Strategies to Enhance the Use of Endophytes as Bioinoculants in Agriculture
Abstract
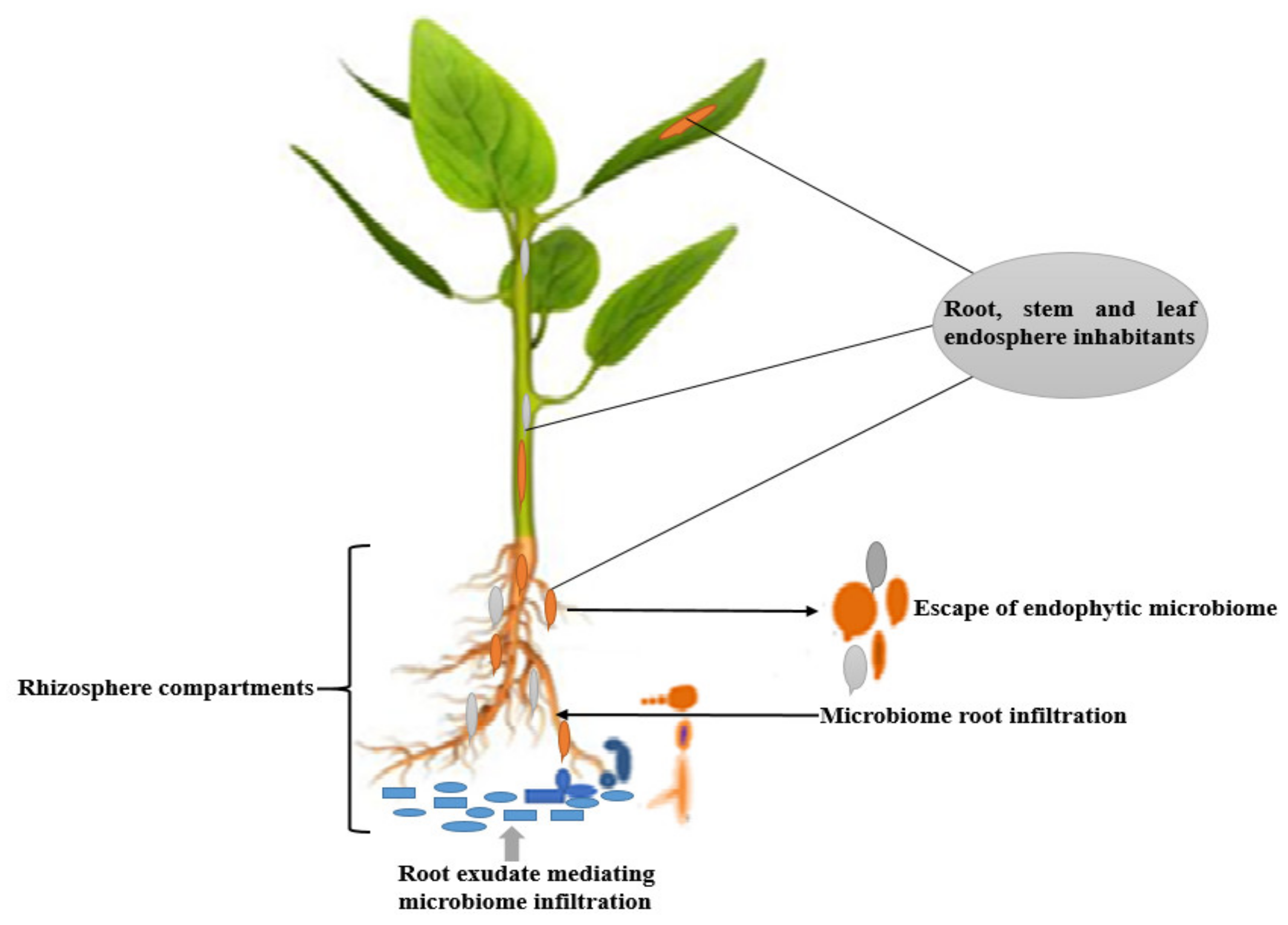
:1. Introduction
2. Endosphere Microbiome
Diverse Endophytic Microbes Associated with Plants
3. Mechanism Involved in Shaping the Endophytic Microbiome for Nutrient Acquisition in Plants
3.1. Inoculation into the Soil and Plant Tissues
3.2. Direct Infection into the Seeds
3.3. Atomization into Plant Tissues
4. Factors Determining Microbial Diversity in the Endosphere
4.1. Plant Organ Factor
4.2. Plant Exudate Secretion
4.3. Soil Factor
4.4. Climatic Factor
4.5. Crop Species
5. Endophytic Microbial Recruitment in Agricultural Management
5.1. Plant Genotype
5.2. Plant Immunity and Signaling
5.3. Plant-Derived Compounds
5.4. Plant Canopy Type
6. Approaches and Strategies for Shaping Endophytic Microbes
6.1. Plant Strategies
6.2. Microbial Strategies
6.3. Meta-Organism Strategies
7. Next-Generation Sequencing (NGS) Technology in the Research of Endophytic Microbial Communities
8. Future Prospects
9. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Omotayo, O.P.; Babalola, O.O. Resident rhizosphere microbiome’s ecological dynamics and conservation: Towards achieving the envisioned Sustainable Development Goals, a review. Int. Soil Water Conserv. Res. 2020, 9, 127–142. [Google Scholar] [CrossRef]
- Lin, W.; Lin, M.; Zhou, H.; Wu, H.; Li, Z.; Lin, W. The effects of chemical and organic fertilizer usage on rhizosphere soil in tea orchards. PLoS ONE 2019, 14, e0217018. [Google Scholar] [CrossRef] [PubMed]
- Yadav, A.; Singh, J.; Singh, C.; Yadav, N.; Ghosh, S.; Bhagwat, T.; Webster, T. Endophytic microbiomes and their plant growth-promoting attributes for plant health. Curr. Trends Microb. Biotechnol. Sustain. Agric. 2020, 11, 245–278. [Google Scholar]
- Mukherjee, A.; Gaurav, A.K.; Patel, A.K.; Singh, S.; Chouhan, G.K.; Lepcha, A.; Pereira, A.P.d.A.; Verma, J.P. Unlocking the potential plant growth-promoting properties of chickpea (Cicer arietinum L.) seed endophytes bio-inoculants for improving soil health and crop production. Land Degrad. Dev. 2021, 32, 4362–4374. [Google Scholar] [CrossRef]
- Dubey, A.; Malla, M.A.; Kumar, A.; Dayanandan, S.; Khan, M.L. Plants endophytes: Unveiling hidden agenda for bioprospecting toward sustainable agriculture. Crit. Rev. Biotechnol. 2020, 40, 1210–1231. [Google Scholar] [CrossRef]
- Santoyo, G.; Moreno-Hagelsieb, G.; del Carmen Orozco-Mosqueda, M.; Glick, B.R. Plant growth-promoting bacterial endophytes. Microbiol. Res. 2016, 183, 92–99. [Google Scholar] [CrossRef]
- Kushwaha, P.; Kashyap, P.L.; Bhardwaj, A.K.; Kuppusamy, P.; Srivastava, A.K.; Tiwari, R.K. Bacterial endophyte mediated plant tolerance to salinity: Growth responses and mechanisms of action. World J. Microbiol. Biotechnol. 2020, 36, 26. [Google Scholar] [CrossRef]
- Ghorbanpour, M.; Omidvari, M.; Abbaszadeh-Dahaji, P.; Omidvar, R.; Kariman, K. Mechanisms underlying the protective effects of beneficial fungi against plant diseases. Biol. Control 2018, 117, 147–157. [Google Scholar] [CrossRef]
- Kidd, P.S.; Alvarez-Lopez, V.; Becerra-Castro, C.; Cabello-Conejo, M.; Prieto-Fernandez, A. Potential role of plant-associated bacteria in plant metal uptake and implications in phytotechnologies. Adv. Bot. Res. 2017, 83, 87–126. [Google Scholar]
- Lopez, A.M.O.; Silva, A.L.D.S.; Maranhão, F.C.D.A.; Ferreira, L.F.R. Plant Growth Promoting Bacteria: Aspects in Metal Bioremediation and Phytopathogen Management; Springer: Cham, Switzerland, 2022; pp. 51–78. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J. Auxin and plant-microbe interactions. Cold Spring Harb. Perspect. Biol. 2011, 3, a001438. [Google Scholar] [CrossRef] [Green Version]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousef, N.M. Capability of plant growth-promoting rhizobacteria (PGPR) for producing indole acetic acid (IAA) under extreme conditions. Europ. J. Biotechnol. Res. 2018, 8, 174–182. [Google Scholar]
- Choudhury, A.R.; Choi, J.; Walitang, D.I.; Trivedi, P.; Lee, Y.; Sa, T. ACC deaminase and indole acetic acid producing endophytic bacterial co-inoculation improves physiological traits of red pepper (Capsicum annum L.) under salt stress. J. Plant Physiol. 2021, 267, 153544. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yu, Z.; Fan, G.; Wang, G.; Liu, X. Isolation and characterization of indole acetic acid producing root endophytic bacteria and their potential for promoting crop growth. J. Agric. Sci. Technol. 2018, 18, 1381–1891. [Google Scholar]
- Jasim, B.; Joseph, A.A.; John, C.J.; Mathew, J.; Radhakrishnan, E. Isolation and characterization of plant growth promoting endophytic bacteria from the rhizome of Zingiber officinale. Biotechnology 2014, 4, 197–204. [Google Scholar] [CrossRef] [Green Version]
- Naveed, M.; Mitter, B.; Yousaf, S.; Pastar, M.; Afzal, M.; Sessitsch, A. The endophyte Enterobacter sp. FD17: A maize growth enhancer selected based on rigorous testing of plant beneficial traits and colonization characteristics. Biol. Fertil. Soils 2014, 50, 249–262. [Google Scholar] [CrossRef]
- Ku, Y.; Xu, G.; Tian, X.; Xie, H.; Yang, X.; Cao, C. Root colonization and growth promotion of soybean, wheat and Chinese cabbage by Bacillus cereus YL6. PLoS ONE 2018, 13, e0200181. [Google Scholar]
- Wang, X.; Xie, H.; Ku, Y.; Yang, X.; Chen, Y.; Yang, N.; Mei, X.; Cao, C. Chemotaxis of Bacillus cereus YL6 and its colonization of Chinese cabbage seedlings. Plant Soil 2020, 447, 413–430. [Google Scholar] [CrossRef]
- Wani, S.H.; Kumar, V.; Shriram, V.; Sah, S.K. Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop J. 2016, 4, 162–176. [Google Scholar] [CrossRef] [Green Version]
- Shore, S.; Sathisha, G. Screening of endophytic colonizing bacteria for cytokinin-like compounds: Crude cell-free broth of endophytic colonizing bacteria is unsuitable in cucumber cotyledon bioassay. World J. Agric. Sci. 2010, 6, 345–352. [Google Scholar]
- Bhore, S.J.; Ravichantar, N.; Loh, C.Y. Screening of endophytic bacteria isolated from leaves of Sambung Nyawa [Gynura procumbens (Lour.) Merr.] for cytokinin-like compounds. Bioinformation 2010, 5, 191. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jaroszuk-Ściseł, J.; Tyśkiewicz, R.; Nowak, A.; Ozimek, E.; Majewska, M.; Hanaka, A.; Tyśkiewicz, K.; Pawlik, A.; Janusz, G. Phytohormones (auxin, gibberellin) and ACC deaminase in vitro synthesized by the mycoparasitic Trichoderma DEMTkZ3A0 strain and changes in the level of auxin and plant resistance markers in wheat seedlings inoculated with this strain conidia. Int. J. Mol. Sci. 2019, 20, 4923. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khan, M.; Asaf, S.; Khan, A.; Adhikari, A.; Jan, R.; Ali, S.; Imran, M.; Kim, K.M.; Lee, I.J. Plant growth-promoting endophytic bacteria augment growth and salinity tolerance in rice plants. Plant Biol. 2020, 22, 850–862. [Google Scholar] [CrossRef] [PubMed]
- Dar, N.A.; Amin, I.; Wani, W.; Wani, S.A.; Shikari, A.B.; Wani, S.H.; Masoodi, K.Z. Abscisic acid: A key regulator of abiotic stress tolerance in plants. Plant Gene 2017, 11, 106–111. [Google Scholar] [CrossRef]
- Vishwakarma, K.; Upadhyay, N.; Kumar, N.; Yadav, G.; Singh, J.; Mishra, R.K.; Kumar, V.; Verma, R.; Upadhyay, R.; Pandey, M. Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Front. Plant Sci. 2017, 8, 161. [Google Scholar] [CrossRef] [Green Version]
- Forchetti, G.; Masciarelli, O.; Alemano, S.; Alvarez, D.; Abdala, G. Endophytic bacteria in sunflower (Helianthus annuus L.): Isolation, characterization, and production of jasmonates and abscisic acid in culture medium. Appl. Microbiol. Biotechnol. 2007, 76, 1145–1152. [Google Scholar] [CrossRef]
- Rostami, S.; Azhdarpoor, A. The application of plant growth regulators to improve phytoremediation of contaminated soils: A review. Chemosphere 2019, 220, 818–827. [Google Scholar] [CrossRef]
- Gamalero, E.; Favale, N.; Bona, E.; Novello, G.; Cesaro, P.; Massa, N.; Glick, B.R.; Orozco-Mosqueda, M.d.C.; Berta, G.; Lingua, G. Screening of bacterial endophytes able to promote plant growth and increase salinity tolerance. Appl. Sci. 2020, 10, 5767. [Google Scholar] [CrossRef]
- Audipudi, A.; Chakicherla, B.; Bhore, S. Bacterial endophytes as biofertilizers and biocontrol agents for sustainable agriculture. Biotechnol. Sustain. 2017, 1, 223–247. [Google Scholar]
- Samson, R.; Shah, M.; Yadav, R.; Sarode, P.; Rajput, V.; Dastager, S.G.; Dharne, M.S.; Khairnar, K. Metagenomic insights to understand transient influence of Yamuna River on taxonomic and functional aspects of bacterial and archaeal communities of River Ganges. Sci. Total Environ. 2019, 674, 288–299. [Google Scholar] [CrossRef]
- Battu, L.; Ulaganathan, K. Whole genome sequencing and identification of host-interactive genes in the rice endophytic Leifsonia sp. KU-LS. Funct. Integr. Genom. 2020, 20, 237–243. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; Van Overbeek, L.S.; Berg, G.; Pirttilä, A.M.; Compant, S.; Campisano, A.; Döring, M.; Sessitsch, A. The hidden world within plants: Ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 2015, 79, 293–320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brader, G.; Compant, S.; Vescio, K.; Mitter, B.; Trognitz, F.; Ma, L.-J.; Sessitsch, A. Ecology and genomic insights into plant-pathogenic and plant-nonpathogenic endophytes. Ann. Rev. Phytopathol. 2017, 55, 61–83. [Google Scholar] [CrossRef] [PubMed]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; Van Themaat, E.V.L.; Schulze-Lefert, P. Structure and functions of the bacterial microbiota of plants. Ann. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [Green Version]
- Dang, H.; Zhang, T.; Li, G.; Mu, Y.; Lv, X.; Wang, Z.; Zhuang, L. Root-associated endophytic bacterial community composition and structure of three medicinal licorices and their changes with the growing year. BMC Microbiol. 2020, 20, 291. [Google Scholar] [CrossRef]
- Maropola, M.K.A.; Ramond, J.-B.; Trindade, M. Impact of metagenomic DNA extraction procedures on the identifiable endophytic bacterial diversity in Sorghum bicolor (L. Moench). J. Microbiol. Methods 2015, 112, 104–117. [Google Scholar] [CrossRef] [Green Version]
- Samad, A.; Trognitz, F.; Compant, S.; Antonielli, L.; Sessitsch, A. Shared and host-specific microbiome diversity and functioning of grapevine and accompanying weed plants. Environ. Microbiol. 2017, 19, 1407–1424. [Google Scholar] [CrossRef]
- Adeleke, B.S.; Ayangbenro, A.S.; Babalola, O.O. Bacterial community structure of the sunflower (Helianthus annuus) endosphere. Plant Signal. Behav. 2021, 16, 1974217. [Google Scholar] [CrossRef]
- Correa-Galeote, D.; Bedmar, E.J.; Arone, G.J. Maize endophytic bacterial diversity as affected by soil cultivation history. Front. Microbiol. 2018, 9, 484. [Google Scholar] [CrossRef]
- Fadiji, A.E.; Ayangbenro, A.S.; Babalola, O.O. Metagenomic profiling of the community structure, diversity and nutrient pathways of bacterial endophytes in maize plant. Antonie Van Leeuwenhoek 2020, 113, 1559–1571. [Google Scholar] [CrossRef]
- Lamichhane, J.R.; Debaeke, P.; Steinberg, C.; You, M.P.; Barbetti, M.J.; Aubertot, J.-N. Abiotic and biotic factors affecting crop seed germination and seedling emergence: A conceptual framework. Plant Soil 2018, 432, 1–28. [Google Scholar] [CrossRef]
- Compant, S.; Cambon, M.C.; Vacher, C.; Mitter, B.; Samad, A.; Sessitsch, A. The plant endosphere world-bacterial life within plants. Environ. Microbiol. 2021, 23, 1812–1829. [Google Scholar] [CrossRef] [PubMed]
- Sasse, J.; Martinoia, E.; Northen, T. Feed your friends: Do plant exudates shape the root microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deveau, A.; Bonito, G.; Uehling, J.; Paoletti, M.; Becker, M.; Bindschedler, S.; Hacquard, S.; Hervé, V.; Labbé, J.; Lastovetsky, O.A. Bacterial–fungal interactions: Ecology, mechanisms and challenges. FEMS Microbiol. Rev. 2018, 42, 335–352. [Google Scholar] [CrossRef] [Green Version]
- Fadiji, A.E.; Ayangbenro, A.S.; Babalola, O.O. Organic Farming enhances the diversity and community structure of endophytic archaea and fungi in maize plant: A shotgun approach. J. Soil Sci. Plant Nutr. 2020, 20, 2587–2599. [Google Scholar] [CrossRef]
- Xia, Y.; Sahib, M.R.; Amna, A.; Opiyo, S.O.; Zhao, Z.; Gao, Y.G. Culturable endophytic fungal communities associated with plants in organic and conventional farming systems and their effects on plant growth. Sci. Rep. 2019, 9, 1669. [Google Scholar] [CrossRef]
- Xia, Y.; DeBolt, S.; Dreyer, J.; Scott, D.; Williams, M.A. Characterization of culturable bacterial endophytes and their capacity to promote plant growth from plants grown using organic or conventional practices. Front. Plant Sci. 2015, 6, 490. [Google Scholar] [CrossRef] [Green Version]
- Heckman, J. A history of organic farming: Transitions from Sir Albert Howard’s war in the soil to USDA National Organic Program. Renew. Agric. Food Syst. 2006, 21, 143–150. [Google Scholar] [CrossRef] [Green Version]
- Zarb, J.; Ghorbani, R.; Koocheki, A.; Leifert, C. The importance of microorganisms in organic agriculture. Outlooks Pest Manag. 2005, 16, 52–55. [Google Scholar] [CrossRef] [Green Version]
- Araújo, A.S.; Leite, L.F.; Santos, V.B.; Carneiro, R.F. Soil microbial activity in conventional and organic agricultural systems. Sustainability 2009, 1, 268–276. [Google Scholar] [CrossRef] [Green Version]
- Li, R.; Khafipour, E.; Krause, D.O.; Entz, M.H.; de Kievit, T.R.; Fernando, W.D. Pyrosequencing reveals the influence of organic and conventional farming systems on bacterial communities. PLoS ONE 2012, 7, e51897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harrison, J.G.; Griffin, E.A. The diversity and distribution of endophytes across biomes, plant phylogeny and host tissues: How far have we come and where do we go from here? Environ. Microbiol. 2020, 22, 2107–2123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vacher, C.; Hampe, A.; Porté, A.J.; Sauer, U.; Compant, S.; Morris, C.E. The phyllosphere: Microbial jungle at the plant–climate interface. Ann. Rev. Ecol. Evol. Syst. 2016, 47, 1–24. [Google Scholar] [CrossRef]
- Rana, K.L.; Kour, D.; Yadav, N.; Yadav, A.N. Endophytic microbes in nanotechnology: Current development, and potential biotechnology applications. In Microbial Endophytes; Kumar, A., Singh, K.V., Eds.; Elsevier: Amsterdam, The Netherlands, 2020; pp. 231–262. [Google Scholar]
- Johnston-Monje, D.; Raizada, M.N. Conservation and diversity of seed associated endophytes in Zea across boundaries of evolution, ethnography and ecology. PLoS ONE 2011, 6, e20396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez, C.E.; Antonielli, L.; Mitter, B.; Trognitz, F.; Sessitsch, A. Heritability and functional importance of the Setaria viridis bacterial seed microbiome. Phytobiomes J. 2020, 4, 40–52. [Google Scholar] [CrossRef] [Green Version]
- Barret, M.; Briand, M.; Bonneau, S.; Préveaux, A.; Valière, S.; Bouchez, O.; Hunault, G.; Simoneau, P.; Jacques, M.-A. Emergence shapes the structure of the seed microbiota. Appl. Environ. Microbiol. 2015, 81, 1257–1266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samreen, T.; Naveed, M.; Nazir, M.Z.; Asghar, H.N.; Khan, M.I.; Zahir, Z.A.; Kanwal, S.; Jeevan, B.; Sharma, D.; Meena, V.S. Seed associated bacterial and fungal endophytes: Diversity, life cycle, transmission, and application potential. Appl. Soil Ecol. 2021, 168, 104191. [Google Scholar] [CrossRef]
- Khalaf, E.M.; Raizada, M.N. Bacterial seed endophytes of domesticated cucurbits antagonize fungal and oomycete pathogens including powdery mildew. Front. Microbiol. 2018, 9, 42. [Google Scholar] [CrossRef]
- Sharma, A.; Kaushik, N.; Sharma, A.; Bajaj, A.; Rasane, M.; Shouche, Y.S.; Marzouk, T.; Djébali, N. Screening of tomato seed bacterial endophytes for antifungal activity reveals lipopeptide producing Bacillus siamensis strain NKIT9 as a potential bio-control agent. Front. Microbiol. 2021, 12, 1228. [Google Scholar] [CrossRef]
- Reiter, B.; Pfeifer, U.; Schwab, H.; Sessitsch, A. Response of endophytic bacterial communities in potato plants to infection with Erwinia carotovora subsp. atroseptica. Appl. Environ. Microbiol. 2002, 68, 2261–2268. [Google Scholar] [CrossRef] [Green Version]
- Hardoim, P.R.; van Overbeek, L.S.; van Elsas, J.D. Properties of bacterial endophytes and their proposed role in plant growth. Trends Microbiol. 2008, 16, 463–471. [Google Scholar] [CrossRef] [PubMed]
- Hardoim, P.R.; Hardoim, C.C.; Van Overbeek, L.S.; Van Elsas, J.D. Dynamics of seed-borne rice endophytes on early plant growth stages. PLoS ONE 2012, 7, e30438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mitter, B.; Pfaffenbichler, N.; Flavell, R.; Compant, S.; Antonielli, L.; Petric, A.; Berninger, T.; Naveed, M.; Sheibani-Tezerji, R.; Von Maltzahn, G. A new approach to modify plant microbiomes and traits by introducing beneficial bacteria at flowering into progeny seeds. Front. Microbiol. 2017, 8, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dubey, A.; Saiyam, D.; Kumar, A.; Hashem, A.; Abd_Allah, E.F.; Khan, M.L. Bacterial root endophytes: Characterization of their competence and plant growth promotion in soybean (Glycine max (L.) Merr.) under drought stress. Int. J. Environ. Res. Public Health 2021, 18, 931. [Google Scholar] [CrossRef]
- Adeleke, B.S.; Ayangbenro, A.S.; Babalola, O.O. Genomic analysis of endophytic Bacillus cereus T4S and its plant growth-promoting traits. Plants 2021, 10, 1776. [Google Scholar] [CrossRef]
- Hiruma, K.; Kobae, Y.; Toju, H. Beneficial associations between Brassicaceae plants and fungal endophytes under nutrient-limiting conditions: Evolutionary origins and host–symbiont molecular mechanisms. Curr. Opin. Plant Biol. 2018, 44, 145–154. [Google Scholar] [CrossRef]
- Verma, P.; Yadav, A.N.; Khannam, K.S.; Panjiar, N.; Kumar, S.; Saxena, A.K.; Suman, A. Assessment of genetic diversity and plant growth promoting attributes of psychrotolerant bacteria allied with wheat (Triticum aestivum) from the northern hills zone of India. Ann. Microbiol. 2015, 65, 1885–1899. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, W.; Li, Q.; Cui, R.; Wang, Z.; Wang, Y.; Zhang, Y.-Z.; Ding, W.; Shen, X. Deciphering the root endosphere microbiome of the desert plant Alhagi sparsifolia for drought resistance-promoting bacteria. Appl. Environ. Microbiol. 2020, 86, e02863-19. [Google Scholar] [CrossRef] [PubMed]
- Del Carmen Orozco-Mosqueda, M.; del Carmen Rocha-Granados, M.; Glick, B.R.; Santoyo, G. Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol. Res. 2018, 208, 25–31. [Google Scholar] [CrossRef]
- Arif, I.; Batool, M.; Schenk, P.M. Plant microbiome engineering: Expected benefits for improved crop growth and resilience. Trends Biotechnol. 2020, 38, 1385–1396. [Google Scholar] [CrossRef]
- Dubey, A.; Malla, M.A.; Khan, F.; Chowdhary, K.; Yadav, S.; Kumar, A.; Sharma, S.; Khare, P.K.; Khan, M.L. Soil microbiome: A key player for conservation of soil health under changing climate. Biodivers. Conserv. 2019, 28, 2405–2429. [Google Scholar] [CrossRef]
- Babalola, O.O.; Adeleke, B.S.; Ayangbenro, A.S. Whole genome sequencing of sunflower root-associated Bacillus cereus. Evolut. Bioinf. 2021, 17, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Ma, Y.; Rajkumar, M.; Oliveira, R.S.; Zhang, C.; Freitas, H. Potential of plant beneficial bacteria and arbuscular mycorrhizal fungi in phytoremediation of metal-contaminated saline soils. J. Hazard. Mater. 2019, 379, 120813. [Google Scholar] [CrossRef] [PubMed]
- Syed-Ab-Rahman, S.F.; Xiao, Y.; Carvalhais, L.C.; Ferguson, B.J.; Schenk, P.M. Suppression of Phytophthora capsici infection and promotion of tomato growth by soil bacteria. Rhizosphere 2019, 9, 72–75. [Google Scholar] [CrossRef] [Green Version]
- Chihaoui, S.-A.; Trabelsi, D.; Jdey, A.; Mhadhbi, H.; Mhamdi, R. Inoculation of Phaseolus vulgaris with the nodule-endophyte Agrobacterium sp. 10C2 affects richness and structure of rhizosphere bacterial communities and enhances nodulation and growth. Arch. Microbiol. 2015, 197, 805–813. [Google Scholar] [CrossRef] [PubMed]
- Aleynova, O.A.; Suprun, A.R.; Nityagovsky, N.N.; Dubrovina, A.S.; Kiselev, K.V. The Influence of the grapevine bacterial and fungal endophytes on biomass ccumulation and stilbene production by the in vitro cultivated cells of Vitis amurensis Rupr. Plants 2021, 10, 1276. [Google Scholar] [CrossRef]
- Bielecka, M.; Pencakowski, B.; Nicoletti, R. Using next-generation sequencing technology to explore genetic pathways in endophytic fungi in the syntheses of plant bioactive metabolites. Agriculture 2022, 12, 187. [Google Scholar] [CrossRef]
- Liu, H.; Khan, M.Y.; Carvalhais, L.C.; Delgado-Baquerizo, M.; Yan, L.; Crawford, M.; Dennis, P.G.; Singh, B.; Schenk, P.M. Soil amendments with ethylene precursor alleviate negative impacts of salinity on soil microbial properties and productivity. Sci. Rep. 2019, 9, 6892. [Google Scholar] [CrossRef]
- Danish, S.; Zafar-ul-Hye, M. Co-application of ACC-deaminase producing PGPR and timber-waste biochar improves pigments formation, growth and yield of wheat under drought stress. Sci. Rep. 2019, 9, 5999. [Google Scholar] [CrossRef] [Green Version]
- Pascale, A.; Proietti, S.; Pantelides, I.S.; Stringlis, I.A. Modulation of the root microbiome by plant molecules: The basis for targeted disease suppression and plant growth promotion. Front. Plant Sci. 2020, 10, 1741. [Google Scholar] [CrossRef]
- Rojas-Solís, D.; Zetter-Salmón, E.; Contreras-Pérez, M.; del Carmen Rocha-Granados, M.; Macías-Rodríguez, L.; Santoyo, G. Pseudomonas stutzeri E25 and Stenotrophomonas maltophilia CR71 endophytes produce antifungal volatile organic compounds and exhibit additive plant growth-promoting effects. Biocatal. Agric. Biotechnol. 2018, 13, 46–52. [Google Scholar] [CrossRef]
- Vyas, P.; Kumar, D.; Dubey, A.; Kumar, A. Screening and characterization of Achromobacter xylosoxidans isolated from rhizosphere of Jatropha curcas L. (energy crop) for plant-growth-promoting traits. J. Adv. Res. Biotechnol. 2018, 3, 1–8. [Google Scholar]
- Mendes, L.W.; Raaijmakers, J.M.; de Hollander, M.; Mendes, R.; Tsai, S.M. Influence of resistance breeding in common bean on rhizosphere microbiome composition and function. ISME J. 2018, 12, 212–224. [Google Scholar] [CrossRef] [PubMed]
- George, C.; Kohler, J.; Rillig, M.C. Biochars reduce infection rates of the root-lesion nematode Pratylenchus penetrans and associated biomass loss in carrot. Soil Biol. Biochem. 2016, 95, 11–18. [Google Scholar] [CrossRef]
- Paungfoo-Lonhienne, C.; Rentsch, D.; Robatzek, S.; Webb, R.I.; Sagulenko, E.; Näsholm, T.; Schmidt, S.; Lonhienne, T.G. Turning the table: Plants consume microbes as a source of nutrients. PLoS ONE 2010, 5, e11915. [Google Scholar] [CrossRef] [Green Version]
- Munshi, M.; Sohrab, M.; Begum, M.; Rony, S.R.; Karim, M.; Afroz, F.; Hasan, M. Evaluation of bioactivity and phytochemical screening of endophytic fungi isolated from Ceriops decandra (Griff.) W. Theob, a mangrove plant in Bangladesh. Clin. Phytosci. 2021, 7, 81. [Google Scholar] [CrossRef]
- Mahgoub, H.A.; Fouda, A.; Eid, A.M.; Ewais, E.E.-D.; Hassan, S.E.-D. Biotechnological application of plant growth-promoting endophytic bacteria isolated from halophytic plants to ameliorate salinity tolerance of Vicia faba L. Plant Biotechnol. Rep. 2021, 15, 819–843. [Google Scholar] [CrossRef]
- Khalil, A.M.A.; Hassan, S.E.-D.; Alsharif, S.M.; Eid, A.M.; Ewais, E.E.-D.; Azab, E.; Gobouri, A.A.; Elkelish, A.; Fouda, A. Isolation and characterization of fungal endophytes isolated from medicinal plant Ephedra pachyclada as plant growth-promoting. Biomolecules 2021, 11, 140. [Google Scholar] [CrossRef]
- Patel, J.K.; Gohel, K.; Patel, H.; Solanki, T. Wheat growth dependent succession of culturable endophytic bacteria and their plant growth promoting traits. Curr. Microbiol. 2021, 78, 4103–4114. [Google Scholar] [CrossRef]
- Hussain, A.; Shah, M.; Hamayun, M.; Qadir, M.; Iqbal, A. Heavy metal tolerant endophytic fungi Aspergillus welwitschiae improves growth, ceasing metal uptake and strengthening antioxidant system in Glycine max L. Environ. Sci. Poll. Res. 2021, 29, 15501–15515. [Google Scholar]
- Zaman, N.R.; Chowdhury, U.F.; Reza, R.N.; Chowdhury, F.T.; Sarker, M.; Hossain, M.M.; Akbor, M.A.; Amin, A.; Islam, M.R.; Khan, H. Plant growth promoting endophyte Burkholderia contaminans NZ antagonizes phytopathogen Macrophomina phaseolina through melanin synthesis and pyrrolnitrin inhibition. PLoS ONE 2021, 16, e0257863. [Google Scholar]
- Kumar, V.; Jain, L.; Jain, S.K.; Chaturvedi, S.; Kaushal, P. Bacterial endophytes of rice (Oryza sativa L.) and their potential for plant growth promotion and antagonistic activities. S. Afr. J. Bot. 2020, 134, 50–63. [Google Scholar] [CrossRef]
- Rahman, M.D.M.; Flory, E.; Koyro, H.-W.; Abideen, Z.; Schikora, A.; Suarez, C.; Schnell, S.; Cardinale, M. Consistent associations with beneficial bacteria in the seed endosphere of barley (Hordeum vulgare L.). Syst. Appl. Microbiol. 2018, 41, 386–398. [Google Scholar] [CrossRef] [PubMed]
- Peng, J.; Ma, J.; Wei, X.; Zhang, C.; Jia, N.; Wang, X.; Wang, E.T.; Hu, D.; Wang, Z. Accumulation of beneficial bacteria in the rhizosphere of maize (Zea mays L.) grown in a saline soil in responding to a consortium of plant growth promoting rhizobacteria. Ann. Microbiol. 2021, 71, 40. [Google Scholar] [CrossRef]
- Nascimento, F.X.; Urón, P.; Glick, B.R.; Giachini, A.; Rossi, M.J. Genomic analysis of the 1-aminocyclopropane-1-carboxylate deaminase-producing Pseudomonas thivervalensis sc5 reveals its multifaceted roles in soil and in beneficial interactions with plants. Front. Microbiol. 2021, 12, 752288. [Google Scholar] [CrossRef]
- AlAli, H.A.; Khalifa, A.; Al-Malki, M. Plant growth-promoting rhizobacteria from Ocimum basilicum improve growth of Phaseolus vulgaris and Abelmoschus esculentus. S. Afr. J. Bot. 2021, 139, 200–209. [Google Scholar] [CrossRef]
- Song, Q.; Song, X.; Deng, X.; Luo, J.; Wang, J.; Min, K.; Song, R. Effects of plant growth promoting Rhizobacteria microbial on the growth, rhizosphere soil properties, and bacterial community of Pinus sylvestris var. mongolica seedlings. Scand. J. For. Res. 2021, 36, 249–262. [Google Scholar] [CrossRef]
- Li, W.; Zhang, Z.; Sun, B.; Hu, S.; Wang, D.; Hu, F.; Li, H.; Xu, L.; Jiao, J. Combination of plant-growth-promoting and fluoranthene-degrading microbes enhances phytoremediation efficiency in the ryegrass rhizosphere. Environ. Sci. Poll. Res. 2021, 28, 6068–6077. [Google Scholar] [CrossRef]
- Rasool, A.; Mir, M.I.; Zulfajri, M.; Hanafiah, M.M.; Unnisa, S.A.; Mahboob, M. Plant growth promoting and antifungal asset of indigenous rhizobacteria secluded from saffron (Crocus sativus L.) rhizosphere. Microb. Pathog. 2021, 150, 104734. [Google Scholar] [CrossRef]
- Fahsi, N.; Mahdi, I.; Mesfioui, A.; Biskri, L.; Allaoui, A. Plant growth-promoting rhizobacteria isolated from the Jujube (Ziziphus lotus) plant enhance wheat growth, Zn uptake, and heavy metal tolerance. Agriculture 2021, 11, 316. [Google Scholar] [CrossRef]
- Souza, I.F.; Napoleão, T.H.; de Sena, K.X.; Paiva, P.M.; de Araújo, J.M.; Coelho, L.C. Endophytic microorganisms in leaves of Moringa oleifera collected in three localities at Pernambuco State, Northeastern Brazil. Microbiol. Res. J. Int. 2016, 13, 1–7. [Google Scholar] [CrossRef]
- Singha, K.M.; Singh, B.; Pandey, P. Host specific endophytic microbiome diversity and associated functions in three varieties of scented black rice are dependent on growth stage. Sci. Rep. 2021, 11, 12259. [Google Scholar] [CrossRef] [PubMed]
- Geisen, S.; Kostenko, O.; Cnossen, M.C.; Ten Hooven, F.C.; Vreš, B.; van Der Putten, W.H. Seed and root endophytic fungi in a range expanding and a related plant species. J. Front. Microbiol. 2017, 8, 1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dastogeer, K.M.; Tumpa, F.H.; Sultana, A.; Akter, M.A.; Chakraborty, A. Plant microbiome—An account of the factors that shape community composition and diversity. Curr. Plant Biol. 2020, 23, 100161. [Google Scholar] [CrossRef]
- Mina, D.; Pereira, J.A.; Lino-Neto, T.; Baptista, P. Epiphytic and endophytic bacteria on olive tree phyllosphere: Exploring tissue and cultivar effect. Microb. Ecol. 2020, 80, 145–157. [Google Scholar] [CrossRef]
- Campisano, A.; Albanese, D.; Yousaf, S.; Pancher, M.; Donati, C.; Pertot, I. Temperature drives the assembly of endophytic communities’ seasonal succession. Environ. Microbiol. 2017, 19, 3353–3364. [Google Scholar] [CrossRef]
- Xie, X.-G.; Zhang, F.-M.; Yang, T.; Chen, Y.; Li, X.-G.; Dai, C.-C. Endophytic fungus drives nodulation and N2 fixation attributable to specific root exudates. MBio 2019, 10, e00728-19. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.-W.; Ma, C.-Y.; Xu, F.-J.; Lu, F.; Zhang, W.; Dai, C.-C. Root endophyte-enhanced peanut-rhizobia interaction is associated with regulation of root exudates. Microbiol. Res. 2021, 250, 126765. [Google Scholar] [CrossRef]
- Alufasi, R.; Gere, J.; Chakauya, E.; Lebea, P.; Parawira, W.; Chingwaru, W. Mechanisms of pathogen removal by macrophytes in constructed wetlands. Environ. Technol. Rev. 2017, 6, 135–144. [Google Scholar] [CrossRef]
- Preston, G.M. Profiling the extended phenotype of plant pathogens: Challenges in bacterial molecular plant pathology. Mol. Plant Pathol. 2017, 18, 443–456. [Google Scholar] [CrossRef] [Green Version]
- Olanrewaju, O.S.; Ayangbenro, A.S.; Glick, B.R.; Babalola, O.O. Plant health: Feedback effect of root exudates-rhizobiome interactions. Appl. Microbiol. Biotechnol. 2019, 103, 1155–1166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hou, L.; He, X.; Li, X.; Wang, S.; Zhao, L. Species composition and colonization of dark septate endophytes are affected by host plant species and soil depth in the Mu Us sandland, northwest China. Fungal Ecol. 2019, 39, 276–284. [Google Scholar] [CrossRef]
- Looby, C.I.; Hollenbeck, E.C.; Treseder, K.K. Fungi in the canopy: How soil fungi and extracellular enzymes differ between canopy and ground soils. Ecosystems 2020, 23, 768–782. [Google Scholar] [CrossRef] [Green Version]
- Oita, S.; Ibáñez, A.; Lutzoni, F.; Miadlikowska, J.; Geml, J.; Lewis, L.A.; Hom, E.F.; Carbone, I.; U’Ren, J.M.; Arnold, A.E. Climate and seasonality drive the richness and composition of tropical fungal endophytes at a landscape scale. Commun. Biol. 2021, 4, 313. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez-Blanco, A.; Sicardi, M.; Frioni, L. Plant genotype and nitrogen fertilization effects on abundance and diversity of diazotrophic bacteria associated with maize (Zea mays L.). Biol. Fertil. Soils 2015, 51, 391–402. [Google Scholar] [CrossRef]
- Bulgarelli, D.; Garrido-Oter, R.; Münch, P.C.; Weiman, A.; Dröge, J.; Pan, Y.; McHardy, A.C.; Schulze-Lefert, P. Structure and function of the bacterial root microbiota in wild and domesticated barley. Cell Host Microbe 2015, 17, 392–403. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lindow, S.E.; Brandl, M.T. Microbiology of the phyllosphere. Appl. Environ. Microbiol. 2003, 69, 1875–1883. [Google Scholar] [CrossRef] [Green Version]
- Lundberg, D.S.; Lebeis, S.L.; Paredes, S.H.; Yourstone, S.; Gehring, J.; Malfatti, S.; Tremblay, J.; Engelbrektson, A.; Kunin, V.; Del Rio, T.G. Defining the core Arabidopsis thaliana root microbiome. Nature 2012, 488, 86–90. [Google Scholar] [CrossRef] [Green Version]
- Lakshmanan, V.; Kitto, S.L.; Caplan, J.L.; Hsueh, Y.-H.; Kearns, D.B.; Wu, Y.-S.; Bais, H.P. Microbe-associated molecular patterns-triggered root responses mediate beneficial rhizobacterial recruitment in Arabidopsis. Plant Physiol. 2012, 160, 1642–1661. [Google Scholar] [CrossRef] [Green Version]
- İnceoğlu, Ö.; Al-Soud, W.A.; Salles, J.F.; Semenov, A.V.; van Elsas, J.D. Comparative analysis of bacterial communities in a potato field as determined by pyrosequencing. PLoS ONE 2011, 6, e23321. [Google Scholar] [CrossRef] [Green Version]
- Peiffer, J.A.; Spor, A.; Koren, O.; Jin, Z.; Tringe, S.G.; Dangl, J.L.; Buckler, E.S.; Ley, R.E. Diversity and heritability of the maize rhizosphere microbiome under field conditions. Proc. Natl. Acad. Sci. USA 2013, 110, 6548–6553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, T.R.; James, E.K.; Poole, P.S. The plant microbiome. Genome Biol. 2013, 14, 209. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marques, J.M.; da Silva, T.F.; Vollu, R.E.; Blank, A.F.; Ding, G.-C.; Seldin, L.; Smalla, K. Plant age and genotype affect the bacterial community composition in the tuber rhizosphere of field-grown sweet potato plants. FEMS Microbiol. Ecol. 2014, 88, 424–435. [Google Scholar] [CrossRef] [PubMed]
- Hacquard, S.; Schadt, C.W. Towards a holistic understanding of the beneficial interactions across the Populus microbiome. New Phytol. 2015, 205, 1424–1430. [Google Scholar] [CrossRef] [PubMed]
- Horton, M.W.; Bodenhausen, N.; Beilsmith, K.; Meng, D.; Muegge, B.D.; Subramanian, S.; Vetter, M.M.; Vilhjálmsson, B.J.; Nordborg, M.; Gordon, J.I. Genome-wide association study of Arabidopsis thaliana leaf microbial community. Nat. Commun. 2014, 5, 5320. [Google Scholar] [CrossRef] [PubMed]
- Wagner, M.R.; Lundberg, D.S.; Tijana, G.; Tringe, S.G.; Dangl, J.L.; Mitchell-Olds, T. Host genotype and age shape the leaf and root microbiomes of a wild perennial plant. Nat. Commun. 2016, 7, 12151. [Google Scholar] [CrossRef] [Green Version]
- Cai, F.; Pang, G.; Miao, Y.; Li, R.; Li, R.; Shen, Q.; Chen, W. The nutrient preference of plants influences their rhizosphere microbiome. Appl. Soil Ecol. 2017, 110, 146–150. [Google Scholar] [CrossRef]
- Monaghan, J.; Zipfel, C. Plant pattern recognition receptor complexes at the plasma membrane. Curr. Opin. Plant Biol. 2012, 15, 349–357. [Google Scholar] [CrossRef]
- Erb, M.; Lenk, C.; Degenhardt, J.; Turlings, T.C. The underestimated role of roots in defense against leaf attackers. Trends Plant Sci. 2009, 14, 653–659. [Google Scholar] [CrossRef] [Green Version]
- Akanmu, A.O.; Babalola, O.O.; Venturi, V.; Ayilara, M.S.; Adeleke, B.S.; Amoo, A.E.; Sobowale, A.A.; Fadiji, A.E.; Glick, B.R. Plant disease management: Leveraging on the plant-microbe-soil interface in the biorational use of organic amendments. Front. Plant Sci. 2021, 12, 1590. [Google Scholar] [CrossRef]
- Chen, Y.; Yan, F.; Chai, Y.; Liu, H.; Kolter, R.; Losick, R.; Guo, J.h. Biocontrol of tomato wilt disease by Bacillus subtilis isolates from natural environments depends on conserved genes mediating biofilm formation. Environ. Microbiol. 2013, 15, 848–864. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, B.; Lee, S.; Ryu, C.-M. Foliar aphid feeding recruits rhizosphere bacteria and primes plant immunity against pathogenic and non-pathogenic bacteria in pepper. Ann. Bot. 2012, 110, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Landgraf, R.; Schaarschmidt, S.; Hause, B. Repeated leaf wounding alters the colonization of Medicago truncatula roots by beneficial and pathogenic microorganisms. Plant Cell Environ. 2012, 35, 1344–1357. [Google Scholar] [CrossRef] [PubMed]
- Lebeis, S.; McHardy, A.C.; Dangl, J.L.; Knight, R.; Ley, R.; Schulze-Lefert, P. Microbiota and host nutrition across plant and animal kingdoms. Cell Host Microbe 2015, 17, 603–616. [Google Scholar]
- Kniskern, J.M.; Traw, M.B.; Bergelson, J. Salicylic acid and jasmonic acid signaling defense pathways reduce natural bacterial diversity on Arabidopsis thaliana. Mol. Plant-Microbe Interact. 2007, 20, 1512–1522. [Google Scholar] [CrossRef] [Green Version]
- Bown, A.W.; Shelp, B.J. Plant GABA: Not just a metabolite. Trends Plant Sci. 2016, 21, 811–813. [Google Scholar] [CrossRef]
- Bednarek, P.; Osbourn, A. Plant-microbe interactions: Chemical diversity in plant defense. Science 2009, 324, 746–748. [Google Scholar] [CrossRef]
- Bressan, M.; Roncato, M.-A.; Bellvert, F.; Comte, G.; el Zahar Haichar, F.; Achouak, W.; Berge, O. Exogenous glucosinolate produced by Arabidopsis thaliana has an impact on microbes in the rhizosphere and plant roots. ISME J. 2009, 3, 1243–1257. [Google Scholar] [CrossRef]
- Maizel, J.; Burkhardt, H.; Mitchell, H. Avenacin, an antimicrobial substance isolated from Avena sativa L. Isolation and antimicrobial activity. Biochemistry 1964, 3, 424–426. [Google Scholar] [CrossRef]
- Carter, J.P.; Spink, J.; Cannon, P.F.; Daniels, M.J.; Osbourn, A.E. Isolation, characterization, and avenacin sensitivity of a diverse collection of cereal-root-colonizing fungi. Appl. Environ. Microbiol. 1999, 65, 3364–3372. [Google Scholar] [CrossRef] [Green Version]
- Papadopoulou, K.; Melton, R.; Leggett, M.; Daniels, M.; Osbourn, A. Compromised disease resistance in saponin-deficient plants. Proc. Natl. Acad. Sci. USA 1999, 96, 12923–12928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, T.R.; Ramakrishnan, K.; Walshaw, J.; Heavens, D.; Alston, M.; Swarbreck, D.; Osbourn, A.; Grant, A.; Poole, P.S. Comparative metatranscriptomics reveals kingdom level changes in the rhizosphere microbiome of plants. ISME J. 2013, 7, 2248–2258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Augusto, L.; Ranger, J.; Binkley, D.; Rothe, A. Impact of several common tree species of European temperate forests on soil fertility. Ann. For. Sci. 2002, 59, 233–253. [Google Scholar] [CrossRef] [Green Version]
- Farrar, K.; Bryant, D.; Cope-Selby, N. Understanding and engineering beneficial plant–microbe interactions: Plant growth promotion in energy crops. Plant Biotechnol. J. 2014, 12, 1193–1206. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Acuña, J.J.; Inostroza, N.G.; Mora, M.L.; Radic, S.; Sadowsky, M.J.; Jorquera, M.A. Endophytic bacterial communities associated with roots and leaves of plants growing in Chilean extreme environments. Sci. Rep. 2019, 9, 4950. [Google Scholar] [CrossRef] [Green Version]
- Youseif, S.H. Genetic diversity of plant growth promoting rhizobacteria and their effects on the growth of maize plants under greenhouse conditions. Ann. Agric. Sci. 2018, 63, 25–35. [Google Scholar] [CrossRef]
- Samaras, A.; Nikolaidis, M.; Antequera-Gómez, M.L.; Cámara-Almirón, J.; Romero, D.; Moschakis, T.; Amoutzias, G.D.; Karaoglanidis, G.S. Whole genome sequencing and root colonization studies reveal novel insights in the biocontrol potential and growth promotion by Bacillus subtilis MBI 600 on cucumber. Front. Microbiol. 2020, 11, 600393. [Google Scholar] [CrossRef]
- Neiverth, A.; Delai, S.; Garcia, D.M.; Saatkamp, K.; de Souza, E.M.; de Oliveira Pedrosa, F.; Guimarães, V.F.; dos Santos, M.F.; Vendruscolo, E.C.G.; da Costa, A.C.T. Performance of different wheat genotypes inoculated with the plant growth promoting bacterium Herbaspirillum seropedicae. Eur. J. Soil Biol. 2014, 64, 1–5. [Google Scholar] [CrossRef]
- Baghel, V.; Thakur, J.K.; Yadav, S.S.; Manna, M.C.; Mandal, A.; Shirale, A.O.; Sharma, P.; Sinha, N.K.; Mohanty, M.; Singh, A.B. Phosphorus and potassium solubilization from rock minerals by endophytic Burkholderia sp. strain FDN2-1 in soil and shift in diversity of bacterial endophytes of corn root tissue with crop growth stage. Geomicrobiol. J. 2020, 37, 550–563. [Google Scholar] [CrossRef]
- Kaul, S.; Choudhary, M.; Gupta, S.; Dhar, M.K. Engineering host microbiome for crop improvement and sustainable agriculture. Front. Microbiol. 2021, 12, 1125. [Google Scholar] [CrossRef]
- Munaweera, T.I.K.; Jayawardana, N.U.; Rajaratnam, R.; Dissanayake, N. Modern plant biotechnology as a strategy in addressing climate change and attaining food security. Agric. Food Security 2022, 11, 26. [Google Scholar] [CrossRef]
- Choudhary, A.K.; Sultana, R.; Vales, M.I.; Saxena, K.B.; Kumar, R.R.; Ratnakumar, P. Integrated physiological and molecular approaches to improvement of abiotic stress tolerance in two pulse crops of the semi-arid tropics. Crop J. 2018, 6, 99–114. [Google Scholar] [CrossRef] [Green Version]
- Varshney, R.K.; Bansal, K.C.; Aggarwal, P.K.; Datta, S.K.; Craufurd, P.Q. Agricultural biotechnology for crop improvement in a variable climate: Hope or hype? Trends Plant Sci. 2011, 16, 363–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Zhu, A.; Tan, H.; Cao, L.; Zhang, R. Engineering banana endosphere microbiome to improve Fusarium wilt resistance in banana. Microbiome 2019, 7, 74. [Google Scholar] [CrossRef]
- Rasche, F.; Velvis, H.; Zachow, C.; Berg, G.; Van Elsas, J.D.; Sessitsch, A. Impact of transgenic potatoes expressing anti-bacterial agents on bacterial endophytes is comparable with the effects of plant genotype, soil type and pathogen infection. J. Appl. Ecol. 2006, 43, 555–566. [Google Scholar] [CrossRef]
- Ou, T.; Xu, W.-f.; Wang, F.; Strobel, G.; Zhou, Z.-y.; Xiang, Z.-h.; Liu, J.; Xie, J. A microbiome study reveals seasonal variation in endophytic bacteria among different mulberry cultivars. Comp. Struct. Biotechnol. J. 2019, 17, 1091–1100. [Google Scholar] [CrossRef]
- Ishizawa, H.; Ogata, Y.; Hachiya, Y.; Tokura, K.-I.; Kuroda, M.; Inoue, D.; Toyama, T.; Tanaka, Y.; Mori, K.; Morikawa, M.; et al. Enhanced biomass production and nutrient removal capacity of duckweed via two-step cultivation process with a plant growth-promoting bacterium, Acinetobacter calcoaceticus P23. Chemosphere 2020, 238, 124682. [Google Scholar] [CrossRef]
- Rajini, S.B.; Nandhini, M.; Udayashankar, A.C.; Niranjana, S.R.; Lund, O.S.; Prakash, H.S. Diversity, plant growth-promoting traits, and biocontrol potential of fungal endophytes of Sorghum bicolor. Plant Pathol. 2020, 69, 642–654. [Google Scholar] [CrossRef]
- Annapurna, K.; Govindasamy, V.; Sharma, M.; Ghosh, A.; Chikara, S.K. Whole genome shotgun sequence of Bacillus paralicheniformis strain KMS 80, a rhizobacterial endophyte isolated from rice (Oryza sativa L.). 3 Biotech 2018, 8, 223. [Google Scholar] [CrossRef]
- Singh, P.; Singh, R.K.; Guo, D.-J.; Sharma, A.; Singh, R.N.; Li, D.-P.; Malviya, M.K.; Song, X.-P.; Lakshmanan, P.; Yang, L.-T. Whole genome analysis of sugarcane root-associated endophyte Pseudomonas aeruginosa B18—A plant growth-promoting bacterium with antagonistic potential against Sporisorium scitamineum. Front. Microbiol. 2021, 12, 104. [Google Scholar] [CrossRef]
- Shastry, R.P.; Welch, M.; Rai, V.R.; Ghate, S.D.; Sandeep, K.; Rekha, P. The whole-genome sequence analysis of Enterobacter cloacae strain Ghats1: Insights into endophytic lifestyle-associated genomic adaptations. Arch. Microbiol. 2020, 202, 1571–1579. [Google Scholar] [CrossRef] [PubMed]
- Wicaksono, W.A.; Jones, E.E.; Casonato, S.; Monk, J.; Ridgway, H.J. Biological control of Pseudomonas syringae pv. actinidiae (Psa), the causal agent of bacterial canker of kiwifruit, using endophytic bacteria recovered from a medicinal plant. Biol. Control 2018, 116, 103–112. [Google Scholar] [CrossRef]
- Jasim, B.; Sreelakshmi, K.; Mathew, J.; Radhakrishnan, E. Surfactin, iturin, and fengycin biosynthesis by endophytic Bacillus sp. from Bacopa monnieri. Microb. Ecol. 2016, 72, 106–119. [Google Scholar] [CrossRef] [PubMed]
- Mukherjee, A.; Das, R.; Pal, A.; Paul, A. Antibacterial activity of endophytic Bacillus safensis isolated from Ophioglossum reticulatum L. Microbiol. Res. J. Int. 2017, 18, 1–12. [Google Scholar] [CrossRef]
- Melo, F.M.P.d.; Fiore, M.F.; Moraes, L.A.B.d.; Silva-Stenico, M.E.; Scramin, S.; Teixeira, M.d.A.; Melo, I.S.d. Antifungal compound produced by the cassava endophyte Bacillus pumilus MAIIIM4A. Sci. Agric. 2009, 66, 583–592. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Oh, S.; Anderson, A.; Neiswender, J.; Kim, J.C.; Kim, Y. Production of the antifungal compounds phenazine and pyrrolnitrin from Pseudomonas chlororaphis O6 is differentially regulated by glucose. Lett. Appl. Microbiol. 2011, 52, 532–537. [Google Scholar] [CrossRef]
- Yan, L.; Zhu, J.; Zhao, X.; Shi, J.; Jiang, C.; Shao, D. Beneficial effects of endophytic fungi colonization on plants. Appl. Microbiol. Biotechnol. 2019, 103, 3327–3340. [Google Scholar] [CrossRef]
- Sánchez-Cañizares, C.; Jorrín, B.; Poole, P.S.; Tkacz, A. Understanding the holobiont: The interdependence of plants and their microbiome. Curr. Opin. Microbiol. 2017, 38, 188–196. [Google Scholar] [CrossRef]
- Kumar, A.; Dubey, A. Rhizosphere microbiome: Engineering bacterial competitiveness for enhancing crop production. J. Adv. Res. 2020, 24, 337–352. [Google Scholar] [CrossRef]
- Gouda, S.; Kerry, R.G.; Das, G.; Paramithiotis, S.; Shin, H.-S.; Patra, J.K. Revitalization of plant growth promoting rhizobacteria for sustainable development in agriculture. Microbiol. Res. 2018, 206, 131–140. [Google Scholar] [CrossRef]
- Cong, J.; Yang, Y.; Liu, X.; Lu, H.; Liu, X.; Zhou, J.; Li, D.; Yin, H.; Ding, J.; Zhang, Y. Analyses of soil microbial community compositions and functional genes reveal potential consequences of natural forest succession. Sci. Rep. 2015, 5, 10007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Tender, C. Microbial Community Analysis in Soil (Rhizosphere) and the Marine (Plastisphere) Environment in Function of Plant Health and Biofilm Formation. Ph.D. Thesis, Ghent University, Ghent, Belgium, 2017; pp. 1–274. [Google Scholar]
- Woźniak, M.; Gałązka, A.; Grządziel, J.; Głodowska, M. The identification and genetic diversity of endophytic bacteria isolated from selected crops. J. Agric. Sci. 2018, 156, 547–556. [Google Scholar] [CrossRef]
- Maropola, M.K.A. Identification of Endophytic Bacterial Communities Associated with South African Crops: Sorghum bicolor (L. Moench), Pennisetum glaucum and Arachis villosulicarpa; University of the Western Cape: Cape Town, South Africa, 2014. [Google Scholar]
- Akinsanya, M.A.; Goh, J.K.; Lim, S.P.; Ting, A.S.Y. Metagenomics study of endophytic bacteria in Aloe vera using next-generation technology. Genom. Data 2015, 6, 159–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fadiji, A.E.; Babalola, O.O. Metagenomics methods for the study of plant-associated microbial communities: A review. J. Microbiol. Methods 2020, 170, 105860. [Google Scholar] [CrossRef] [PubMed]
- Muyzer, G.; De Waal, E.C.; Uitterlinden, A. Profiling of complex microbial populations by denaturing gradient gel electrophoresis analysis of polymerase chain reaction-amplified genes coding for 16S rRNA. Appl. Environ. Microbiol. 1993, 59, 695–700. [Google Scholar] [CrossRef] [Green Version]
- Lee, S.I.; Kim, S.; Park, S.H.; Ricke, S.C. Molecular and new-generation techniques for rapid detection of foodborne pathogens and characterization of microbial communities in poultry meat. In Food Safety in Poultry Meat Production; Springer: Berlin/Heidelberg, Germany, 2019; pp. 235–260. [Google Scholar] [CrossRef]
- Garbeva, P.; Van Overbeek, L.; Van Vuurde, J.; Van Elsas, J. Analysis of endophytic bacterial communities of potato by plating and denaturing gradient gel electrophoresis (DGGE) of 16S rDNA based PCR fragments. Microb. Ecol. 2001, 41, 369–383. [Google Scholar] [CrossRef]
- Hanning, I.B.; Ricke, S.C. Prescreening of microbial populations for the assessment of sequencing potential. In High-Throughput Next Generation Sequencing; Springer: Berlin/Heidelberg, Germany, 2011; pp. 159–170. [Google Scholar]
- Gafan, G.P.; Spratt, D.A. Denaturing gradient gel electrophoresis gel expansion (DGGEGE)—An attempt to resolve the limitations of co-migration in the DGGE of complex polymicrobial communities. FEMS Microbiol. Lett. 2005, 253, 303–307. [Google Scholar] [CrossRef]
- Nikolcheva, L.G.; Bärlocher, F. Terminal Restriction Fragment Length Polymorphism (T-RFLP) to Estimate Fungal Diversity. In Methods to Study Litter Decomposition; Springer: Berlin/Heidelberg, Germany, 2020; pp. 311–318. [Google Scholar]
- Liu, W.-T.; Marsh, T.L.; Cheng, H.; Forney, L.J. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 1997, 63, 4516–4522. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.-w.; Ding, W.-p.; Xiong, L.-y.; Guo, L.; Sun, J.-m.; Xiao, P. Recent advancements in intestinal microbiota analyses: A review for non-microbiologists. Curr. Med. Sci. 2018, 38, 949–961. [Google Scholar] [CrossRef]
- Hyman, E.D.J.A.b. A new method of sequencing DNA. Analyt. Chem. 1988, 174, 423–436. [Google Scholar] [CrossRef]
- Harrington, C.T.; Lin, E.I.; Olson, M.T.; Eshleman, J.R. Fundamentals of pyrosequencing. Arch. Pathol. Lab. Med. 2013, 137, 1296–1303. [Google Scholar] [CrossRef] [PubMed]
- Li, O.; Xiao, R.; Sun, L.; Guan, C.; Kong, D.; Hu, X. Bacterial and diazotrophic diversities of endophytes in Dendrobium catenatum determined through barcoded pyrosequencing. PLoS ONE 2017, 12, e0184717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Busato, F.; Dejeux, E.; Gut, I.G.; Tost, J. Quantitative DNA methylation analysis at single-nucleotide resolution by pyrosequencing®. In DNA Methylation Protocols; Springer: Berlin/Heidelberg, Germany, 2018; pp. 427–445. [Google Scholar]
- Cheng, L.; Lopez-Beltran, A.; Massari, F.; MacLennan, G.T.; Montironi, R. Molecular testing for BRAF mutations to inform melanoma treatment decisions: A move toward precision medicine. Mod. Pathol. 2018, 31, 24–38. [Google Scholar] [CrossRef] [PubMed]
- Jose, B.V.; De Leon, D.T.; Malonzo, M.A.; Undan, J.R. Molecular identification of endophytic fungi associated with Cynometra ramiflora L. and Wrightia pubescens (R. Br.) using the Internal Transcribed Spacer (ITS) region of rDNA and its morphotypes. Curr. Appl. Sci. Technol. 2021, 21, 535–544. [Google Scholar]
- Thinh, B.B.; Chac, L.D.; Thu, L.T.M. Application of internal transcribed spacer (ITS) sequences for identifying Anoectochilus setaceus Blume in Thanh Hoa, Vietnam. Proc. Appl. Bot. Genet. Breed. 2020, 181, 108–116. [Google Scholar] [CrossRef]
- Hollingsworth, P.M. Refining the DNA barcode for land plants. Proc. Natl. Acad. Sci. USA 2011, 108, 19451–19452. [Google Scholar] [CrossRef] [Green Version]
- Timpano, E.K.; Scheible, M.K.; Meiklejohn, K.A. Optimization of the second internal transcribed spacer (ITS2) for characterizing land plants from soil. PLoS ONE 2020, 15, e0231436. [Google Scholar] [CrossRef] [Green Version]
- Nelson, M.C.; Morrison, H.G.; Benjamino, J.; Grim, S.L.; Graf, J. Analysis, optimization and verification of Illumina-generated 16S rRNA gene amplicon surveys. PLoS ONE 2014, 9, e94249. [Google Scholar] [CrossRef]
- Wu, L.; Wen, C.; Qin, Y.; Yin, H.; Tu, Q.; Van Nostrand, J.D.; Yuan, T.; Yuan, M.; Deng, Y.; Zhou, J. Phasing amplicon sequencing on Illumina Miseq for robust environmental microbial community analysis. BMC Microbiol. 2015, 15, 125. [Google Scholar] [CrossRef] [Green Version]
- Fadiji, A.E.; Ayangbenro, A.S.; Babalola, O.O. Shotgun metagenomics reveals the functional diversity of root-associated endophytic microbiomes in maize plant. Curr. Plant Biol. 2021, 25, 100195. [Google Scholar] [CrossRef]
- Bouchez, T.; Blieux, A.; Dequiedt, S.; Domaizon, I.; Dufresne, A.; Ferreira, S.; Godon, J.-J.; Hellal, J.; Joulian, C.; Quaiser, A. Molecular microbiology methods for environmental diagnosis. Environ. Chem. Lett. 2016, 14, 423–441. [Google Scholar] [CrossRef]
- Hong, C.E.; Kim, J.U.; Lee, J.W.; Bang, K.H.; Jo, I.H. Metagenomic analysis of bacterial endophyte community structure and functions in Panax ginseng at different ages. 3 Biotech 2019, 9, 300. [Google Scholar] [CrossRef] [PubMed]
- Basha, H.; Ramanujam, B. Growth promotion effect of Pichia guilliermondii in chilli and biocontrol potential of Hanseniaspora uvarum against Colletotrichum capsici causing fruit rot. Biocontrol Sci. Technol. 2015, 25, 185–206. [Google Scholar] [CrossRef]
- Berg, G.; Grube, M.; Schloter, M.; Smalla, K. The plant microbiome and its importance for plant and human health. Front. Microbiol. 2014, 5, 491. [Google Scholar] [CrossRef] [Green Version]
- Tian, B.-Y.; Cao, Y.; Zhang, K.-Q. Metagenomic insights into communities, functions of endophytes and their associates with infection by root-knot nematode, Meloidogyne incognita, in tomato roots. Sci. Rep. 2015, 5, 17087. [Google Scholar] [CrossRef] [Green Version]
- Delmotte, N.; Knief, C.; Chaffron, S.; Innerebner, G.; Roschitzki, B.; Schlapbach, R.; von Mering, C.; Vorholt, J.A. Community proteogenomics reveals insights into the physiology of phyllosphere bacteria. Proc. Natl. Acad. Sci. USA 2009, 106, 16428–16433. [Google Scholar] [CrossRef] [Green Version]
- Rastogi, G.; Coaker, G.L.; Leveau, J.H. New insights into the structure and function of phyllosphere microbiota through high-throughput molecular approaches. FEMS Microbiol. Lett. 2013, 348, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Kaul, S.; Sharma, T.; K Dhar, M. “Omics” tools for better understanding the plant–endophyte interactions. Front. Plant Sci. 2016, 7, 955. [Google Scholar] [CrossRef] [Green Version]
- Felitti, S.; Shields, K.; Ramsperger, M.; Tian, P.; Sawbridge, T.; Webster, T.; Logan, E.; Erwin, T.; Forster, J.; Edwards, D. Transcriptome analysis of Neotyphodium and Epichloë grass endophytes. Fungal Genet. Biol. 2006, 43, 465–475. [Google Scholar] [CrossRef]
- Wang, Y.; Ohara, Y.; Nakayashiki, H.; Tosa, Y.; Mayama, S. Microarray analysis of the gene expression profile induced by the endophytic plant growth-promoting rhizobacteria, Pseudomonas fluorescens FPT9601-T5 in Arabidopsis. Mol. Plant-Microbe Interact. 2005, 18, 385–396. [Google Scholar] [CrossRef] [Green Version]
- Barnett, M.J.; Toman, C.J.; Fisher, R.F.; Long, S.R. A dual-genome Symbiosis Chip for coordinate study of signal exchange and development in a prokaryote–host interaction. Proc. Natl. Acad. Sci. USA 2004, 101, 16636–16641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Aharwal, R.P.; Shukla, H.; Rajak, R.; Sandhu, S.S. Endophytic fungi: As a source of antimicrobials bioactive compounds. World J. Pharm. Pharm. Sci. 2014, 3, 1179–1197. [Google Scholar]
- Martinez-Klimova, E.; Rodríguez-Peña, K.; Sánchez, S. Endophytes as sources of antibiotics. Biochem. Pharmacol. 2017, 134, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Cruz, R.; Tpia Vázquez, I.; Batista-García, R.A.; Méndez-Santiago, E.W.; Sánchez-Carbente, M.d.R.; Leija, A.; Lira-Ruan, V.; Hernández, G.; Wong-Villarreal, A.; Folch-Mallol, J.L. Isolation and characterization of endophytes from nodules of Mimosa pudica with biotechnological potential. Microbiol. Res. 2019, 218, 76–86. [Google Scholar] [CrossRef]
- Shahzad, R.; Waqas, M.; Khan, A.L.; Al-Hosni, K.; Kang, S.-M.; Seo, C.-W.; Lee, I.-J. Indoleacetic acid production and plant growth promoting potential of bacterial endophytes isolated from rice (Oryza sativa L.) seeds. Acta Biol. Hung. 2017, 68, 175–186. [Google Scholar] [CrossRef] [Green Version]
- Martínez-Hidalgo, P.; Flores-Félix, J.D.; Sánchez-Juanes, F.; Rivas, R.; Mateos, P.F.; Santa Regina, I.; Peix, Á.; Martínez-Molina, E.; Igual, J.M.; Velázquez, E. Identification of Canola roots endophytic bacteria and analysis of their potential as biofertilizers for Canola crops with special emphasis on sporulating bacteria. Agronomy 2021, 11, 1796. [Google Scholar] [CrossRef]
- Basu, A.; Prasad, P.; Das, S.N.; Kalam, S.; Sayyed, R.; Reddy, M.; El Enshasy, H. Plant growth promoting rhizobacteria (PGPR) as green bioinoculants: Recent developments, constraints, and prospects. Sustainability 2021, 13, 1140. [Google Scholar] [CrossRef]
- Lin, G.-Y.; Lin, C.-Y.; Chang, S.-J.; Lin, W.-Y. The dynamics of endophytic bacterial community structure in rice roots under different field management systems. Agronomy 2020, 10, 1623. [Google Scholar] [CrossRef]
- Kuźniar, A.; Włodarczyk, K.; Grządziel, J.; Goraj, W.; Gałązka, A.; Wolińska, A. Culture-independent analysis of an endophytic core microbiome in two species of wheat: Triticum aestivum L. (cv. ‘Hondia’) and the first report of microbiota in Triticum spelta L. (cv. ‘Rokosz’). Syst. Appl. Microbiol. 2019, 43, 126025. [Google Scholar] [CrossRef]
- Bikel, S.; Valdez-Lara, A.; Cornejo-Granados, F.; Rico, K.; Canizales-Quinteros, S.; Soberón, X.; Del Pozo-Yauner, L.; Ochoa-Leyva, A. Combining metagenomics, metatranscriptomics and viromics to explore novel microbial interactions: Towards a systems-level understanding of human microbiome. Comput. Struct. Biotechnol. J. 2015, 13, 390–401. [Google Scholar] [CrossRef] [Green Version]
- Adeleke, B.S.; Babalola, O.O. Meta-omics of endophytic microbes in agricultural biotechnology. Biocatal. Agric. Biotechnol. 2022, 42, 102332. [Google Scholar] [CrossRef]
- Fadiji, A.E.; Ayangbenro, A.S.; Babalola, O.O. Unveiling the putative functional genes present in root-associated endophytic microbiome from maize plant using the shotgun approach. J. Appl. Genet. 2021, 62, 339–351. [Google Scholar] [CrossRef] [PubMed]
- Adeleke, B.S.; Ayangbenro, A.S.; Babalola, O.O. Genomic assessment of Stenotrophomonas indicatrix for improved sunflower plant. Curr. Genet. 2021, 67, 891–907. [Google Scholar] [CrossRef] [PubMed]
- Adeleke, B.S.; Ayangbenro, A.; Babalola, O.O. Effect of endophytic bacterium, Stenotrophomonas maltophilia JVB5 on sunflowers. Plant Prot. Sci. 2022, 58, 185–198. [Google Scholar] [CrossRef]
| Microbial Niche | Host Plant | Beneficial Traits | References |
|---|---|---|---|
| Endosphere | Ceriops decandra | Biocontrol activity | [88] |
| Vicia faba, Ephedra pachyclada | Plant growth-promoting traits | [89,90] | |
| Capsicum annum | Salt stress tolerance | [14] | |
| Triticum aestivum | IAA, ammonia, HCN, an enzyme production | [91] | |
| Glycine max | Bioremediation of heavy metals | [92] | |
| Corchorus olitorius | Nitrogen fixation, 1-aminocyclopropane-1-carboxylate (ACC) deaminase activity, IAA, and siderophore production | [93] | |
| Oryza sativa | IAA, phosphate solubilization, siderophore, HCN, DNase activity | [94] | |
| Hordeum vulgare | IAA, phosphate and potassium solubilization, nitrogen fixation, siderophore, ACC deaminase activity | [95] | |
| Rhizosphere | Zea mays | Salt stress tolerance | [96] |
| Cucumis sativus | Biocontrol activity, mineral solubilization, siderophore and enzyme production, IAA and polyamine biosynthesis, ACC deaminase activity | [97] | |
| Ocimum basilicum | Plant growth-promoting traits | [98] | |
| Pinus sylvestris | Phosphate solubilization, siderophore production, and IAA synthesis | [99] | |
| Lolium perenne | Phytoremediation of environmental pollutants | [100] | |
| Crocus sativus | Plant growth-promoting traits and biocontrol potential | [101] | |
| Ziziphus lotus | Heavy metal tolerance | [102] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Adeleke, B.S.; Fadiji, A.E.; Ayilara, M.S.; Igiehon, O.N.; Nwachukwu, B.C.; Babalola, O.O. Strategies to Enhance the Use of Endophytes as Bioinoculants in Agriculture. Horticulturae 2022, 8, 498. https://doi.org/10.3390/horticulturae8060498
Adeleke BS, Fadiji AE, Ayilara MS, Igiehon ON, Nwachukwu BC, Babalola OO. Strategies to Enhance the Use of Endophytes as Bioinoculants in Agriculture. Horticulturae. 2022; 8(6):498. https://doi.org/10.3390/horticulturae8060498
Chicago/Turabian StyleAdeleke, Bartholomew Saanu, Ayomide Emmanuel Fadiji, Modupe Stella Ayilara, Ozede Nicholas Igiehon, Blessing Chidinma Nwachukwu, and Olubukola Oluranti Babalola. 2022. "Strategies to Enhance the Use of Endophytes as Bioinoculants in Agriculture" Horticulturae 8, no. 6: 498. https://doi.org/10.3390/horticulturae8060498
APA StyleAdeleke, B. S., Fadiji, A. E., Ayilara, M. S., Igiehon, O. N., Nwachukwu, B. C., & Babalola, O. O. (2022). Strategies to Enhance the Use of Endophytes as Bioinoculants in Agriculture. Horticulturae, 8(6), 498. https://doi.org/10.3390/horticulturae8060498






