functions [80]. Additionally, from our results, we found that this gene did not show any clear expression level in any tomato tissue, while being highly expressed in the cytosol as well as observed in all root cell types, endodermis and cortex, epidermis and lateral root cap under 35Spro, SIPEPpro, and under AtWER promoters.
Additionally, the CT114 SSR marker was linked with the protein suppressor of the PHYA-105 1 (SPA1) gene (Solyc10g011690). This gene contains six domains such as IPR000719 (protein kinase domain), IPR017986 (WD40 repeat-containing domain), IPR002290 (serine/threonine/dual-specificity protein kinase, catalytic domain), IPR001680 (WD40 repeat), IPR015943 (WD40/YVTN repeat-like-containing domain) and IPR011009 (protein kinase-like domain). These domains play important roles in many cellular processes, including division, proliferation, cellular activities, apoptosis, differentiation, plant-specific developmental events, and protect cells from extreme environments [81,82]. Thus, we found that this gene was highly expressed in different tissues, presented in the nucleus, and observed in all root cell types, columella and cortex, exodermis and cortex under 35Spro, SICO2pro, and SIPEP promoters. Furthermore, the protein suppressor of PHYA-105 1 (SPA1) is involved in regulating the circadian cycle and flowering time in plants, and SPA1 has worked as a negative regulator of phytochrome A-mediated de-etiolation in seed germination and seedlings of Arabidopsis [83]. Many researchers have recently described links between anthocyanin accumulation and the CONSTITUTIVELY PHOTOMORPHOGENIC1/SUPPRESSOR OF PHYA-105 (COP1/SPA) in plants [84]. These relationships are complex and they have a positive correlation between the increased anthocyanidin content during drought. Cirillo et al. [85] reported that in tobacco, the anthocyanin content is considered the key regulator for drought stress tolerance by playing various roles in osmotic balance, scavenging of ROS, re-assimilation of the excess of ammonium, biochemical pH-stat, and regulation of leaf gas exchange [86].
Finally, the Asr2 SSR marker was related to ABA/WDS-induced protein (ABA_WDS) gene (Solyc10g011690), and this gene contains Interpro domain IPR003496 (ABA/WDS induced protein). This domain is caused by water deficit stress (WDS) or abscisic acid (ABA) stress. We found that this gene was highly expressed in different tissues, presented in the nucleus, and observed in all root cell types, exodermis and cortex under SIPEPpro and AtPEP promoters. Moreover, this gene is involved in the tolerance of various abiotic stresses such as dehydration, heat, and salinity for different plant species such as durum wheat, barley, and Pinus taeda L. [87,88].
5. Conclusions
Conclusively, this study assessed the genetic variation among tomato accessions using the SSR markers to detect the diversity of Jordanian tomato accessions. Moreover, tomato response to increasing drought stress was apparent through a significant reduction in morphological traits in addition to physiological and biochemical alternation, e.g., increasing the proline concentration. A wide range of variations were detected among tomato accessions that are important in selection for adaptation and yield improvement. Here, we demonstrated that the SSR method effectively discovers the genetic diversity of tomato accessions, which is vital for germplasm classification, management, and further molecular and breeding utilization. The bioinformatics analysis provides excellent information for predicting the function of candidate genes in tomatoes. Furthermore, the evaluation of these accessions under different water regimes could be helpful in producing new tomato varieties coping with drought stress conditions. Further molecular and genetic validation of the candidate genes would help understand the molecular mechanisms of drought stress tolerance in tomatoes.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae8070600/s1. Table S1: Analysis of variance (ANOVA) for all the studied parameters related to seedling tomato accessions for three treatments (control, 40%FC, 70%FC). Table S2: Natural variation for all the studied parameters of tomato accessions under drought stress at the seedling stage. Table S3: Mean value of fresh root weight, fresh shoot weight, dry root weight, dry shoot weight, root growth rate, shoot growth rate, relative fresh root weight, relative fresh shoot weight, relative shoot growth rate, and relative root growth rate. Table S4: Phenotypic variations for 20 morphological traits of tomato accessions. Figure S1: Coefficients of genetic similarity [45] for 36 local tomato accessions by using SSR markers. Figure S2: SSR patterns using primer Asr2 for 41 tomato accessions. M = molecular weight marker (100 bp). Figure S3: SSR pattern using primer LEat018 for 41 tomato accessions. M = molecular weight marker (100 bp). Figure S4: SSR patterns using primer LEct004 for 41 tomato accessions. M = molecular weight marker (100 bp). Figure S5: SSR patterns using primer LEa014 for 41 tomato accessions. M = molecular weight marker (100 bp). Figure S6: SSR patterns using primer LEta020 for 41 tomato accessions. M = molecular weight marker (100 bp). Figure S7: SSR patterns using primer CT114 for 41 tomato accessions. M = molecular weight marker (1 kp, 100 bp) using 3% Agarose gel.
Author Contributions
Conceptualization, I.M.; methodology, I.M. and S.J.; data analysis, W.A.D., S.G.T., M.A. and A.M.A.; investigation, I.M., W.A.D., S.J., S.G.T., M.A. and A.M.A.; writing—review and editing, I.M., W.A.D., S.G.T., M.A., A.A.A. and A.M.A. All authors have read and agreed to the published version of the manuscript.
Funding
The research leading to these results received funding from JUST Deanship of Research under grant agreement No188/2010.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Not applicable.
Acknowledgments
The authors would like to thank everyone who supported this work.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Knapp, S.; Bohs, L.; Nee, M.; Spooner, D.M. Solanaceae—A model for linking genomics with biodiversity. Comp. Funct. Genomics 2004, 5, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Siddiqui, M.H.; Alamri, S.; Alsubaie, Q.D.; Ali, H.M.; Khan, M.N.; Al-Ghamdi, A.; Ibrahim, A.A.; Alsadon, A. Exogenous nitric oxide alleviates sulfur deficiency-induced oxidative damage in tomato seedlings. Nitric Oxide 2020, 94, 95–107. [Google Scholar] [CrossRef] [PubMed]
- Gonias, E.D.; Ganopoulos, I.; Mellidou, I.; Bibi, A.C.; Kalivas, A.; Mylona, P.V.; Osanthanunkul, M.; Tsaftaris, A.; Madesis, P.; Doulis, A.G. Exploring genetic diversity of tomato (Solanum lycopersicum L.) germplasm of genebank collection employing SSR and SCAR markers. Genet. Resour. Crop Evol. 2019, 66, 1295–1309. [Google Scholar] [CrossRef]
- Foolad, M.R. Genome mapping and molecular breeding of tomato. Int. J. Plant Genom. 2007, 2007, 64358. [Google Scholar] [CrossRef] [PubMed]
- Naeem, M.; Shahzad, K.; Saqib, S.; Shahzad, A.; Nasrullah; Younas, M.; Afridi, M. The Solanum melongena COP1LIKE manipulates fruit ripening and flowering time in tomato (Solanum lycopersicum). J. Plant Growth Regul. 2022, 96, 14. [Google Scholar] [CrossRef]
- Reynolds, M.; Dreccer, F.; Trethowan, R. Drought-adaptive traits derived from wheat wild relatives and landraces. J. Exp. Bot. 2007, 58, 177–186. [Google Scholar] [CrossRef]
- Casanas, F.; Simo, J.; Casals, J.; Prohens, J. Toward an Evolved Concept of Landrace. Front. Plant Sci. 2017, 8, 145. [Google Scholar] [CrossRef]
- Makhadmeh, I.M.; Thabet, S.G.; Ali, M.; Alabbadi, B.; Albalasmeh, A.; Alqudah, A.M. Exploring genetic variation among Jordanian Solanum lycopersicon L. landraces and their performance under salt stress using SSR markers. J. Genet. Eng. Biotechnol. 2022, 20, 45. [Google Scholar] [CrossRef]
- Brake, M.H.; Al-Gharaibeh, M.A.; Hamasha, H.R.; Sakarneh, N.S.A.; Alshomali, I.A.; Migdadi, H.M.; Qaryouti, M.M.; Haddad, N.J. Assessment of genetic variability among Jordanian tomato landrace using inter-simple sequence repeats markers. Jordan J. Biol. Sci. 2021, 14, 91–95. [Google Scholar]
- Foolad, M.R.; Zhang, L.P.; Subbiah, P. Genetics of drought tolerance during seed germination in tomato: Inheritance and QTL mapping. Genome 2003, 46, 536–545. [Google Scholar] [CrossRef]
- Cattivelli, L.; Rizza, F.; Badeck, F.W.; Mazzucotelli, E.; Mastrangelo, A.M.; Francia, E.; Mare, C.; Tondelli, A.; Stanca, A.M. Drought tolerance improvement in crop plants: An integrated view from breeding to genomics. Field Crops Res. 2008, 105, 1–14. [Google Scholar] [CrossRef]
- Bargali, K.; Tewari, A. Growth and water relation parameters in drought-stressed Coriaria nepalensis seedlings. J. Arid. Environ. 2004, 58, 505–512. [Google Scholar] [CrossRef]
- Thabet, S.G.; Alqudah, A.M. Crops and Drought. In eLS; John Wiley & Sons, Ltd., Ed.; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2019; pp. 1–8. [Google Scholar]
- Takele, A. Seedling emergence and of growth of sorghum genotypes under variable soil moisture deficit. Acta Agron. Hung. 2000, 48, 95–102. [Google Scholar] [CrossRef]
- Dhanda, S.S.; Sethi, G.S.; Behl, R.K. Indices of drought tolerance in wheat genotypes at early stages of plant growth. J. Agron. Crop Sci. 2004, 190, 6–12. [Google Scholar] [CrossRef]
- Kashiwagi, J.; Krishnamurthy, L.; Upadhyaya, H.D.; Krishna, H.; Chandra, S.; Vadez, V.; Serraj, R. Genetic variability of drought-avoidance root traits in the mini-core germplasm collection of chickpea (Cicer arietinum L.). Euphytica 2005, 146, 213–222. [Google Scholar] [CrossRef]
- Ali, M.A.; Jabran, K.; Awan, S.I.; Abbas, A.; Ehsanullah; Zulkiffal, M.; Acet, T.; Farooq, J.; Rehman, A. Morpho-physiological diversity and its implications for improving drought tolerance in grain sorghum at different growth stages. Aust. J. Crop Sci. 2011, 5, 308–317. [Google Scholar]
- Wahb-Allah, M.A.; Alsadon, A.A.; Ibrahim, A.A. Drought tolerance of several tomato genotypes under greenhouse conditions. World Appl. Sci. J. 2011, 15, 933–940. [Google Scholar]
- Ovesna, J.; Poláková, K.; Leišová, L. DNA analyses and their applications in plant breeding. Czech J. Genet. Plant Breed. 2002, 38, 29. [Google Scholar] [CrossRef]
- Kumar, P.; Gupta, V.K.; Misra, A.K.; Modi, D.R.; Pandey, B.K. Potential of Molecular Markers in Plant Biotechnology. Plant Omics 2009, 2, 141–162. [Google Scholar]
- Ezekiel, C.N.; Nwangburuka, C.C.; Ajibade, O.A.; Odebode, A.C. Genetic diversity in 14 tomato (Lycopersicon esculentum Mill.) varieties in Nigerian markets by RAPD-PCR technique. Afr. J. Biotechnol. 2011, 10, 4961–4967. [Google Scholar]
- Tautz, D.; Renz, M. Simple sequences are ubiquitous repetitive components of eukaryotic genomes. Nucleic Acids Res. 1984, 12, 4127–4138. [Google Scholar] [CrossRef]
- Soranzo, N.; Provan, J.; Powell, W. An example of microsatellite length variation in the mitochondrial genome of conifers. Genome 1999, 42, 158–161. [Google Scholar] [CrossRef]
- Farooq, S.; Azam, F. Molecular markers in plant breeding-I: Concepts and characterization. Pak. J. Biol. Sci. 2002, 5, 1135–1140. [Google Scholar] [CrossRef]
- Pearson, C.E.; Sinden, R.R. Trinucleotide repeat DNA structures: Dynamic mutations from dynamic DNA. Curr. Opin. Struct. Biol. 1998, 8, 321–330. [Google Scholar] [CrossRef]
- Zhou, R.; Wu, Z.; Cao, X.; Jiang, F.L. Genetic diversity of cultivated and wild tomatoes revealed by morphological traits and SSR markers. Genet. Mol. Res. 2015, 14, 13868–13879. [Google Scholar] [CrossRef]
- EL-Mansy, A.B.; Abd El-Moneim, D.; ALshamrani, S.M.; Alsafhi, F.A.; Abdein, M.A.; Ibrahim, A.A. Genetic Diversity Analysis of Tomato (Solanum lycopersicum L.) with Morphological, Cytological, and Molecular Markers under Heat Stress. Horticulturae 2021, 7, 65. [Google Scholar] [CrossRef]
- Cotti, C. Molecular Markers for the Assessment of Genetic Variability in Threatened Plant Species. Ph.D Thesis, University of Bologna, Department of Experimental Evolutionary Biology, Bologna, Italy, 2008. [Google Scholar] [CrossRef]
- Sardaro, M.L.; Marmiroli, M.; Maestri, E.; Marmiroli, N. Genetic characterization of Italian tomato varieties and their traceability in tomato food products-Sardaro-2012-Food Science & Nutrition-Wiley Online Library. Food Sci. Nutr 2013, 1, 54–62. [Google Scholar] [CrossRef]
- Sacco, A.; Ruggieri, V.; Parisi, M.; Festa, G.; Rigano, M.M.; Picarella, M.E.; Mazzucato, A.; Barone, A. Exploring a Tomato Landraces Collection for Fruit-Related Traits by the Aid of a High-Throughput Genomic Platform. PLoS ONE 2015, 10, e0137139. [Google Scholar] [CrossRef]
- Wang, T.; Zou, Q.D.; Qi, S.Y.; Wang, X.F.; Wu, Y.Y.; Liu, N.; Zhang, Y.M.; Zhang, Z.J.; Li, H.T. Analysis of genetic diversity and population structure in a tomato (Solanum lycopersicum L.) germplasm collection based on single nucleotide polymorphism markers. Genet. Mol. Res. 2016, 15, 1–12. [Google Scholar] [CrossRef]
- Liu, S.; Cantrell, R.G.; McCarty, J.C.; Stewart, J.M. Simple sequence repeat-based assessment of genetic diversity in cotton race stock accessions. Crop Sci. 2000, 40, 1459–1469. [Google Scholar] [CrossRef]
- Raveendren, T.J.B.; Reviews, M.B. Molecular marker technology in cotton. Biotechnol. Mol. Biol. Rev. 2008, 3, 32–45. [Google Scholar]
- Shiri, M. Identification of informative simple sequence repeat (SSR) markers for drought tolerance in maize. Afr. J. Biotechnol. 2011, 10, 16414–16420. [Google Scholar] [CrossRef]
- Tam, S.M.; Mhiri, C.; Vogelaar, A.; Kerkveld, M.; Pearce, S.R.; Grandbastien, M.A. Comparative analyses of genetic diversities within tomato and pepper collections detected by retrotransposon-based SSAP, AFLP and SSR. Theor. Appl. Genet. 2005, 110, 819–831. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I. Rapid determination of free proline for water-stress studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Senthilkumar, M.; Amaresan, N.; Sankaranarayanan, A. Estimation of Proline Content in Plant Tissues. In Plant-Microbe Interactions: Laboratory Techniques; Senthilkumar, M., Amaresan, N., Sankaranarayanan, A., Eds.; Springer: New York, NY, USA, 2021; pp. 95–98. [Google Scholar]
- He, C.; Poysa, V.; Yu, K.J.T. Development and characterization of simple sequence repeat (SSR) markers and their use in determining relationships among Lycopersicon esculentum cultivars. Theor. Appl. Genet. 2003, 106, 363–373. [Google Scholar] [CrossRef]
- Asif, M.; Rahman, M.; Mirza, J.; Zafar, Y. High resolution metaphor agarose gel electrophoresis for genotyping with microsatellite markers. Pak. J. Agric. Sci. 2008, 45, 75–79. [Google Scholar]
- Anderson, J.A.; Churchill, G.A.; Autrique, J.E.; Tanksley, S.D.; Sorrells, M.E. Optimizing parental selection for genetic linkage maps. Genome 1993, 36, 181–186. [Google Scholar] [CrossRef]
- Dice, L.R. Measures of the Amount of Ecologic Association between Species. Ecology 1945, 26, 297–302. [Google Scholar] [CrossRef]
- Rohlf, F.J. NTSYSpc Numerical Taxonomy and Multivariate Analysis System Version 2.0 User Guide; Exeter Software Publishers Ltd.: Setauket, NY, USA, 1998. [Google Scholar]
- Fucile, G.; Di Biase, D.; Nahal, H.; La, G.; Khodabandeh, S.; Chen, Y.; Easley, K.; Christendat, D.; Kelley, L.; Provart, N.J. ePlant and the 3D data display initiative: Integrative systems biology on the world wide web. PLoS ONE 2011, 6, e15237. [Google Scholar] [CrossRef]
- Kimura, M.; Crow, J.F. The number of alleles that can be maintained in a finite population. Genetics 1964, 49, 725. [Google Scholar] [CrossRef]
- Nei, M. Analysis of gene diversity in subdivided populations. Proc. Natl. Acad. Sci. USA 1973, 70, 3321–3323. [Google Scholar] [CrossRef]
- Lewontin, R.C. Testing the theory of natural selection. Nature 1972, 236, 181–182. [Google Scholar] [CrossRef]
- Luo, C.; He, X.H.; Chen, H.; Ou, S.J.; Gao, M.P. Analysis of diversity and relationships among mango cultivars using Start Codon Targeted (SCoT) markers. Biochem. Syst. Ecol. 2010, 38, 1176–1184. [Google Scholar] [CrossRef]
- Vurayai, R.; Emongor, V.; Moseki, B. Effect of water stress imposed at different growth and development stages on morphological traits and yield of bambara groundnuts (Vigna subterranea L. Verdc). Am. J. Plant Physiol. 2011, 6, 17–27. [Google Scholar] [CrossRef]
- Yücel, D.; Anlarsal, A.; Mart, D.; Yücel, C. Effects of drought stress on early seedling growth of chickpea (Cicer arietinum L.) genotypes. World Appl. Sci. J. 2010, 11, 478–485. [Google Scholar]
- Vasquez-Robinet, C.; Mane, S.P.; Ulanov, A.V.; Watkinson, J.I.; Stromberg, V.K.; De Koeyer, D.; Schafleitner, R.; Willmot, D.B.; Bonierbale, M.; Bohnert, H.J.; et al. Physiological and molecular adaptations to drought in Andean potato genotypes. J. Exp. Bot. 2008, 59, 2109–2123. [Google Scholar] [CrossRef]
- Mastrangelo, A.; Rascio, A.; Mazzucco, L.; Russo, M.; Cattivelli, L.; Di Fonzo, N. Series A. Molecular aspects of abiotic stress resistance in durum wheat. Options Méditerranéennes Ser. A 2000, 40, 207–213. [Google Scholar]
- Ranc, N.; Munos, S.; Santoni, S.; Causse, M. A clarified position for Solanum lycopersicum var. cerasiforme in the evolutionary history of tomatoes (solanaceae). BMC Plant Biol. 2008, 8, 130. [Google Scholar] [CrossRef]
- Brdar-Jokanović, M.; Girek, Z.; Pavlović, S.; Ugrinović, M.; Zdravković, J. Traits related to drought tolerance in tomato accessions of different growth type and fruit size. J. Anim. Plant Sci. 2017, 27, 869–876. [Google Scholar]
- Korir, N.K.; Diao, W.; Tao, R.; Li, X.; Kayesh, E.; Li, A.; Zhen, W.; Wang, S. Genetic diversity and relationships among different tomato varieties revealed by EST-SSR markers. Genet. Mol. Res. 2014, 13, 43–53. [Google Scholar] [CrossRef]
- Abd El-Hady, E.A.; Haiba, A.A.; Abd El-Hamid, N.R.; Rizkalla, A.A. Phylogenetic diversity and relationships of some tomato varieties by electrophoretic protein and RAPD analysis. J. Am. Sci. 2010, 6, 434–441. [Google Scholar]
- Benor, S.; Zhang, M.Y.; Wang, Z.F.; Zhang, H.S. Assessment of genetic variation in tomato (Solanum lycopersicum L.) inbred lines using SSR molecular markers. J. Genet. Genom. 2008, 35, 373–379. [Google Scholar] [CrossRef]
- Smulders, M.J.M.; Bredemeijer, G.; RusKortekaas, W.; Arens, P.; Vosman, B. Use of short microsatellites from database sequences to generate polymorphisms among Lycopersicon esculentum cultivars and accessions of other Lycopersicon species. Theor. Appl. Genet. 1997, 94, 264–272. [Google Scholar] [CrossRef]
- Rajput, S.G.; Plyler-Harveson, T.; Santra, D.K.J.A.J.o.P.S. Development and characterization of SSR markers in proso millet based on switchgrass genomics. Am. J. Plant Sci. 2014, 5, 175–186. [Google Scholar] [CrossRef]
- Smith, D.; Devey, M.E. Occurrence and inheritance of microsatellites in Pinus radiata. Genome 1994, 37, 977–983. [Google Scholar] [CrossRef]
- Vargas, J.E.E.; Aguirre, N.C.; Coronado, Y.M. Study of the genetic diversity of tomato (Solanum spp.) with ISSR markers. Revista Ceres 2020, 67, 199–206. [Google Scholar] [CrossRef]
- Alzahib, R.H.; Migdadi, H.M.; Al Ghamdi, A.A.; Alwahibi, M.S.; Afzal, M.; Elharty, E.H.; Alghamdi, S.S. Exploring Genetic Variability among and within Hail Tomato Landraces Based on Sequence-Related Amplified Polymorphism Markers. Diversity 2021, 13, 135. [Google Scholar] [CrossRef]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Reactive oxygen gene network of plants. Trends Plant Sci. 2004, 9, 490–498. [Google Scholar] [CrossRef]
- Dominguez, R.; Holmes, K.C. Actin structure and function. Annu. Rev. Biophys. 2011, 40, 169–186. [Google Scholar] [CrossRef]
- Nie, W.F.; Wang, J. Actin-Related Protein 4 Interacts with PIE1 and Regulates Gene Expression in Arabidopsis. Genes 2021, 12, 520. [Google Scholar] [CrossRef]
- McKinney, E.C.; Kandasamy, M.K.; Meagher, R.B. Arabidopsis contains ancient classes of differentially expressed actin-related protein genes. Plant Physiol. 2002, 128, 997–1007. [Google Scholar] [CrossRef] [PubMed]
- Mittler, R.; Vanderauwera, S.; Gollery, M.; Van Breusegem, F. Questions and future challenges. Trends Plant Sci. 2004, 10, 490–498. [Google Scholar] [CrossRef] [PubMed]
- Nagasaki, H.; Sakamoto, T.; Sato, Y.; Matsuoka, M. Functional analysis of the conserved domains of a rice KNOX homeodomain protein, OSH15. Plant Cell 2001, 13, 2085–2098. [Google Scholar] [CrossRef] [PubMed]
- Ito, Y.; Hirochika, H.; Kurata, N. Organ-specific alternative transcripts of KNOX family class 2 homeobox genes of rice. Gene 2002, 288, 41–47. [Google Scholar] [CrossRef]
- Clark, S.E.; Jacobsen, S.E.; Levin, J.Z.; Meyerowitz, E.M. The CLAVATA and SHOOT MERISTEMLESS loci competitively regulate meristem activity in Arabidopsis. Development 1996, 122, 1567–1575. [Google Scholar] [CrossRef]
- Bhattacharjee, A.; Khurana, J.P.; Jain, M. Characterization of Rice Homeobox Genes, OsHOX22 and OsHOX24, and Over-expression of OsHOX24 in Transgenic Arabidopsis Suggest Their Role in Abiotic Stress Response. Front Plant Sci. 2016, 7, 627. [Google Scholar] [CrossRef]
- Sakamoto, T.; Nishimura, A.; Tamaoki, M.; Kuba, M.; Tanaka, H.; Iwahori, S.; Matsuoka, M. The conserved KNOX domain mediates specificity of tobacco KNOTTED1-type homeodomain proteins. Plant Cell 1999, 11, 1419–1432. [Google Scholar] [CrossRef][Green Version]
- Liscum, E.; Reed, J.W. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol. Biol. 2002, 49, 387–400. [Google Scholar] [CrossRef]
- Kulaeva, O.N.; Prokoptseva, O.S. Recent advances in the study of mechanisms of action of phytohormones. Biochemistry 2004, 69, 233–247. [Google Scholar] [CrossRef]
- Kang, C.; He, S.; Zhai, H.; Li, R.; Zhao, N.; Liu, Q. A Sweetpotato Auxin Response Factor Gene (IbARF5) Is Involved in Carotenoid Biosynthesis and Salt and Drought Tolerance in Transgenic Arabidopsis. Front Plant Sci. 2018, 9, 1307. [Google Scholar] [CrossRef]
- Bouzroud, S.; Gouiaa, S.; Hu, N.; Bernadac, A.; Mila, I.; Bendaou, N.; Smouni, A.; Bouzayen, M.; Zouine, M. Auxin Response Factors (ARFs) are potential mediators of auxin action in tomato response to biotic and abiotic stress (Solanum lycopersicum). PLoS ONE 2018, 13, e0193517. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhu, X.; Liu, X.; Wu, C.; Yu, C.; Hu, G.; Chen, L.; Chen, R.; Bouzayen, M.; Zouine, M.; et al. Knockout of Auxin Response Factor SlARF4 Improves Tomato Resistance to Water Deficit. Int. J. Mol. Sci. 2021, 22, 3347. [Google Scholar] [CrossRef] [PubMed]
- Salehin, M.; Li, B.; Tang, M.; Katz, E.; Song, L.; Ecker, J.R.; Kliebenstein, D.J.; Estelle, M. Auxin-sensitive Aux/IAA proteins mediate drought tolerance in Arabidopsis by regulating glucosinolate levels. Nat. Commun. 2019, 10, 4021. [Google Scholar] [CrossRef] [PubMed]
- Maeda, K.; Tsugita, A.; Dalzoppo, D.; Vilbois, F.; Schurmann, P. Further characterization and amino acid sequence of m-type thioredoxins from spinach chloroplasts. Eur. J. Biochem. 1986, 154, 197–203. [Google Scholar] [CrossRef]
- Daie, J. Cytosolic fructose-1,6-bisphosphatase: A key enzyme in the sucrose biosynthetic pathway. Photosynth Res. 1993, 38, 5–14. [Google Scholar] [CrossRef] [PubMed]
- Jacquot, J.J.; Lancelin, J.M.; Meyer, Y. Thioredoxins: Structure and function in plant cells. New Phytol. 1997, 136, 543–570. [Google Scholar] [CrossRef]
- Li, D.; Roberts, R.J.C. Human Genome and Diseases: WD-repeat proteins: Structure characteristics, biological function, and their involvement in human diseases. Cell. Mol. Life Sci. CMLS 2001, 58, 2085–2097. [Google Scholar] [CrossRef]
- Datta, S.; Ikeda, T.; Kano, K.; Mathews, F.S. Structure of the phenylhydrazine adduct of the quinohemoprotein amine dehydrogenase from Paracoccus denitrificans at 1.7 Å resolution. Acta Crystallogr. Sect. D Biol. Crystallogr. 2003, 59, 1551–1556. [Google Scholar] [CrossRef]
- Ishikawa, M.; Kiba, T.; Chua, N.H. The Arabidopsis SPA1 gene is required for circadian clock function and photoperiodic flowering. Plant J. 2006, 46, 736–746. [Google Scholar] [CrossRef]
- Maier, A.; Schrader, A.; Kokkelink, L.; Falke, C.; Welter, B.; Iniesto, E.; Rubio, V.; Uhrig, J.F.; Hulskamp, M.; Hoecker, U. Light and the E3 ubiquitin ligase COP1/SPA control the protein stability of the MYB transcription factors PAP1 and PAP2 involved in anthocyanin accumulation in Arabidopsis. Plant J. 2013, 74, 638–651. [Google Scholar] [CrossRef]
- Cirillo, V.; D’Amelia, V.; Esposito, M.; Amitrano, C.; Carillo, P.; Carputo, D.; Maggio, A. Anthocyanins are Key Regulators of Drought Stress Tolerance in Tobacco. Biology 2021, 10, 139. [Google Scholar] [CrossRef] [PubMed]
- Hinojosa-Gomez, J.; San Martin-Hernandez, C.; Heredia, J.B.; Leon-Felix, J.; Osuna-Enciso, T.; Muy-Rangel, M.D. Anthocyanin Induction by Drought Stress in the Calyx of Roselle Cultivars. Molecules 2020, 25, 1555. [Google Scholar] [CrossRef] [PubMed]
- Yacoubi, I.; Hamdi, K.; Fourquet, P.; Bignon, C.; Longhi, S. Structural and Functional Characterization of the ABA-Water Deficit Stress Domain from Wheat and Barley: An Intrinsically Disordered Domain behind the Versatile Functions of the Plant Abscissic Acid, Stress and Ripening Protein Family. Int. J. Mol. Sci. 2021, 22, 2314. [Google Scholar] [CrossRef]
- Hamdi, K.; Brini, F.; Kharrat, N.; Masmoudi, K.; Yakoubi, I. Abscisic Acid, Stress, and Ripening (TtASR1) Gene as a Functional Marker for Salt Tolerance in Durum Wheat. Biomed. Res. Int. 2020, 2020, 7876357. [Google Scholar] [CrossRef] [PubMed]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).