Abstract
Powdery mildew disease caused by Oidium neolycopersici is one of the major diseases affecting tomato production in South Africa. Interestingly, limited studies exist on how this disease affects the community structure microbial communities associated with tomato plants employing shotgun metagenomics. In this study, we assess how the health status of a tomato plant affects the diversity of the rhizosphere microbial community. We collected soil samples from the rhizosphere of healthy (HR) and diseased (DR; powdery mildew infected) tomatoes, alongside bulk soil (BR), extracted DNA, and did sequencing using shotgun metagenomics. Our results demonstrated that the rhizosphere microbiome alongside some specific functions were abundant in HR followed by DR and bulk soil (BR) in the order HR > DR > BR. We found eighteen (18) bacterial phyla abundant in HR, including Actinobacteria, Acidobacteria, Aquificae, Bacteroidetes, etc. The dominant fungal phyla include; Ascomycota and Basidiomycota, while the prominent archaeal phyla are Thaumarchaeota, Crenarchaeota, and Euryarchaeota. Three (3) bacteria phyla dominated the DR samples; Bacteroidetes, Gemmatimonadetes, and Thermotoga. Our result also employed the SEED subsystem and revealed that the metabolic pathways involved were abundant in HR. The α-diversity demonstrates that there is no significant difference among the rhizosphere microbiomes across the sites, while β-diversity demonstrated a significant difference.
1. Introduction
Microorganisms inhabiting soil are associated with the living ecosystem, but once the nature of the environment changes, the microbial species change [1,2]. These microorganisms are diverse and live in extreme conditions in the ecosystem [3]. Various crop plant species can bring together microorganisms present in their endosphere, phyllosphere, and rhizosphere [4,5]. Several studies have reported how the microbiomes interacting with the plant are essential in observing the plant’s health status and productivity. As a result, some microorganisms are critical for developing plants and preventing environmental stresses [4,6,7,8].
Plant growth-promoting microorganisms present in crop plants’ rhizosphere have been extensively studied to establish their roles and potential in plant growth promotion, abundance, and diversity [9,10,11]. Solubilization of nutrients, stimulation of phytohormone, and fixation of nitrogen are some of the mechanisms rhizosphere microbes employ to improve plant growth and health [12]. The indirect means used by these microbes for plant growth promotion include the reduction of soil-borne phytopathogens by antagonistic activity [13].
Soil microbes enable balance and stable agrobiodiversity below ground level for improved crop production [14]. However, pathogenic microbes in the ecosystem have caused considerable losses in most economical crops, resulting in food shortage and yield loss [15]. Exploring soil-inhabiting microbes can help mitigate the surge of disease resurgence in many plants [16,17]. Findings have demonstrated the advantages of soil microbes in controlling phytopathogens [18,19]. Nevertheless, their full exploration is limited due to the uncultivable nature of the envisaged agriculturally important microbes in the soil [20,21].
Globally, one of the most planted vegetables is tomato (Solanum lycopersicum), with more than 177 million tons worldwide [22]. Complex microbial communities inhabit and perform particular functions in the plant’s healthy living, growth, and development [23]. Among the microbial communities colonizing the plant and environments are phytopathogens prone to causing diseases on the plant. In contrast, others can improve the healthy living of the plant by enhancing nutrient acquisition and tolerance to both biotic and abiotic stresses [24].
Tomato plants are often cultivated in fields and greenhouses [25]. Despite its importance, the invasion of pests and diseases cause production problems [26], such as early blight caused by Alternaria solani [27], tomato late blight caused by Phytophthora infestans [28], bacterial wilt (Ralstonia solanacearum) [29], powdery mildew (Oidium neolycopersici) [30], and soft rot (Xanthomonas spp and Fusarium oxysporum) [31,32,33], among others. Powdery mildew disease caused by Oidium neolycopersici limits the cultivation of tomatoes [34,35]. Symptoms of the disease include pale yellow spots on leaves that later become covered with white spores, which make the leaves appear as if they have been dusted with flour [34,35]. This spoilage organism infects all other parts of the plant but not the tomato fruit. Premature aging of the tomato plant leaves and demotion of the fruits are symptoms of severe infection [26].
Metagenomics is a promising technique for unraveling the microbiome associated with most crop plants [36]. Interestingly, research on the microbial communities associated with the rhizosphere of the diseased and healthy plant has employed both culture-dependent and independent techniques such as Pyrosequencing, including 16SrRNA and ITS [37,38,39]. Previous researchers have explained much on the bacterial [40,41,42] and fungal [22,43,44] communities present in healthy and diseased tomato plants but did not demonstrate other microbes, such as archaea. To the best of our knowledge, no current study has explored the entire microbiome associated with the healthy and diseased rhizosphere of the tomato plant, employing shotgun metagenomic sequencing. That is why, in this study, we examined the community structure and functional diversity of microbiomes present in the rhizosphere of healthy and powdery mildew diseased tomato plants using shotgun metagenomic sequencing. We hypothesized that microbes would be more abundant in the healthy rhizosphere than the diseased rhizosphere.
2. Materials and Methods
2.1. The Study Area
We collected the rhizosphere soils of the tomato plant used in this research from the teaching research farm of the North-West University School, Mafikeng, South Africa (26°019′36.9″ S, 26°053′19.0″ E; 25°47′19.1″ S, 25°37′05.1″ E, 25°47′17.0″ S, 25°37′03.2″; altitude, 159 km).
2.2. Field Description and Sampling
We conducted this research in the Northwest Province of South Africa. This region experiences summer weather with slight rain showers between August and March. The average annual rainfall is 300 to 600 mm and temperatures range from 25 to 38 °C, which shows how extreme the temperature is during the summer. However, during winter temperatures drop below 10 °C, and this takes place from May to July every year. We planted the Roma tomato cultivar. The healthy, powdery mildew diseased rhizosphere and bulk soils were collected 40 m away from one another. This farming site has been used to cultivate tomatoes for over seven years. The farm has a history of inorganic (NPK) fertilizer application to improve soil fertility continuously.
We sampled the soil in March 2021, collecting the rhizosphere soils from the rhizosphere of the tomato plant at a depth of 4–15 cm using a 5 cm diameter drill. The region has an optimum for an average temperature of 28 °C and annual precipitation of 450 mm. We collected the soil samples from the root zones of healthy tomato plants (HR), powdery mildew diseased tomato plants (DR), and bulk soil (BR) [3]. We selected the powdery mildew diseased plants based on the bright yellow spots 1.27 cm in diameter on the lower leaves. The spots magnify and change to the brown and powdery appearance on the leaves and stems of the tomato plant [34,35]. We divided the site into three regions. In each area, we collected soil samples from the rhizosphere soil of five (5) tomato plants (40–50 cm high) for healthy and diseased and the bulk soil site without tomato plantation. The soil collected from the five plant samples was pooled together to replicate HR, DR, and BR. Three (3) replicates from the three regions were created for the soil samples from the HR, DR, and BR sites. All the samples were collected into sterile polythene bags and conveyed to the laboratory at 4 °C. The soil sample was further sieved with a 2 mm sieve and kept at −20 °C for further analysis.
2.3. Extraction and Shotgun Sequencing of DNA Obtained from Soil Samples
We extracted DNA using 5 g of the soil samples measured using a calibrated weighing machine for each collected soil and the NucleoSpin Soil kit (Macherey-Nagel, Duren, Nordrhein-Westfalen, Germany). We assessed the qualities of the extracted DNA using the Nanodrop. The libraries that we set up with the Nextra DNA Flex kit were prepared with 50 ng DNA and later underwent fragmentation and removal of adapter sequences. The final concentrations of the libraries were measured using the Qubit double-stranded DNA (dsDNA) HS assay kit (Life Technologies Carlsbad, CA, United states of America), and we ascertained the mean length of the DNA fragments using a 2100 Bioanalyzer (Agilent Technologies Santa Clara, California). Furthermore, the libraries were then monitored, combined at 0.6 nM, and sequenced using the NovaSeq 6000 system Illumina (300 cycles).
2.4. Data Analysis
Each metagenome obtained was uploaded on a metagenomics rapid annotation online server (MG-RAST) [45]. We subjected the sequences to quality control analysis on the MG-RAST database, including removing artificial sequences (dereplication). They emanate as sequencing artifacts, sequence length filtering, and filtering of the ambiguous base. Sequences with length >2 from the mean were removed. The BLAST-like alignment tool (BLAT) was used to annotate sequences algorithm after quality control (QC) [46] against the M5NR database [47], thereby providing no excess database integration. The SEED Subsystem carried out taxonomic profiling of archaeal, bacterial, and fungal taxa, while the SEED Subsystem level 3 functions database followed the method of Mitra, et al. [48] with an absolute parameter of the maximum e-value 1 × 10−5 cut off and 60% minimum sequence resemblance to the subsystem was used for profiling the functional categories of the rhizosphere microbiomes. Since we focus on bacteria, archaea, and fungi, we manually removed other sequences unrelated to these, alongside viruses. Normalization of data was carried out on MG-RAST. For a single taxon, derived tables were joined to avoid problems with the experiment. We kept the taxonomical values of unclassified sequences for statistical analysis, after which we calculated the relative abundance in percentages. The mean of the relative abundance for the soil samples (HR, DR, and BR) was employed for further statistical analysis. The sequences used in this study have been deposited on the NCBI SRA dataset with the bio-project number PRJNA766489 for the microbiome in the rhizosphere of tomato and bulk soil samples.
2.5. Statistical Analysis
We used the shiny heatmap to plot the relative abundance of the microbial communities at phylum to genus levels following the method of Khomtchouk, et al. [49]. Statistical analysis was carried out with the aid of PAST version 3.20, Oslo, Norway which was used to assess Simpson, Shannon, and Evenness’ diversity indices [50]. The Bray–Curtis-based principal coordinates analysis (PCoA) and ANOSIM were used to determine the differences in the microbial community structure [51]. However, we plotted PCA and PCoA graphs using the CANOCO 5.0 version Ithaca, USA.
3. Results
3.1. Shotgun Sequencing of the Microbiome in Tomato Rhizosphere and Bulk Soil
The respective mean values of the uploaded sequences were 13,739,258 for healthy tomato rhizosphere soil (HR), 19,765,082 for diseased rhizosphere soil (DR), and 138,145,859 for bulk soil (BR). After QC analysis on MG-RAST, the retained sequences’ mean values were 12,665,143 for HR, 11,966,279 for DR, and 5,247,459 for BR (Supplementary Table S1; Figure S1).
3.2. Distribution of Major Rhizosphere Soil Microbiome Phyla across the Tomato Plant Sites
The 18 prevalent bacteria phyla found in the soil samples were: Proteobacteria, Actinobacteria, Acidobacteria, Bacteroidetes, Planctomycetes, Chloroflexi, Firmicutes, Verrucomicrobia, Cyanobacteria, Gemmatimonadetes, unclassified (Bacteria), Nitrospirae, Deinococcus-Thermus, Chlorobi, Spirochaetes, Aquificae, Synergistetes, and Thermotogae. The predominant five phyla of Archaea were: Thaumarchaeota, Crenarchaeota, Euryarchaeota, Korarchaeota, and Nanoarchaeota. The dominant three phyla of fungi were: Ascomycota, Basidiomycota, and Blastocladiomycota.
However, the relative abundance of the taxa is highly prevalent in a healthy rhizosphere, followed by the diseased rhizosphere soil of tomato plants (DR) and bulk soil (BR) following the order HR > DR > BR (Figure 1). Still, the difference observed in the community composition does not differ significantly (p > 0.05) (Supplementary Table S2). We used the principal component analysis to investigate the distribution of significant phyla obtained in the tomato rhizosphere and the bulk rhizosphere, with the rhizosphere of the healthy tomato plant showing the highest abundance of the microbiome (Figure 2).
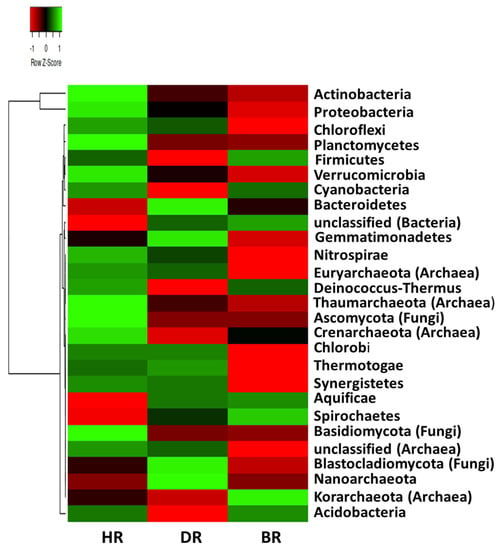
Figure 1.
Phyla heatmap of soil microorganism affiliated with the tomato plants. Each plot with particular color functioned as a saturation gradient employing the mean value with a z-score. HR = rhizosphere soil of healthy tomato plant; DR = rhizosphere soil of diseased tomato plant; and BR = bulk soil obtained from the soil without tomato plantation.
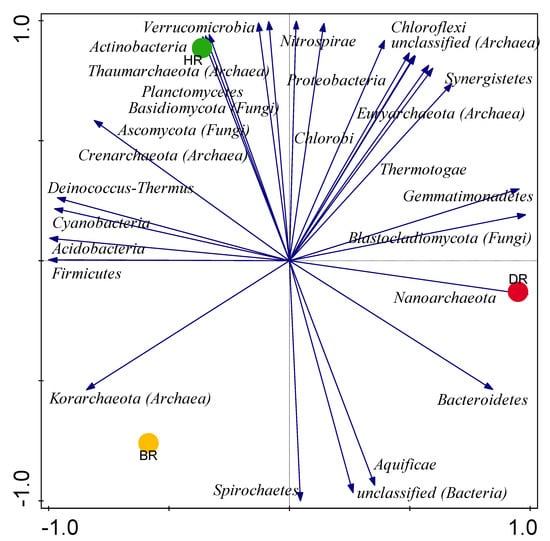
Figure 2.
PCA graph of phyla of the community of microbes present in the rhizosphere and bulk soils. Each arrow shows the impact of the microbial metagenomes, axis 1 (59%) and axis 2 (41%) explain the variations, employing the Bray–Curtis dissimilarity matrix. HR = rhizosphere soil of healthy tomato plant; DR = rhizosphere soil of diseased tomato plant; and BR = bulk soil obtained from the soil without tomato plantation.
The bacterial phyla dominant in HR are Proteobacteria (54.0%), Actinobacteria (35.5%), Planctomycetes (3.4%), Chloroflexi (2.8%), Verrucomicrobia (2.3%), Cyanobacteria (2%), Ntrospirae (0.4%), Deinococcus-Thermus (0.44%), Chlorobi (0.15%), and Synergistetes (0.05%). Meanwhile the fungal phyla dominant in the HR site are Ascomycota and Basidiomycota, with a mean abundance of 0.8 and 0.03%, respectively. The dominant archaeal phyla in HR includes; Euryarchaeota, Crenarchaeota, Thaumarchaeota (0.3%), and unclassified (Archaea) with a mean abundance of 0.43%, 0s.13%, and 0.03%, respectively. The dominant phyla obtained in the DR samples are the Bacteroidetes (4.0%), Gemmatimonadetes (1.6%), and Thermotoga (0.05%). The archaeal phyla include Nanoarchaeota (0.0003%), and fungal phyla are Blastocladiomycota (0.0003%).
Moreover, the dominant bacterial phyla identified in the BR are: Acidobacteria (5.2%), Firmicutes (2.4%), unclassified (Bacteria) (0.9%), Spirochaetes (0.08%), and Aquificae (0.06%). The archaeal phylum is Korarchaeota (0.004%), while no fungal phylum was identified.
The prevalent class of microbial communities with the highest mean of abundance in HR include the bacterial class: Actinobacteria (37.6%), Alphaproteobacteria (18%), Solibacteres (9%), Betaproteobacteria (4.8%), Gammaproteobacteria (3.4%), Gemmatimonadetes (1.7%), Chloroflexi (1.2%), Sphingobacteria (0.9%), Acidobacteria (0.8%), Cytophagia (0.6%), Thermomicrobia (0.6%), Flavobacteria (0.4%), and Gloeobacteria (0.2%). The archaeal class includes Methanomicrobia (0.2%) and Halobacteria (0.2%). The fungal class includes Sordariomycetes (0.15%), Eurotiomycetes (0.1%), Dothideomycetes (0.08%), Saccharomycetes (0.01%), and Ustilaginomycetes (0.03%). The bacterial class dominant in the DR samples includes Deltaproteobacteria (6%), Planctomycetacia (2.7%), Deinococci (0.5%), and Bacteroidia (0.3%). Only Methanococci (0.008%) was identified as the archaeal class, while no fungal genera were identified. The microbial communities dominant in the bulk soil are bacteria, which are Clostridia (1%) and unclassified (bacteria) (0.9%) (Figure 3).
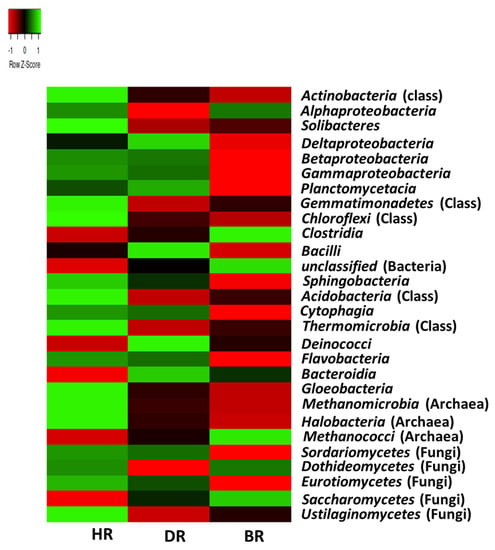
Figure 3.
The class heatmap of soil microorganisms affiliated with the tomato plants. Each plot with unique color functioned as a saturation gradient employing the mean value with a z-score. HR = rhizosphere soil of healthy tomato plant; DR = rhizosphere soil of diseased tomato plant; and BR = bulk soil obtained from the soil without tomato plantation.
The predominant bacterial order in HR include Rhizobiales (12%), Burkholderiales (6.4%), Myxococcales (5.5%), Sphingomonadales (5.1%), Solibacterales (3.2%), Gemmatimonadales (2%), Xanthomonadales (1.6%), Caulobacterales (1.8%), Rhodospirillales (1.7%), Chloroflexales (1.1%), Acidobacteriales (1.0%), Clostridiales (0.9%), and Pseudomonadales (0.7%). The archaeal orders are Halobacteriales (0.2%) and Methanosarcinales (0.1%). The fungal orders are Eurotiales (0.07%) and Hypocreales (0.2%). The predominant bacterial order in the DR samples include Planctomycetales (4.8%), Solirubrobacterales (2.9%), Rhodobacterales (2.0%), Sphingobacteriales (1.3%), and Bacillales (1.1%). The fungal order is Sordariales (0.05%). The predominant bacterial order of microbial communities with the highest mean in BR samples are Actinomycetales (35.6%), and Rubrobacterales (3.6%) with the fungus Pleosporales (0.2%), while the most abundant archaeal order is the Methanmicrobiales (0.06%) (Figure 4)

Figure 4.
The order heatmap of soil microorganisms affiliated with the tomato plants. Each plot with unique color functioned as a saturation gradient employing the mean value with a z-score. HR = rhizosphere soil of healthy tomato plant; DR = rhizosphere soil of diseased tomato plant; and BR = bulk soil obtained from the soil without tomato plantation.
The prevalent family of microbial communities with the highest abundance mean in HR include the bacterial family; Bradyrhizobiaceae (5.2%), Planctomycetaceae (6.3%), Solibacteraceae (4.2%), Streptomycetaceae (5.1%), Rubrobacteraceae (4.9%), Conexibacteraceae (4.0%), Gemmatimonadaceae (2.8%), Polyangiaceae (2.28%), Burkholderiaceae (2.2%), Mycobacteriaceae (2.4%), Frankiaceae (2.0%), Rhodobacteraceae (2.0%), Acidobacteriaceae (1.4%), Chloroflexaceae (1.1%), Cytophagaceae (1.1%), and Tthermoplasmataceae (0.06%). The fungal family is Euphorbiaceae (0.09%), Trichocomaceae (0.2%), and Sordariaceae (0.03%). The predominant family of microbial communities with the highest mean of abundance in DR includes Micromonosporaceae (2.9%) and Pseudonocardiaceae (1.4%). The only archaeal family present in the diseased rhizosphere is Methanosarcinaceae (0.12%). The only fungal family in the diseased rhizosphere is Nectriaceae (0.13%). The predominant family of microbial communities with the highest mean of abundance in BR includes the bacterial family, namely Sphingomonadaceae (4.0%) and Xanthomonadaceae (2.1%). The archaea family includes Halobacteriaceae (0.24%) and Thermococcaceae (0.05%) (Figure 5).
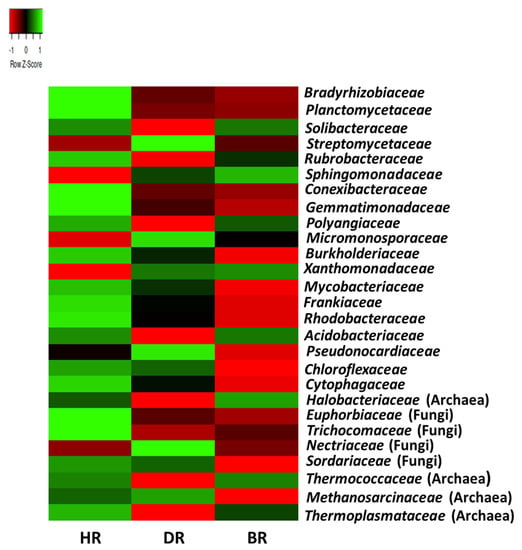
Figure 5.
The family heatmap of soil microorganisms affiliated with the tomato plants. Each plot with unique color functioned as a saturation gradient employing the mean value with a z-score. HR = rhizosphere soil of healthy tomato plant; DR = rhizosphere soil of diseased tomato plant; and BR = bulk soil obtained from the soil without tomato plantation.
The prevalent genus of microbial communities with highest mean of abundance in HR include the bacterial genus Rubrobacter (5.5%), Candidatus solibacter (4.7%), Conexibacter (4.4%), Geodermatophilus (3.3%), Gemmatimonas (3.1%), Frankia (2.2%), Methylobacterium (1.8%), Burkholderia (1.5%), Candidatus koribacter (1.4%), Planctomyces (1.3%), Acidobacterium (1.0%), and Anaeromyxobacter (1.0%). The genus archaea are Haloterrigena (0.03%), and fungi genera present in the healthy rhizosphere are Pyrenophora (0.05%), Sordaria (0.03%), and Penicillium (0.03%). The prevalent genera of microbial communities with the highest mean of abundance in DR includes bacterial genera Streptomyces (5.2%), Bradyrhizobium (1.74%), Pseudomonas (0.8%), and Bacillus (0.56%). The fungal genera found in the diseased rhizosphere were Nectria (0.06%), Gibberella (0.05%), Aspergillus (0.06%), and Fusarium (0.001%). The only archaeal genus was Methanocella (Archaea) (0.03%), found in the diseased tomato rhizosphere. The prevalent genera of microbial communities with the highest mean of abundance in BR include bacterial genera Mycobacterium (2.8%) and Sorangium (2.7%), while Methanosarcina (0.1%) and Candida (0.02%) are the only archaea and fungus in the BR (Figure 6).
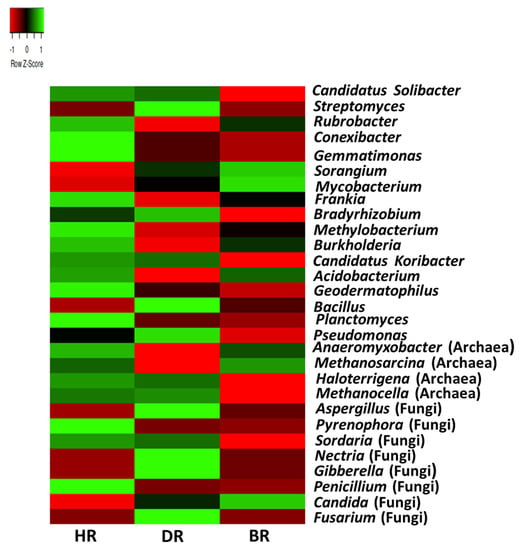
Figure 6.
The genus heatmap of soil microorganisms affiliated with the tomato plants. Each plot with unique color functioned as a saturation gradient employing the mean value with a z-score. HR = rhizosphere soil of healthy tomato plant; DR = rhizosphere soil of diseased tomato plant; and BR = bulk soil obtained from the soil without tomato plantation.
3.3. Alpha (α) and Beta (β) Diversity
The diversity assessment carried out using Shannon, Simpson, and Evenness indexes calculated for the soil microbial communities do not differ significantly (p > 0.05) across the tomato rhizosphere and bulk soil. We used the PCoA graph in analyzing the soil microbial community composition in the rhizosphere and bulk soil sample, which demonstrated that samples from HR differ significantly from DR and BR (Figure 7, Table 1). ANOSIM demonstrated that the differences in the diversity of the soil microbial communities across the soil sample differed significantly (p = 0.01 and R = 0.67).
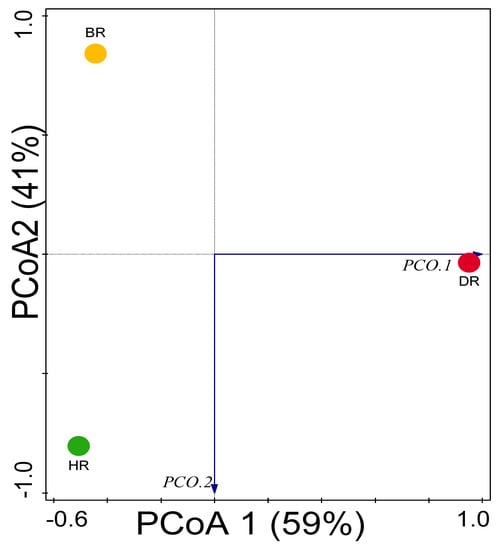
Figure 7.
PCoA graph of the community of microbes in the soil based on Bray–Curtis differences. HR = rhizosphere soil of healthy tomato plant; DR = rhizosphere soil of diseased tomato plant; and BR = bulk soil obtained from the soil without tomato plantation.

Table 1.
Diversity and evenness of microbial communities present in the rhizosphere and bulk soil samples.
3.4. Major Metabolic Pathways of Microbial Communities Associated with the Tomato Rhizosphere and Bulk Soils
Employing level 3 of the SEED subsystem, the metagenome study of tomato rhizosphere and bulk soil revealed the microbial functions. The functional diversity of the microbial metagenomes of HR, DR, and BR sites were compared. The major metabolic pathways identified are linked to functional metabolism such as carbohydrate metabolism, nitrogen metabolism, sulfur metabolism, phosphorus metabolism, and secondary metabolism were greater HR compared to DR and BR as observed in Table 2.

Table 2.
Relative abundance of important functional pathways obtained from the metagenomic sequencing of microbiomes present in tomato rhizosphere and bulk soil employing SEED subsystem.
4. Discussion
The diversity and composition of microbes in the soil contribute special functions in conserving soil health, process, and quality [20]. Microbial communities, including bacteria, fungi, and archaea, are present in rhizosphere soils of tomatoes [3]. However, not all of these microorganisms are known to protect crop plants from phytopathogens. Some microbes play an essential role in enhancing crop productivity and improving soil fertility [52].
The shotgun sequencing procedure used in this research helped surmount restrictions and obtained thousand counts sequences in other to confirm the diversity of microbes in the soil samples. However, leveraging on the strength of this novel sequencing method, we profile the composition of the microbial community present in the rhizospheric soils of the healthy, diseased tomato plant and bulk soil. Aside from assessing the diversity of the rhizosphere microbiome using the plant’s health status, other factors such as soil types and secretion of root exudates have been considered prominent factors contributing to the diversity of microbial species in the tomato plant rhizosphere [53,54,55].
Prominent bacteria phyla observed in this study include Proteobacteria, Actinobacteria, Planctomycetes, Chloroflexi, Verrucomicrobia, Cyanobacteria, Ntrospirae, Deinococcus-Thermus, Chlorobi, and Synergistetes, which were more abundant in HR. The phyla of bacteria, as mentioned, have been previously reported to improve the plant’s health and also confirm to promote the tomato plant’s growth and cultivation [22,23]. Bacteroidetes, Gemmatimonadetes, and Thermotoga were observed to be dominant in the DR. This finding is in line with the study of Wei, et al. [20], which reported that Bacteroidetes, Gemmatimonadetes, Nitrospirae, and Cyanobacteria were increased to varying degrees in the rusty root-affected Panax ginseng soils compared with those in healthy soil. However, Acidobacteria, Firmicutes, unclassified (derived bacteria), Spirochaetes, and Aquificae were observed in the BR. This result is in line with the study of Nessner Kavamura, et al. [56] that reported an abundance of similar phyla for bulk soil while comparing the study of the structure of bulk soil and bacterial diversity in the rhizosphere of a cactus.
Moreover, the dominant bacteria genera in HR are: Rubrobacter, Candidatus solibacter, Conexibacter, Gemmatimonas, Frankia, Methylobacterium, Burkholderia, Candidatus koribacter, Planctomyces, Acidobacterium, Anaeromyxobacter, and Geodermatophilus. Pang, et al. [57] also reported these genera in assessing rhizosphere soil associated with the sugarcane plant. We observed Streptomyces, Bradyrhizobium, Pseudomonas, and Bacillus in DR, which was in line with the report of Wei, et al. [20] and identified similar genera in the rhizosphere soil of rusty Panax ginseng.
Furthermore, the dominant archaeal phyla in HR samples are Euryarchaeota, Crenarchaeota, and Thaumarchaeota. This result coincides with earlier studies, where archaea were reported to improve plant health and form a beneficial relationship in ecosystems [58]. The production of root exudates by tomato plants was also enhanced by archaea, as reported by [59]. Main fungal phyla also present in the HR samples are Ascomycota and Basidiomycota. Our result aligns with recent research on similar phyla obtained in tomato, maize, and pepper rhizosphere soil [60]. The fungi, Ascomycota, and Basidiomycota benefit the plant’s health. They have also been a favorite biological agent in controlling Fusarium wilt (Fol diseases) of tomatoes and other crops [22,61,62].
The fungi genera present in the HR are Pyrenophora, Sordaria, and Penicillium. Omomowo, et al. [63] reported Penicillium chrysogenum and Trichoderma viride for their ability to control the spoilage organism and phytopathogen on sweet oranges. Nectria, Gibberella, Aspergillus, and Fusarium are fungi genera found in DR. These fungi have previously been reported to destroy fruits and vegetable crops such as tomato, pepper, potato, banana, and orange [64]. We also found that genus archaea Haloterrigena is in the HR, which agrees with Ayangbenro and Babalola [65], who reported the function of growth-promoting archaea in soil health promotion.
Moreover, as observed on the PCA graph, the abundance is much celebrated in healthy rhizosphere soil with 59% variance (Figure 2). The bars occupied by each microbe explained the sequence composition concerning the phylum. Utilizing this method makes it possible to know where the microbial diversity is higher in the rhizosphere soil sample compared to bulk soil. It has been reported that there are more microbial communities in the rhizosphere of tomatoes [22,40], while we observed fewer microbial communities in bulk soil [37,66].
The microbial community abundance in the healthy rhizosphere of S. lycopersicum can boost the production of its fruits for sustainable agriculture [37]. Higher microbial diversity in HR illustrated the role of plant growth-promoting rhizobacteria (PGPR), and plant growth-promoting fungi (PGPF) in the healthy rhizosphere of tomatoes and other crops. This coincides with the study of PGPR as reported by Abbasi, et al. [67] and the inhibition of Pseudomonas syringae by PGPF as reported by Elsharkawy, et al. [68], which act as biological control agents, thereby improving the wellbeing of tomatoes’ production. According to Kong, et al. [69], the microbiota has harmed tomato plant growth, limiting its production and resulting in a hungry populace. Yet, others reported how microbiota functioned in the soil to inhibit the potentiality of the soil pathogens on S. lycopersicum [18,70].
Alpha diversity revealed that microbial communities and functions were not significantly different (p-value > 0.05) between the soil samples (HR, DR, and BR) employed in this study. Shannon, Simpson, and Evenness indices were confirmed at phylum and genus levels, revealing no significant difference (p > 0.05) in the soil, as also observed in class, order and family level. Microbial communities were evenly distributed and more abundant in the healthy rhizosphere of tomatoes (HR) than compared with the diseased rhizosphere of tomatoes (DR) and bulk soil (BR) (Table 1). However, our results confirmed our hypothesis: the abundance and diversity of rhizosphere microbial communities are greater in the healthy rhizosphere (HR) than in the diseased rhizosphere (DR). It primarily indicates that a plant’s health status affects the microbial diversity of rhizosphere microbiota, as reported in this study. The PCoA graph demonstrated that the diversity of microbes in HR is higher than DR and BR, as illustrated in Figure 7. This Euclidean dissimilarity matrix-based plot explained the distribution of the microbial communities in healthy rhizosphere soil of tomato plants compared to diseased rhizosphere and bulk soil.
The major metabolic pathways obtained for the microbiome in the rhizosphere of the tomato were linked to carbohydrate metabolism, nitrogen metabolism, sulfur metabolism, phosphorus metabolism, and secondary metabolism. Each site contains special functional categories from the study represented as HR >DR >BR. The result obtained from this study coincides with the research of Zheng, et al. [71] on the bacterial community structure and their functions on the rhizosphere of continuous tomato cropping in the greenhouse. The result was different when compared to the study of Taffner, et al. [72] that demonstrated results obtained from a metagenomic analysis of archaeal functional categories employing shotgun and 16S rRNA in the phyllosphere and rhizosphere soil of Eruca sativa.
The pathways linked to carbohydrate metabolism that includes Trehalose biosynthesis, TCA Cycle, Glycolysis and Gluconeogenesis, Glycogen Metabolism, Glycerol, and Glycerol-3-phosphate Uptake and Utilization are found dominating the HR (Table 2). Among the macro-elements biologically required for tomato plant growth is nitrogen, which is generally available in the atmosphere. Tomato plants fix nitrogen into the soil for metabolic activities by nitrogen-fixing bacteria in the form of fixed molecules such as amino acids, urea, nitrate, and ammonia [73]. Sufficient N in the soil improves abundant tomato plant produce and other physiological characteristics [74]. This study revealed six (6) predominant pathways of nitrogen metabolism, which includes Allantoin Utilization, Ammonia assimilation, Nitrate and nitrite ammonification, Nitric oxide synthase, Nitrogen fixation, and Denitrification, all abundant in HR. Allantoin is made up of nitrogenous components that are derived from purine ureides. These nitrogenous components were transported through root nodules to the aerial of the plant. Allantoin usage in plants contributes to the nitrogen cycle, influences the purpose of nutrient cycling and endeavors toleration of various stresses in nature [75]. Allantoin utilization is well represented in our metagenome study and has been reported to play a major role in nutrient cycling and stress tolerance [76]. Oher pathways involved in nitrogen metabolism are nitrate and nitrite ammonification, nitric oxide (NO) synthase, nitrogen fixation, and denitrification. According to Mendes, et al. [77], the aforementioned nitrogenous pathways were also reported to be presented in the rhizosphere and bulk soil of soya beans, which also agrees with our report, as obtained for the HR samples.
Pathways involved in sulfur metabolism from the microbiomes in the healthy rhizosphere soil of tomato plant was created at lower levels with special metabolic processes, such as inorganic sulfur assimilation, sulfur oxidation, alkanesulfonates’ utilization, and alkanesulfonate assimilation, in this study (Table 2). This result coincides with the research conducted by Santana, et al. [78] that reveals how microbial communities in the soil perform notable function as for enzymatic sulfur metabolism. Phosphorus (P) is one of the macro-elements required for tomato plant growth [79]. P is required as form of biomolecule involved in energy metabolism, namely, nucleic acids, ATP, phospholipids, and pyrophosphate. This element is frequently required in the soil and can be absorbed by crop plants, thereby improving the extensive application of phosphorus fertilizer for the development of the plant [80]. The pathways of phosphorus metabolism abundantly observed in the HR are six (6) pathways and they include Phosphate metabolism, Pentose phosphate pathway, P-uptake (cyanobacteria), Alkylphosphonate utilization, Phosphonate metabolism, and Phosphoenolpyruvate phosphomutase. A similar result was obtained by Akinola, et al. [81] that reported the abundance of P uptake by cyanobacteria, alkylphosphonate, alkylphosphonate utilization, and phosphate and phosphonate metabolism in the rhizosphere soil of maize plant. Other functions discussed above and observed in the HR sites, demonstrated an improved plant P-availability through mineralization, reduced soil pH to lower level, and P-solubilization.
The pathways involved in secondary metabolism include, auxin biosynthesis, phytoalexin biosynthesis, alkaloid biosynthesis from L-lysine, tannin biosynthesis, heme, and siroheme biosynthesis, and pyridoxin (Vitamin B6) biosynthesis dominated the HR (Table 2). This is because of adequate domination of beneficial plant growth-promoting (PGP) organisms in the soil that frequently produces plant hormones. Plant hormones are specially required for plant growth promotion via root and shoot elongation [82].
5. Conclusions
This study investigated the rhizosphere microbial communities and nutrient pathways of healthy and diseased tomato plants and bulk soil, employing shotgun metagenomic sequencing. Our results demonstrated that the tomato plant’s health status affects its rhizosphere microbial community. We also proved that microbial communities were more abundant in the rhizosphere of the healthy tomato plant than other sites, thereby contributing a positive impact on its growth and development of functional pathways produced. The functions of these microbial communities at the root of the tomatoes will promote beneficial interactions between rhizosphere microbiota and the root of tomato plants. Therefore, they can improve plant health, enhance the production of tomato fruits, and improve microbiome engineering. In the future, it would be interesting to explore the metabolic abilities of the microbial community in the rhizosphere of the tomato plant through metabolomic analysis.
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/horticulturae8050404/s1, Table S1: Analysis of sequenced data and diversity evaluation of the shotgun metagenome from the rhizosphere and bulk soil of the tomato plant; Table S2: Evaluation of major phyla in the rhizosphere of the tomato plant, and bulk soil (BR); Figure S1: The abundance and species richness of the microorganisms in the rhizosphere were investigated through rarefaction analysis with MG-RAST.
Author Contributions
A.A.A. did the laboratory work, analyzed and interpreted the results with the help of A.E.F., and wrote the first draft of the manuscript. O.O.B. supervised all the listed coauthors, helped structure the research, verified the analytical methods, and edited/commented on the document at all stages. All authors have read and agreed to the published version of the manuscript.
Funding
The National Research Foundation of South Africa grants (UID123634 and UID132595).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are publicly available on the NCBI database with bio-project number PRJNA766489 for all the samples and controls.
Acknowledgments
We acknowledge the National Research Foundation, South Africa grants (UID123634 and UID132595) to O.O.B. that support research in our laboratory.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Liu, Y.; Zhu, A.; Tan, H.; Cao, L.; Zhang, R. Engineering Banana Endosphere Microbiome to improve Fusarium wilt resistance in Banana. Microbiome 2019, 7, 74. [Google Scholar] [CrossRef] [PubMed]
- Tian, L.; Lin, X.; Tian, J.; Ji, L.; Chen, Y.; Tran, L.-S.P.; Tian, C. Research Advances of Beneficial Microbiota Associated with Crop Plants. Int. J. Mol. Sci. 2020, 21, 1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Babalola, O.O.; Adedayo, A.A.; Fadiji, A.E. Metagenomic Survey of Tomato Rhizosphere Microbiome Using the Shotgun Approach. Microbiol. Resour. Announc. 2022, 11, e01131-21. [Google Scholar] [CrossRef]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.; Jiao, X.-Y.; Chen, Q.; Wu, A.-L.; Zheng, Y.; Lin, Y.-X.; He, J.-Z.; Hu, H.-W. Microbial communities in crop phyllosphere and root endosphere are more resistant than soil microbiota to fertilization. Soil Biol. Biochem. 2021, 153, 108113. [Google Scholar] [CrossRef]
- Vaishnav, A.; Shukla, A.K.; Sharma, A.; Kumar, R.; Choudhary, D.K. Endophytic Bacteria in Plant Salt Stress Tolerance: Current and Future Prospects. J. Plant Growth Regul. 2019, 38, 650–668. [Google Scholar] [CrossRef]
- de Vries Franciska, T.; Griffiths Rob, I.; Knight Christopher, G.; Nicolitch, O.; Williams, A. Harnessing rhizosphere microbiomes for drought-resilient crop production. Science 2020, 368, 270–274. [Google Scholar] [CrossRef]
- Porter, S.S.; Bantay, R.; Friel, C.A.; Garoutte, A.; Gdanetz, K.; Ibarreta, K.; Moore, B.M.; Shetty, P.; Siler, E.; Friesen, M.L. Beneficial microbes ameliorate abiotic and biotic sources of stress on plants. Funct. Ecol. 2020, 34, 2075–2086. [Google Scholar] [CrossRef] [Green Version]
- Lyu, D.; Backer, R.; Robinson, W.G.; Smith, D.L. Plant Growth-Promoting Rhizobacteria for Cannabis Production: Yield, Cannabinoid Profile and Disease Resistance. Front. Microbiol. 2019, 10, 1761. [Google Scholar] [CrossRef] [Green Version]
- Konappa, N.; Krishnamurthy, S.; Arakere, U.C.; Chowdappa, S.; Ramachandrappa, N.S. Efficacy of indigenous plant growth-promoting rhizobacteria and Trichoderma strains in eliciting resistance against bacterial wilt in a tomato. Egypt. J. Biol. Pest Control 2020, 30, 106. [Google Scholar] [CrossRef]
- Saia, S.; Aissa, E.; Luziatelli, F.; Ruzzi, M.; Colla, G.; Ficca, A.G.; Cardarelli, M.; Rouphael, Y. Growth-promoting bacteria and arbuscular mycorrhizal fungi differentially benefit tomato and corn depending upon the supplied form of phosphorus. Mycorrhiza 2020, 30, 133–147. [Google Scholar] [CrossRef] [PubMed]
- Alori, E.T.; Babalola, O.O. Microbial Inoculants for Improving Crop Quality and Human Health in Africa. Front. Microbiol. 2018, 9, 2213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dukare, A.; Paul, S. Biological control of Fusarium wilt and growth promotion in pigeon pea (Cajanus cajan) by antagonistic rhizobacteria, displaying multiple modes of pathogen inhibition. Rhizosphere 2021, 17, 100278. [Google Scholar] [CrossRef]
- Sahu, P.K.; Singh, D.P.; Prabha, R.; Meena, K.K.; Abhilash, P.C. Connecting microbial capabilities with the soil and plant health: Options for agricultural sustainability. Ecol. Indic. 2019, 105, 601–612. [Google Scholar] [CrossRef]
- Fones, H.N.; Bebber, D.P.; Chaloner, T.M.; Kay, W.T.; Steinberg, G.; Gurr, S.J. Threats to global food security from emerging fungal and oomycete crop pathogens. Nat. Food 2020, 1, 332–342. [Google Scholar] [CrossRef]
- Wei, Z.; Gu, Y.; Friman, V.-P.; Kowalchuk, G.A.; Xu, Y.; Shen, Q.; Jousset, A. Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 2019, 5, eaaw0759. [Google Scholar] [CrossRef] [Green Version]
- Soumare, A.; Boubekri, K.; Lyamlouli, K.; Hafidi, M.; Ouhdouch, Y.; Kouisni, L. From Isolation of Phosphate Solubilizing Microbes to Their Formulation and Use as Biofertilizers: Status and Needs. Front. Bioeng. Biotechnol. 2020, 7, 425. [Google Scholar] [CrossRef]
- De Corato, U. Soil microbiota manipulation and its role in suppressing soil-borne plant pathogens in organic farming systems under the light of microbiome-assisted strategies. Chem. Biol. Technol. Agric. 2020, 7, 17. [Google Scholar] [CrossRef]
- Adeleke, B.S.; Babalola, O.O. The endosphere microbial communities, a great promise in agriculture. Int. Microbiol 2021, 24, 1–17. [Google Scholar] [CrossRef]
- Wei, X.; Wang, X.; Cao, P.; Gao, Z.; Chen, A.J.; Han, J. Microbial Community Changes in the Rhizosphere Soil of Healthy and Rusty Panax ginseng and Discovery of Pivotal Fungal Genera Associated with Rusty Roots. BioMed Res. Int. 2020, 2020, 8018525. [Google Scholar] [CrossRef] [Green Version]
- Wei, F.; Feng, H.; Zhang, D.; Feng, Z.; Zhao, L.; Zhang, Y.; Deakin, G.; Peng, J.; Zhu, H.; Xu, X. Composition of Rhizosphere Microbial Communities Associated With Healthy and Verticillium Wilt Diseased Cotton Plants. Front. Microbiol. 2021, 12, 618169. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Wang, J.-T.; Wang, W.-H.; Tsui, C.K.; Cai, L. Changes in Bacterial and Fungal Microbiomes Associated with Tomatoes of Healthy and Infected by Fusarium oxysporum f. sp. lycopersici. Microb. Ecol. 2021, 81, 1004–1017. [Google Scholar] [CrossRef] [PubMed]
- Dong, C.-J.; Wang, L.-L.; Li, Q.; Shang, Q.-M. Bacterial communities in the rhizosphere, phyllosphere and endosphere of tomato plants. PLoS ONE 2019, 14, e0223847. [Google Scholar] [CrossRef] [PubMed]
- Ajilogba, C.F.; Babalola, O.O. Integrated Management Strategies for Tomato Fusarium Wilt. Biocontrol Sci. 2013, 18, 117–127. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ajilogba, C.F.; Babalola, O.O.; Ahmad, F. Antagonistic Effects of Bacillus species in Biocontrol of Tomato Fusarium Wilt. Stud. Ethno-Med. 2013, 7, 205–216. [Google Scholar] [CrossRef]
- Ahmad, F.; Babalola, O.O.; Tak, H.I. Potential of MALDI-TOF mass spectrometry as a rapid detection technique in plant pathology: Identification of plant-associated microorganisms. Anal. Bioanal. Chem 2012, 404, 1247–1255. [Google Scholar] [CrossRef]
- Akhtar, K.P.; Ullah, N.; Saleem, M.Y.; Iqbal, Q.; Asghar, M.; Khan, A.R. Evaluation of tomato genotypes for early blight disease resistance caused by Alternaria solani in Pakistan. Plant Pathol. 2019, 101, 1159–1170. [Google Scholar] [CrossRef]
- Mazumdar, P.; Singh, P.; Kethiravan, D.; Ramathani, I.; Ramakrishnan, N. Late blight in tomato: Insights into the pathogenesis of the aggressive pathogen Phytophthora infestans and future research priorities. Planta 2021, 253, 119. [Google Scholar] [CrossRef]
- Zhang, Y.; Hu, A.; Zhou, J.; Zhang, W.; Li, P. Comparison of bacterial communities in soil samples with and without tomato bacterial wilt caused by Ralstonia solanacearum species complex. BMC Microbiol. 2020, 20, 89. [Google Scholar] [CrossRef]
- Sun, G.; Feng, C.; Guo, J.; Zhang, A.; Xu, Y.; Wang, Y.; Day, B.; Ma, Q. The tomato Arp2/3 complex is required for resistance to the powdery mildew fungus Oidium neolycopersici. Plant Cell Environ. 2019, 42, 2664–2680. [Google Scholar] [CrossRef]
- Khalil Bagy, H.M.M.; Abo-Elyousr, K.A.M. Antibacterial activity of some essential oils on bacterial spot disease of tomato plant caused by Xanthomonas axonopodis pv. vesicatoria. Int. J. Plant Pathol. 2019, 8, 53–61. [Google Scholar] [CrossRef] [Green Version]
- Oh, J.-W.; Chun, S.C.; Chandrasekaran, M. Preparation and In Vitro Characterization of Chitosan Nanoparticles and Their Broad-Spectrum Antifungal Action Compared to Antibacterial Activities against Phytopathogens of Tomato. Agronomy 2019, 9, 21. [Google Scholar] [CrossRef] [Green Version]
- Safari, Z.S.; Ding, P.; Nakasha, J.J.; Yusoff, S.F. Controlling Fusarium oxysporum Tomato Fruit Rot under Tropical Condition Using Both Chitosan and Vanillin. Coatings 2021, 11, 367. [Google Scholar] [CrossRef]
- Istifadah, N.; Firman, A.R.; Desiana, M.F. Effectiveness of compost and microbial-enriched compost to suppress powdery mildew and early blight diseases in tomato. J. Anim. Plant Sci. 2020, 30, 377–383. [Google Scholar] [CrossRef] [Green Version]
- Jahnová, J.; Činčalová, L.; Sedlářová, M.; Jedelská, T.; Sekaninová, J.; Mieslerová, B.; Luhová, L.; Barroso, J.B.; Petřivalský, M. Differential modulation of S-nitrosoglutathione reductase and reactive nitrogen species in wild and cultivated tomato genotypes during development and powdery mildew infection. Plant Physiol. Biochem. 2020, 155, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Fadiji, A.E.; Babalola, O.O. Metagenomics methods for the study of plant-associated microbial communities: A review. J. Microbiol. Methods 2020, 170, 105860. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.A.; Kim, Y.; Kim, J.M.; Chu, B.; Joa, J.-H.; Sang, M.K.; Song, J.; Weon, H.-Y. A preliminary examination of bacterial, archaeal, and fungal communities inhabiting different rhizocompartments of tomato plants under real-world environments. Sci. Rep. 2019, 9, 9300. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Xu, X.; Huo, Y.; Xiao, Y. Trichoderma-Inoculation and Mowing Synergistically Altered Soil Available Nutrients, Rhizosphere Chemical Compounds and Soil Microbial Community, Potentially Driving Alfalfa Growth. Front. Microbiol. 2019, 9, 3241. [Google Scholar] [CrossRef]
- Liu, L.; Huang, X.; Zhang, J.; Cai, Z.; Jiang, K.; Chang, Y. Deciphering the relative importance of soil and plant traits on the development of rhizosphere microbial communities. Soil Biol. Biochem. 2020, 148, 107909. [Google Scholar] [CrossRef]
- López, S.M.Y.; Pastorino, G.N.; Fernández-González, A.J.; Franco, M.E.E.; Fernández-López, M.; Balatti, P.A. The endosphere bacteriome of diseased and healthy tomato plants. Arch. Microbiol. 2020, 202, 2629–2642. [Google Scholar] [CrossRef]
- Zheng, X.; Liu, B.; Zhu, Y.; Wang, J.; Zhang, H.; Wang, Z. Bacterial community diversity associated with the severity of bacterial wilt disease in tomato fields in Southeast China. Can. J. Microbiol. 2019, 65, 538–549. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.-M.; Kong, H.G.; Song, G.C.; Ryu, C.-M. Disruption of Firmicutes and Actinobacteria abundance in tomato rhizosphere causes the incidence of bacterial wilt disease. ISME J. 2021, 15, 330–347. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Xiong, W.; Zhang, R.; Hang, X.; Wang, D.; Li, R.; Shen, Q. Continuous application of different organic additives can suppress tomato disease by inducing the healthy rhizospheric microbiota through alterations to the bulk soil microflora. Plant Soil 2018, 423, 229–240. [Google Scholar] [CrossRef] [Green Version]
- Su, L.; Zhang, L.; Nie, D.; Kuramae, E.E.; Shen, B.; Shen, Q. Bacterial Tomato Pathogen Ralstonia solanacearum Invasion Modulates Rhizosphere Compounds and Facilitates the Cascade Effect of Fungal Pathogen Fusarium solani. Microorganisms 2020, 8, 806. [Google Scholar] [CrossRef]
- Meyer, F.; Paarmann, D.; D’Souza, M.; Olson, R.; Glass, E.; Kubal, M.; Paczian, T.; Rodriguez, A.; Stevens, R.; Wilke, A.; et al. The metagenomics RAST server—A public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinform. 2008, 9, 386. [Google Scholar] [CrossRef] [Green Version]
- Kent, W.J. BLAT—The BLAST-Like Alignment Tool. Genome Res. 2002, 12, 656–664. [Google Scholar] [CrossRef] [Green Version]
- Wilke, A.; Harrison, T.; Wilkening, J.; Field, D.; Glass, E.M.; Kyrpides, N.; Mavrommatis, K.; Meyer, F. The M5nr: A novel non-redundant database containing protein sequences and annotations from multiple sources and associated tools. BMC Bioinform. 2012, 13, 141. [Google Scholar] [CrossRef] [Green Version]
- Mitra, S.; Rupek, P.; Richter, D.C.; Urich, T.; Gilbert, J.A.; Meyer, F.; Wilke, A.; Huson, D.H. Functional analysis of metagenomes and metatranscriptomes using SEED and KEGG. BMC Bioinform. 2011, 12, S21. [Google Scholar] [CrossRef] [Green Version]
- Khomtchouk, B.B.; Hennessy, J.R.; Wahlestedt, C. Shinyheatmap: Ultra fast low memory heatmap web interface for big data genomics. PLoS ONE 2017, 12, e0176334. [Google Scholar] [CrossRef] [Green Version]
- Hamel, C.; Gan, Y.; Sokolski, S.; Bainard, L.D. High frequency cropping of pulses modifies soil nitrogen level and the rhizosphere bacterial microbiome in 4-year rotation systems of the semiarid prairie. Agric. Ecosyst. Environ. Appl. Soil Ecol. 2018, 126, 47–56. [Google Scholar] [CrossRef]
- Carrell, A.; Frank, C. Bacterial endophyte communities in the foliage of coast redwood and giant sequoia. Front. Microbiol. 2015, 6, 1008. [Google Scholar] [CrossRef] [PubMed]
- Raimi, A.; Adeleke, R.; Roopnarain, A. Soil fertility challenges and Biofertiliser as a viable alternative for increasing smallholder farmer crop productivity in sub-Saharan Africa. Cogent Food Agric. 2017, 3, 1400933. [Google Scholar] [CrossRef]
- Rolfe, S.A.; Griffiths, J.; Ton, J. Crying out for help with root exudates: Adaptive mechanisms by which stressed plants assemble health-promoting soil microbiomes. Curr. Opin. Microbiol. 2019, 49, 73–82. [Google Scholar] [CrossRef] [PubMed]
- Tang, M.; Li, L.; Wang, X.; You, J.; Li, J.; Chen, X. Elevational is the main factor controlling the soil microbial community structure in alpine tundra of the Changbai Mountain. Sci. Rep. 2020, 10, 12442. [Google Scholar] [CrossRef]
- Fadiji, A.E.; Kanu, J.O.; Babalola, O.O. Metagenomic profiling of rhizosphere microbial community structure and diversity associated with maize plant as affected by cropping systems. Int. Microbiol. 2021, 24, 325–335. [Google Scholar] [CrossRef]
- Nessner Kavamura, V.; Taketani, R.G.; Lançoni, M.D.; Andreote, F.D.; Mendes, R.; Soares De Melo, I. Water Regime Influences Bulk Soil and Rhizosphere of Cereus jamacaru Bacterial Communities in the Brazilian Caatinga Biome. PLoS ONE 2013, 8, e73606. [Google Scholar] [CrossRef]
- Pang, Z.; Dong, F.; Liu, Q.; Lin, W.; Hu, C.; Yuan, Z. Soil Metagenomics Reveals Effects of Continuous Sugarcane Cropping on the Structure and Functional Pathway of Rhizospheric Microbial Community. Front. Microbiol. 2021, 12, 369. [Google Scholar] [CrossRef]
- Jung, J.; Kim, J.-S.; Taffner, J.; Berg, G.; Ryu, C.-M. Archaea, tiny helpers of land plants. Comput. Struct. Biotechnol. J. 2020, 18, 2494–2500. [Google Scholar] [CrossRef]
- Hussain, H.I.; Kasinadhuni, N.; Arioli, T. The effect of seaweed extract on tomato plant growth, productivity and soil. J. Appl. Phycol. 2021, 33, 1305–1314. [Google Scholar] [CrossRef]
- Wu, A.-L.; Jiao, X.-Y.; Fan, F.-F.; Wang, J.-S.; Guo, J.; Dong, E.-W.; Wang, L.-G.; Shen, X.-M. Effect of continuous sorghum cropping on the rhizosphere microbial community and the role of Bacillus amyloliquefaciens in altering the microbial composition. Plant Growth Regul. 2019, 89, 299–308. [Google Scholar] [CrossRef]
- Enebe, M.C.; Babalola, O.O. Effects of inorganic and organic treatments on the microbial community of maize rhizosphere by a shotgun metagenomics approach. Ann. Microbiol. 2020, 70, 49. [Google Scholar] [CrossRef]
- Dong, C.; Wang, L.; Li, Q.; Shang, Q. Epiphytic and Endophytic Fungal Communities of Tomato Plants. Hortic. Plant J. 2021, 7, 38–48. [Google Scholar] [CrossRef]
- Omomowo, I.O.; Adedayo, A.A.; Omomowo, O.I. Biocontrol potential of rhizospheric fungi from Moringa oleifera, their phytochemicals and secondary metabolite assessment against spoilage fungi of sweet orange (Citrus sinensis). Asian J. Appl. Sci. 2020, 8, 1–14. [Google Scholar] [CrossRef]
- Rampersad, S. Pathogenomics and Management of Fusarium Diseases in Plants. Pathogens 2020, 9, 340. [Google Scholar] [CrossRef] [PubMed]
- Ayangbenro, A.S.; Babalola, O.O. Reclamation of arid and semi-arid soils: The role of plant growth-promoting archaea and bacteria. Curr. Plant Biol. 2021, 25, 100173. [Google Scholar] [CrossRef]
- Schmidt, J.E.; Kent, A.D.; Brisson, V.L.; Gaudin, A.C.M. Agricultural management and plant selection interactively affect rhizosphere microbial community structure and nitrogen cycling. Microbiome 2019, 7, 146. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abbasi, S.; Sadeghi, A.; Omidvari, M.; Tahan, V. The stimulators and responsive genes to induce systemic resistance against pathogens: An exclusive focus on tomato as a model plant. Biocatal. Agric. Biotechnol. 2021, 33, 101993. [Google Scholar] [CrossRef]
- Elsharkawy, M.M.; Khedr, A.A.; Mehiar, F.; El-Kady, E.M.; Baazeem, A.; Shimizu, M. Suppression of Pseudomonas syringae pv. tomato infection by rhizosphere fungi. Pest Manag. Sci. 2021, 77, 4350–4356. [Google Scholar] [CrossRef]
- Kong, H.G.; Song, G.C.; Sim, H.-J.; Ryu, C.-M. Achieving similar root microbiota composition in neighbouring plants through airborne signalling. ISME J. 2021, 15, 397–408. [Google Scholar] [CrossRef]
- Nwachukwu, B.C.; Babalola, O.O. Perspectives for sustainable agriculture from the microbiome in plant rhizosphere. Plant Biotechnol. Rep. 2021, 15, 259–278. [Google Scholar] [CrossRef]
- Zheng, X.; Wang, Z.; Zhu, Y.; Wang, J.; Liu, B. Effects of a microbial restoration substrate on plant growth and rhizosphere bacterial community in a continuous tomato cropping greenhouse. Sci. Rep. 2020, 10, 13729. [Google Scholar] [CrossRef] [PubMed]
- Taffner, J.; Cernava, T.; Erlacher, A.; Berg, G. Novel insights into plant-associated archaea and their functioning in arugula (Eruca sativa Mill.). J. Adv. Res. 2019, 19, 39–48. [Google Scholar] [CrossRef] [PubMed]
- Mayak, S.; Tirosh, T.; Glick, B.R. Plant growth-promoting bacteria confer resistance in tomato plants to salt stress. Plant Physiol. Biochem. 2004, 42, 565–572. [Google Scholar] [CrossRef]
- Goswami, L.; Nath, A.; Sutradhar, S.; Bhattacharya, S.S.; Kalamdhad, A.; Vellingiri, K.; Kim, K.-H. Application of drum compost and vermicompost to improve soil health, growth, and yield parameters for tomato and cabbage plants. J. Environ. Manag. 2017, 200, 243–252. [Google Scholar] [CrossRef]
- Bielsa, B.; Hewitt, S.; Reyes-Chin-Wo, S.; Dhingra, A.; Rubio-Cabetas, M.J. Identification of water use efficiency related genes in ‘Garnem’ almond-peach rootstock using time-course transcriptome analysis. PLoS ONE 2018, 13, e0205493. [Google Scholar] [CrossRef]
- Dawood, M.F.A.; Tahjib-Ul-Arif, M.; Sohag, A.A.M.; Abdel Latef, A.A.H.; Ragaey, M.M. Mechanistic Insight of Allantoin in Protecting Tomato Plants Against Ultraviolet C Stress. Plants 2020, 10, 11. [Google Scholar] [CrossRef] [PubMed]
- Mendes, L.W.; Kuramae, E.E.; Navarrete, A.A.; van Veen, J.A.; Tsai, S.M. Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 2014, 8, 1577–1587. [Google Scholar] [CrossRef] [PubMed]
- Santana, M.M.; Dias, T.; Gonzalez, J.M.; Cruz, C. Transformation of organic and inorganic sulfur– adding perspectives to new players in soil and rhizosphere. Soil Biol. Biochem. 2021, 160, 108306. [Google Scholar] [CrossRef]
- Bodale, I.; Mihalache, G.; Achiţei, V.; Teliban, G.-C.; Cazacu, A.; Stoleru, V. Evaluation of the Nutrients Uptake by Tomato Plants in Different Phenological Stages Using an Electrical Conductivity Technique. Agriculture 2021, 11, 292. [Google Scholar] [CrossRef]
- Stigter, K.A.; Plaxton, W.C. Molecular Mechanisms of Phosphorus Metabolism and Transport during Leaf Senescence. Plants 2015, 4, 773–798. [Google Scholar] [CrossRef] [Green Version]
- Akinola, S.; Ayangbenro, A.; Babalola, O. Metagenomic Insight into the Community Structure of Maize-Rhizosphere Bacteria as Predicted by Different Environmental Factors and Their Functioning within Plant Proximity. Microorganisms 2021, 9, 1419. [Google Scholar] [CrossRef] [PubMed]
- Emenecker, R.J.; Strader, L.C. Auxin-Abscisic Acid Interactions in Plant Growth and Development. Biomolecules 2020, 10, 281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).