Abstract
Osmanthus fragrans Lour. is a popular and traditional Chinese decorative plant. Salinity is one of the major abiotic stresses affecting the growth and development of O. fragrans. However, the involvement of the SQUAMOSA PROMOTER BINDING PROTEIN-like (SPL) gene in salt stress response is little understood. To elucidate the role of the OfSPL genes in salt stress resistance, we isolated a candidate gene, OfSPL11, from the O. fragrans genotype ‘Yanhong Gui’. OfSPL11 is a transcriptional activator that is located in the nucleus. OfSPL11 is a salt-inducible gene that is highly expressed in young leaves and shoots, according to tissue-specific expression and external treatment. The promoter activity of OfSPL11 is activated by salt treatments in the leaves of tobacco and callus of O. fragrans. The OfSPL11 transgenic lines exhibited better growth and physiological performance; under salt stress, transgenic lines have a faster germination rate, longer roots, and less leaf withering than the wild type (WT). In addition, OfSPL11 overexpression protected the leaves from oxidative damage by suppressing the accumulation of malondialdehyde (MDA) and reactive oxygen species (ROSs) in Arabidopsis. OfSPL11 overexpression can promote the expression of some genes in response to abiotic stresses, including AtCBL1, AtCOR15A, AtCOR6.6, AtRD29A, AtSOS2 and AtSOS3. Yeast one-hybrid assays and transient expression assays showed that OfZAT12 (homologous to Arabidopsis AtRHL41 gene) specifically binds to the OfSPL11 promoter and positively regulates its expression. This study sheds fresh light on the role of OfSPL11 in enhancing salt tolerance in O. fragrans by promoting growth and reducing oxidative damage.
1. Introduction
Environmental (abiotic) stresses, such as salt, cold, drought, and heat stress, have a negative impact on plant growth and development. Among them, salt stress is a common adverse environmental condition. Salt stress causes salt ion imbalance in plants, which is manifested by dehydration, imbalance of osmotic potential and production of excess reactive oxygen species (ROSs). This seriously affects the ornamental and economic value of plants [1,2,3]. However, plants have evolved diverse molecular, biochemical, physiological, and cellular processes, including modification of antioxidant activities and control of the exclusion of ions, enabling them to survive the potentially harmful effects of salt stress [4,5,6]. Some transcription factors regulate salt stress response by regulating one or more target genes essential for salt stress response. For example, VvSNAT1 enhances salt tolerance by modulating melatonin accumulation, plant growth, and oxidative damage reduction [7]. Heterologous overexpression of the Citrus sinensis CsMYB30 gene improves salt tolerance in Arabidopsis [8].
SPL genes are transcription factors specific to green plants containing a highly conserved SBP structural domain, which contains a nuclear localization signal and two zinc finger structures. The SBP-box genes, SBP1 and SBP2, were the first to be identified in Antirrhinum majus because of their ability to bind specifically to the promoter of the SQUAMOSA gene. Since then, SBP-box genes have been found in major plant taxa as members of relatively big gene families [9,10,11]. SPL genes are a group of transcription factors that are involved in a variety of plant developmental processes [12]. However, only a few SPL genes were found to be involved in abiotic stress responses. In Arabidopsis, AtSPL1 and AtSPL12 enhanced the heat stress resistance [13]. The birch (Betula platyphylla) BpSPL9 gene imparts drought tolerance by improving scavenging of ROSs [14].
Osmanthus fragrans Lour., is a common stiff-leaved evergreen large shrub or small tree belonging to the Oleaceae family. It is among the top ten most cultivated traditional flowers in China. However, abiotic stresses, such as salt, drought, pathogens, high temperature, and heavy metals, frequently affect its growth and development. Plant responses to salt stress are regulated by a number of transcription factors, which have been identified. The genetic mechanisms behind OfSPLs’ protective role in the O. fragrans response to salt stress, however, have yet to be revealed. In this study, we obtained the OfSPL11 transcript from the transcriptome sequence of O. fragrans and cloned its coding sequence. Its phylogenetic relationships, bioinformatics, subcellular localization, transactivation, and tissue-specific expression were analyzed. In addition, we generated several transgenic Arabidopsis lines overexpressing OfSPL11 to investigate its role in response to salt stress in vitro. The transgenic plants were then used to explore the relationship between the function of OfSPL11 and salt tolerance. Validation of the interaction between abiotic stress-regulated genes and OfSPL11 using yeast one-hybrid assays and transient expression assay. This study provides new insights into the regulatory role of OfSPL11 in plant salt resistance.
2. Materials and Methods
2.1. Plant Material and Salt Treatment
During the vegetative growth stage (from 24 April 2020 to 25 May 2020), different organs were collected from the O. fragrans plants, including mature leaves, young leaves, young stems, shoots, flowers, roots, and mature stems. On 28 June 2020, shoots were collected during shoot development. Floral samples were collected during flower opening stage. Twelve O. fragrans plants with consistent growth were transferred to a growth chamber maintained at a relative humidity of 55–65%, a constant temperature of 24 °C, and a 12 h light/12 h dark photoperiod with illumination of 10,000 xl. After one week, 100 mL of 200 mM NaCl solution was used to treat O. fragrans at 3, 6, 12, 24, and 48 h time points under uniform environmental conditions. The young leaves from these plants were collected. All samples were quickly placed in liquid nitrogen and then stored at −80 °C for RNA extractions. Total RNA was extracted using the RNAprep Pure Plant Plus Kit (Tiangen, Beijing, China). T3 transgenic lines and wild-type (ecotype Columbia-0, A. thaliana) were cultivated at 23 °C, 65% relative humidity, and under long-day (16 h light and 8 h dark) conditions. Tobacco (Nicotiana benthamiana) was grown at a temperature of 25 °C, a relative humidity of 70%, and under long-day conditions (12 h of light and 12 h of darkness).
2.2. Gene Cloning and Bioinformatics Analysis
The coding sequence (CDS) from the cDNA of O. fragrans was amplified using two primers (OfSPL11-F and OfSPL11-R). Using the chromosome walking method, two pairs of gene primers (OfSPL11-GSP1, OfSPL11-GSP2, AP1, and AP2) (Table S1) were utilized to employ Taq DNA polymerase (TaKaRa Biotechnology, Dalian, China) to amplify the anticipated OfSPL11 promoter from the DNA library. The PlantCARE program was used to analyze the cis-elements of the OfSPL11 promoter. The theoretical pI, molecular weight, and atoms number of the translated protein were determined using ExPASy (http://web.expasy.org, accessed on 24 March 2021). MEGA6.0 software was used to construct a phylogenetic tree using the neighbor-joining method. Secondary structure predictions were conducted on the PRABI (http://www.prabi.fr/spip.php?page=sommaire, accessed on 26 March 2021) protein structure prediction server. The softberry (www.softberry.com, accessed on 11 April 2021) website is able to predict gene subcellular localization.
2.3. Subcellular Localization and Transcriptional Autoactivation
The OfSPL11 CDS was ligated into the pORER4-GFP vector (Tsingke, Hangzhou, China). The heat shock approach was used to introduce the OfSPL11-GFP expression plasmid into the Agrobacterium tumefaciens strain GV3101. Subsequently, the Agrobacterium suspension was injected into the abaxial surface of four-week-old tobacco (Nicotiana benthamiana) leaves. Subcellular localization of OfSPL11-GFP was observed three days after transient expression, using a laser confocal microscope (Zeiss LSM 880, Germany). The coding regions of OfSPL11 were ligated into the empty vector pGBKT7. The resulting pGBKT7 vector and plasmid pGBKT7-OfSPL11 were transformed into yeast AH109 cells (Saccharomyces cerevisiae). To observe yeast growth, the cells were streaked on SD/-Trp and SD/-Trp-Ade-His-Leu + X-α-Gal screening mediums and incubated at 30 °C for 3–4 days.
2.4. Transient Transformation in Leaves of Tobacco and Callus of O. fragrans
The promoter of OfSPL11 was ligated into the pGreen0800II-LUC vector. The heat shock method was used to transform fusion vector OfSPL11pro:LUC into A. tumefaciens GV3101 (adding pSoup) and then the leaves of tobacco plants were infiltrated. After 1 day of dark culture, the same leaves of tobacco were sprayed with different concentrations of NaCl (0 mM and 200 mM). Additionally, the tobaccos were then transferred to normal culture conditions with 12 h/12 h of light/dark at 25 °C for 2 days. Unlike tobacco leaves, calli of O. fragrans after instantaneous conversion by the vacuum infiltration method were cultured in the symbiotic medium (including 0 mM and 200 mM NaCl, respectively) under dark culture conditions for 3 days. Symbiotic medium was prepared by weighing 4.414 g of MS, 20 g of sucrose, 10 g of glucose, and 7 g of agar into a conical flask, and sterile water was fixed to 1 L. The medium was sterilized in an autoclave for 30 min, and the temperature was lowered to 55 °C. To this, 100 μL of naphthalene acetic acid (0.1 mg L−1), 500 μL of zeatin (1 mg L−1), 1 mL of folic acid (1 mg L−1), 1 mL of 6-BA (2 mg L−1), and 500 μL of acetosyringone (0.1 mol L−1) were added. All the tobacco leaves and calli were taken out after 3 days of culture to measure the activities of fluorescein enzyme via GLOMAX® multifunctional instrument (Promega, Madison, WI, USA).
2.5. Generation of Transgenic Arabidopsis Plants
Agrobacterium tumefaciens suspensions containing the plant expression vector OfSPL11-GFP were used to generate transgenic Arabidopsis seedlings using the floral dip method [15]. MS agar medium supplemented with 50 mg·L−1 kanamycin and 150 mM NaCl was used to select T3 transgenic lines. Three OfSPL11 overexpression lines with the highest levels of OfSPL11 expression and obvious phenotypes were selected for further analysis.
2.6. Salt Tolerance Assay
WT plants and three T3-generation pure transgenic lines, with 80–100 seeds of each line, were selected and sown on MS medium containing 0 mM NaCl and 150 mM NaCl for seed germination experiments, respectively, and vernalized at 4 °C for 3 days. The seeds were then grown in an artificial climate chamber at 22 °C and 70% humidity and 100 µmol m−2s−1 irradiation. The number of seedlings with cotyledons emerging within 10 days of sowing as a percentage of the total number of seeds sown was used to indicate the germination rate of seeds [16]. Three replications were carried out for each experiment. Seedlings of transgenic and wild-type Arabidopsis were transplanted vertically into MS medium containing 0 mM NaCl and 150 mM NaCl. After ten days of seedling growth, the seedlings were observed for root length and fibrous root number. In addition to culturing in the medium, seedlings of wild-type plants and transgenic strains were transplanted into nutrient soil. The 20-day-old transgenic strain and wild type were treated with 200 mM NaCl for one week with watering every 2 days. Additionally, Arabidopsis leaves were collected at 0 and 6 h after treatment for RNA extraction.
2.7. Measurement of Plant Physiological Index
Excised leaves were incubated in HEPES buffer (pH 7.6) containing 6 mM nitro blue tetrazolium (NBT) for 2.5 h to detect O2− production and immersed in 1 mg·mL−1 diaminobenzidine (DAB) solution for 7.5 h to monitor H2O2 production. The samples were immersed in 10 percent (v/v) glycerol for observations after being incubated in 80 percent (v/v) ethanol at 80 °C for 2 h to eliminate chlorophyll [17,18]. The concentrations of malondialdehyde (MDA) were determined using the MDA assay kit (Jiancheng Bioengineering Institute, Nanjing, China) according to the instructions of the manufacturer. Each experiment was carried out three times.
2.8. Yeast One-Hybrid Assays
The promoter of OfSPL11 was constructed on the pHIS2 vector. The recombinant plasmid OfSPL11pro-pHIS2 and the yeast library plasmid were cotransferred into the receptor cells according to the yeast Y187 receptor strain preparation and transformation kit (Coolaber, Beijing, China), and the upstream gene was screened. OfZAT12 was constructed into the pGADT7 vector. Using the cotransformed null pGADT7-GUS as a negative control, pGADT7-OfZAT12 and OfSPL11pro-pHIS2 were cotransformed into yeast cells and grown on S/-His-Leu-Trp medium containing 50 mM 3-AT.
2.9. Transient Expression Assay
The coding sequence of OfZAT12 was constructed into the pORER4 vector with recombinant plasmid 35Spro:OfZAT12. OfSPL11pro:LUC and 35Spro:OfZAT12 were cotransferred into Agrobacterium GV3101 using the cotransformed empty vector pORER4 as a negative control, and then the Agrobacterium suspension was injected into the abaxial surface of 25-day-old tobacco leaves. Dark incubation was carried out for one day and normal incubation for two days. Fluorescence signal intensity and enzyme activity assay were observed using a CCD cryoluminescence meter and luminescence detector. The GloMax 20/20 luminometer and Dual-Luciferase Assay Reagents (Promega) were used to measure the Renilla luciferase (REN) and Firefly luciferase (LUC) activities. Three biological replicates per experiment were carried out.
2.10. Quantitative Real-Time PCR and Statistical Analysis
On the basis of coding sequence, gene-specific primers for qRT-PCR were designed (Table S1). SYBR Premix Ex Taq was used in qRT-PCR on the Applied Biosystems 7300 Realtime PCR instrument. OfACT was used as an internal reference gene, and three biological re-reads were performed for each reaction. Calculation of relative gene expression was carried out using the 2−ΔΔCt method [19]. Excel 2010 (Microsoft, USA) and IBM SPSS Statistics 21 (IBM, USA) were used to analyze the data. All data are provided as the average of three biological replicates with standard deviation (SD). * p < 0.05 and ** p < 0.01 were used to determine statistical significance.
3. Results
3.1. OfSPL11 Clone and Sequence Analysis
OfSPL11 (Genbank: QTM66379.1) has a full-length CDS of 1185 bp, which was isolated from the O. fragrans cultivar ‘Yanhong Gui’ (Figure 1A). OfSPL11 is a 394-amino-acid protein with a molecular mass of 44.179 kDa, a theoretical pI of 8.66, and an amino acid formula of C1920H3010N560O598S21. The highest amino acid score at position 223 of OfSPL11 is 1.889. Amino acid 179 is the lowest, with −3.067. Meanwhile, the number of hydrophilic regions is greater than that of hydrophobic regions, indicating that the OfSPL11 protein is hydrophilic (Figure 1B). The secondary structure of the OfSPL11 protein is mainly composed of α-helix (25.13%), β-turn (5.08%), extended chain (13.71%), and irregular coil (56.09%) (Figure 1C). The predicted OfSPL11 amino acid sequence contains a conserved SBP structural domain, including a nuclear localization signal and two C2HCH-type zinc binding sites (Figure S1). Comparison of OfSPL11 amino acid sequences with those of homologous genes in other plant species showed that they are highly similar and both contain the same conserved structural domains (Figure 2A). The amino acid sequences of OfSPL11 and its homologous genes were used to construct a phylogenetic tree using the software MEGA 6.0. The findings reveal that OfSPL11 protein has the closest evolutionary relationship with OeSPL12 from Olea europaea (Figure 2B). The OfSPL11 protein exhibits 84% sequence identity with OeSPL12, demonstrating that the OfSPL11 gene is evolutionarily conserved; thus its functions may be similar to those of SPLs found in other plant species.
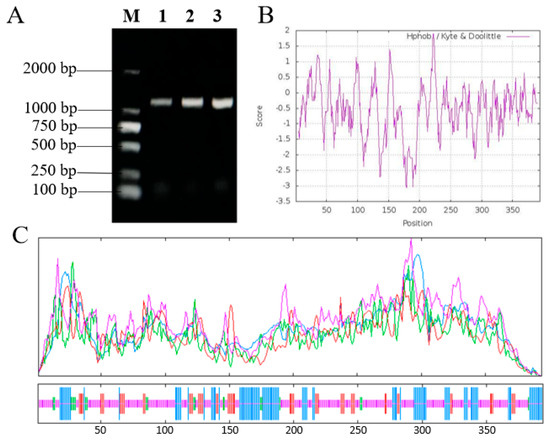
Figure 1.
Sequence cloning and analysis of OfSPL11. (A) CDS cloning of the OfSPL11; (B) hydrophilicity and hydrophobicity of the OfSPL11; (C) protein secondary structure of OfSPL11. Blue line indicates α−helix, red line indicates extended chain, green line indicates β−turn, and purple line indicates irregular coil.
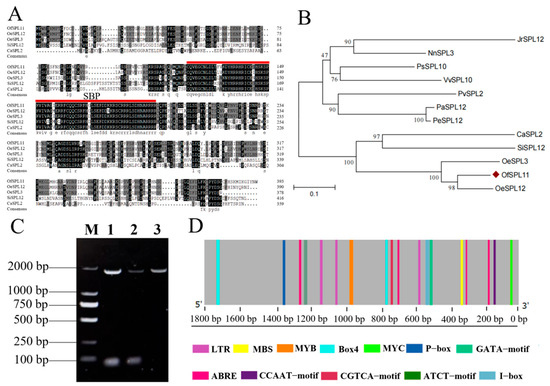
Figure 2.
Multiple sequence alignment, phylogenetic analysis, and promoter cis-acting element analysis of OfSPL11. (A) OfSPL11 and homologous gene amino acid sequence comparison. Black represents identical amino acids, gray represents similar amino acids. Conserved structural domains of SBP have been underlined in red; (B) phylogenetic tree of OfSPL11 and homologous genes. Detailed information is as follows: Osmanthus fragrans, OfSPL11, QTM66379.1; Olea europaea, OlSPL12, XP_022888134.1; Olea europaea, OlSPL3, XP_022883063.1; Coffea arabica, CaSPL2, XP_027066228.1; Paeonia suffruticosa, PsSPL10, QMV47673.1; Sesamum indicum, SiSPL12, XP_011093932.1; Juglans regia, JrSPL12, XP_011014590.1; Vitis vinifera, VvSPL10, XP_018836770.1; Pistacia vera, PvSPL2, XP_031264374.1; Populus alba, PaSPL12, XP_034888712.1; Populus euphratica, PeSPL12, XP_011014590.1; Nelumbo nucifera, NnSPL3, XP_019055864.1; (C) cloning of the OfSPL11 promoter; (D) putative cis-acting elements in the promoter of the OfSPL11.
3.2. Cis-Acting Element Analysis of OfSPL11 Promoter
Analysis of the cis-acting element of the OfSPL11 promoter to probe the molecular mechanism of its function was performed. The OfSPL11 promoter with a length of 1800 bp was obtained using the chromosome walking method (Figure 2C). Multiple cis-elements implicated in hormone and stress responses are found in the promoter region of the OfSPL11 gene, including ABREs, P-box, MBS, CGTCA-motif, MYB, MYC, and LTRs (Figure 2D).
3.3. Subcellular Localization and Transcriptional Autoactivation
The results of the predicted subcellular localization of the Softberry (www.softberry.com, accessed on 11 April 2021) website indicate that OfSPL11 is localized in the nucleus. A suspension of the A. rhizogenes strain GV3101 containing the pORER4-OfSPL11-GFP recombinant plasmid was aspirated using a 1 mL-range syringe (needle not included), and the suspension was gently injected into the abaxial surface of four-week-old tobacco leaves, avoiding the leaf veins. This result reveals that the green fluorescent signal of OfSPL11 and the DAPI nuclear localization signal overlapped, indicating that OfSPL11 was localized in the nucleus (Figure 3A), consistent with the results predicted on the website. Yeast cells transformed by the empty vector of pGBKT7 were grown on the SD/-Trp medium only. However, yeast cells transformed by the OfSPL11 plasmid were not only able to grow on SD/-Trp and SD/-Trp-Ade-His-Leu media, but the yeast strain turned blue in SD/-Trp-Ade-His-Leu + X-α-Gal media (Figure 3B). The results indicate that OfSPL11 has transcriptional activation activity.
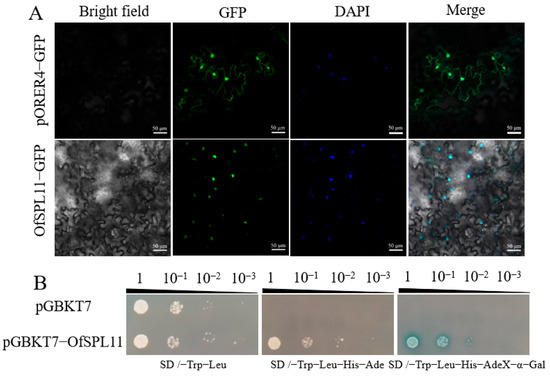
Figure 3.
Subcellular localization and transcriptional autoactivation analysis of OfSPL11. (A) Subcellular localization of OfSPL11. Green indicates green fluorescent protein, and blue indicates DAPI nuclear localization signal. Scale bars = 50 μm; (B) transcriptional autoactivation analysis of OfSPL11.
3.4. Tissue and Salt Stress Expression Profiling of OfSPL11
The RT-qPCR of OfSPL11 in various tissues of O. fragrans showed that the OfSPL11 gene was expressed in all tissues, and its expression in shoots and young leaves was significantly higher than that in other tissues (Figure 4A). The expression of the OfSPL11 gene in young leaves is approximately 20 times greater than that in flowers, stems and roots. In addition, salt treatment promoted the expression of OfSPL11, and its transcription rose considerably after 3 h of treatment with 200 mM NaCl and peaked at 24 h. Overall, the OfSPL11 expression exhibited a general trend of first increasing and then decreasing during the 48 h (Figure 4B), suggesting that OfSPL11 is induced by salt stress and may be involved in salt tolerance.
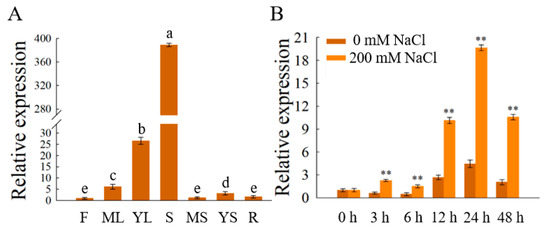
Figure 4.
Expression of OfSPL11. (A) Expression of OfSPL11 in various Osmanthus fragrans tissues. F indicates flowers, ML indicates mature leaves, YL indicates young leaves, S indicates shoots, MS indicates mature stems, YS indicates young stems, and R indicates roots. Different lowercase letters represent the significant differences as assessed by Duncan's test at the 0.01 level. (B) OfSPL11 expression levels in Osmanthus fragrans young leaves after 200 mM NaCl treatment. The internal reference gene for O. fragrans is OfACT and three biological re-reads were performed for each reaction. ** p < 0.01 were used to determine statistical significance.
3.5. The Promoter of OfSPL11 Was Sensitive to Salinity
To explore the way in which salt treatment promotes the expression of OfSPL11, we isolated the promoter of OfSPL11 with 1800 bp. The promoter was ligated into the pGreen0800II-LUC vector (Figure 5A), the transformation of the fusion vector OfSPL11pro:LUC into A. tumefaciens GV3101 (adding pSoup) receptor cells occurred, and the leaves of tobacco plants were infiltrated. Subsequently, the leaves were treated with 0 mM and 200 mM NaCl. The results show that relative LUC activity was significantly different under 200 mM NaCl (Figure 5B,C), compared to the leaves that had not undergone salt treatment. It indicates that 200 mM NaCl could directly promote the activity of the promoter of OfSPL11 in the leaves of tobacco. In addition, A. tumefaciens suspensions containing the fusion vector OfSPL11pro:LUC were transiently transformed into O. fragrans calli using vacuum infiltration, and then the O. fragrans calli were cultured in a symbiotic medium containing 0 mM and 200 mM NaCl. After 3 days of dark culture, the calli were taken out for observation, and the results suggest that compared to the control, relative LUC activity increased under 200 mM NaCl (Figure 5D,E). In conclusion, all the results mean that salt treatment could activate the expression of OfSPL11 directly through its promoter.
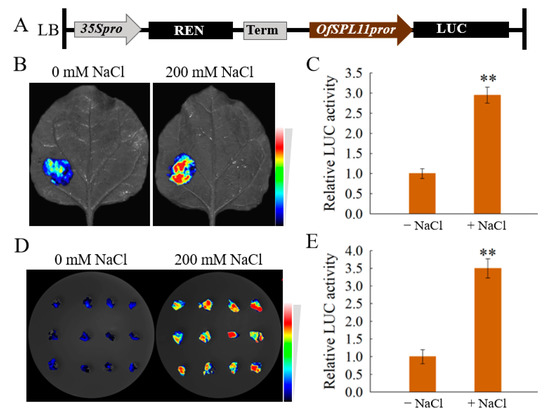
Figure 5.
Salinity directly activates the activity of proOfSPL11. (A) Schematic representation of OfSPL11pro:LUC; (B,C) the activity of proOfSPL11 was activated in tobacco leaves; (D,E) the activity of proOfSPL11 was activated in callus of Osmanthus fragrans. Each experiment had three biological replicates. ** p < 0.01 were used to determine statistical significance.
3.6. Phenotypic Characteristics of Arabidopsis Lines Overexpressing OfSPL11
Transgenic Arabidopsis lines overexpressing the OfSPL11 gene were generated to examine the function of OfSPL11 in salt tolerance. For further investigation, the three T3 transgenic lines (OE4, OE8, and OE9) with the highest OfSPL11 expression level were chosen. Under the same conditions, the three OfSPL11 overexpression lines exhibited similar growth characteristics to the wild-type (WT) plants at the vegetative stage (Figure S2A). In three repeated batch cultivations, the three transgenic lines did not differ in rosette leaf number or shape compared to the WT plants (Table S2). At the reproductive development stage, the three transgenic lines were taller and flowered earlier than wild-type plants (Figure S2B,C).
3.7. OfSPL11 Enhances Salt Tolerance in Arabidopsis
We cultured transgenic lines OE4, OE8, and OE9 on an MS medium containing 0 mM NaCl and 150 mM NaCl to further confirm the role of OfSPL11 in the salt stress response. The germination rates of the transgenic lines were similar to those of WT seedlings on a 0 mM NaCl MS medium (Figure 6A). However, under 150 mM NaCl treatment, the germination rates of OE4, OE8, and OE9 lines were significantly higher than those of wild-type (WT) plants (Figure 6B). Specifically, the germination rates of OE4, OE8, and OE9 lines were 70.83%, 89.58%, and 85.42%, respectively, under salt conditions, compared with 47.92% in the WT (Figure 6C). However, the root lengths of transgenic seedlings and wild-type seedlings were similar in the MS medium (Figure 6D). Notably, the transgenic seedlings developed more lateral roots and longer roots than the WT seedlings in the MS medium with 150 mM NaCl (Figure 6E,F). These experimental results suggest that OfSPL11 participates in salt stress resistance. Subsequently, we examined mature transgenic lines and wild-type plants for resistance to salt stress. We found that the WT plant leaves exhibited wilted and etiolated symptoms after one week of salt treatment. The transgenic lines’ leaves, in contrast, stayed green after being exposed to 200 mM NaCl (Figure 7A).
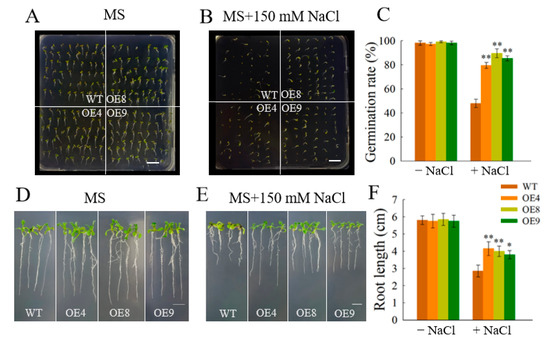
Figure 6.
Seed germination and seedlings’ root length of OfSPL11−overexpressed Arabidopsis. (A,B) seed germination phenotypes of Arabidopsis in MS medium containing different concentrations of NaCl; (C) germination rate of Arabidopsis seeds; (D,E) root phenotypes of Arabidopsis seedlings in MS medium containing different concentrations of NaCl. Scale bars = 1 cm; (F) root length of Arabidopsis seedlings. Each experiment had three biological replicates. * p < 0.05 and ** p < 0.01 were used to determine statistical significance.
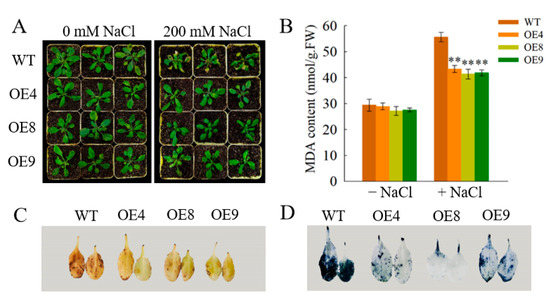
Figure 7.
Effect of salt treatment on OfSPL11−overexpressed lines and WT Arabidopsis. (A) Effect of different concentrations of NaCl treatment on Arabidopsis plants in soil; (B) malondialdehyde (MDA) content of Arabidopsis plants; (C) detection of H2O2 levels in Arabidopsis; and (D) detection of O2− levels in Arabidopsis. Each experiment had three biological replicates. ** p < 0.01 were used to determine statistical significance.
To better understand the physiological mechanism of OfSPL11 in response to salt stress, we further measured the physiological indexes of abiotic stresses, such as malondialdehyde (MDA) and reactive oxygen species (ROS) accumulation. Salt treatment was able to induce the accumulation of MDA in both wild-type and transgenic plants. The MDA content of wild-type plants reached 55.6 nmol g−1 FW, which was significantly higher than that of the transgenic lines (Figure 7B). Similarly, we performed DAB and NBT histochemical staining analysis on plant leaves grown under 200 mM NaCl treatment to measure the levels of H2O2 and O2−, respectively. Leaves of wild-type(WT) plants accumulated much higher levels of H2O2 and O2− than leaves of transgenic lines (Figure 7C,D). These results indicate that the OfSPL11 transgenic lines reduced the accumulation of MDA and ROSs, thus reducing cell membrane damage from salt stress; this facilitates the enhancement of salt tolerance in transgenic lines.
3.8. Expression of Abiotic Stress Response Genes in OfSPL11 Overexpression Lines
To better understand why OfSPL11 transgenic lines exhibit enhanced abiotic stress tolerance, we analyzed the expression of known abiotic stress response genes in 3-week-old wild-type and transgenic Arabidopsis (Figure 8), such as CALCINEURIN B-LIKE PROTEIN 1 (CBL1), COLD-REGULATED 15A (COR15A), DESICCATION-RESPONSIVE PROTEIN 29A (RD29A), SALT OVERLY SENSITIVE 3 (SOS3), COLD-RESPONSIVE 6.6 (COR6.6), and SALT OVERLY SENSITIVE 2 (SOS2). After 6 h under 200 mM NaCl treatment, compared to wild-type plants, all abiotic stress genes were significantly up-regulated in OfSPL11 overexpression strains. The presence of this expression difference suggests that OfSPL11 responds to salt stress and improves salt resistance.
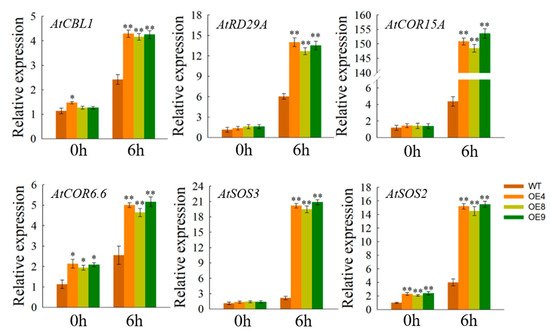
Figure 8.
Expression of abiotic stress response genes in OfSPL11-overexpressed lines and WT Arabidopsis. Each experiment had three biological replicates. * p < 0.05 and ** p < 0.01 were used to determine statistical significance.
3.9. OfZAT12 Regulated the Expression of OfSPL11
To further verify how OfSPL11 improves salt tolerance in plants, A 1800 bp promoter fragment of OfSPL11 was constructed on the pHIS2 vector to produce the OfSPL11pro-pHIS2 construct. Then, yeast one-hybrid library screening was performed using OfSPL11pro-pHIS2. The results reveal that OfZAT12(homologous to the Arabidopsis AtRHL41 gene) may be able to bind to the promoter of OfSPL11. To verify whether there is an interaction between OfZAT12 and OfSPL11, we performed a yeast single-hybridization analysis. The recombinant plasmids of OfSPL11pro-pHIS2 and pGADT7-OfZAT12 were cotransformed into Y187 yeast receptor cells and cultured on a screening medium for 4–5 days. Compared to the control group, yeast cells cotransfected with recombinant plasmids OfSPL11pro-pHIS2 and pGADT7-OfZAT12 were able to grow normally on the SD/-Trp-Leu-His medium containing 50 mM 3-AT, indicating that OfZAT12 was able to bind directly and specifically to the promoter of OfSPL11 and regulate its expression (Figure 9A). We further verified the regulatory relationship of OfSPL11 and OfZAT12 with a tobacco transient expression assay. Compared with the 35Spro control, Agrobacterium GV3101 suspensions containing recombinant plasmids OfSPL11pro:LUC and 35Spro:OfZAT12 injected into the back of tobacco revealed a significantly enhanced fluorescent signal and transient coexpression of OfSPL11 and OfZAT12 in tobacco. The results indicate that OfZAT12 can directly activate OfSPL11 transcriptional expression (Figure 9B–D). The expression of OfZAT12 under salt stress assessed by RT-qPCR is shown in Figure 9E. Salt treatment promoted the expression of OfZAT12, and its transcription rose considerably after 3 h of treatment with 200 mM NaCl and peaked at 12 h. The expression of OfZAT12 changed earlier than that of OfSPL11, indicating that OfZAT12 is an upstream regulatory gene of OfSPL11. These results suggest that the induction of OfZAT12 by salt stress causes the expression of OfZAT12 to rise; thus, OfZAT12 promotes the expression of the downstream target gene OfSPL11.
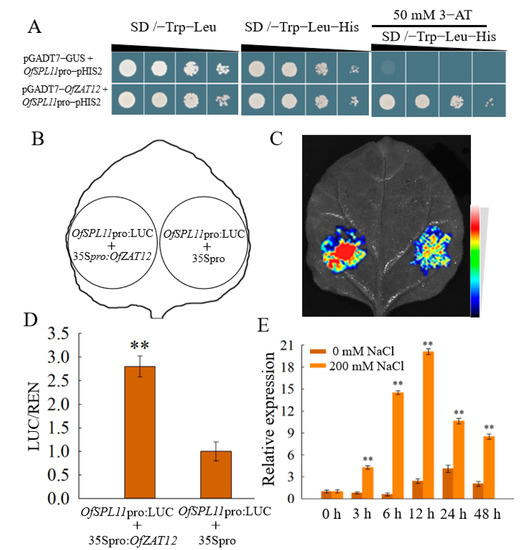
Figure 9.
OfZAT12 regulated the expression of OfSPL11. (A) Yeast one-hybrid assay to verify the interaction between OfZAT12 and OfSPL11; (B–D) the Dual-Luciferase Assay Reagents were used to measure the activities of firefly LUC and REN; (E) OfSPL11 expression levels in Osmanthus fragrans young leaves after 200 mM NaCl treatment. The internal reference gene for O. fragrans is OfACT, and three biological re-reads were performed for each reaction. ** p < 0.01 were used to determine statistical significance.
4. Discussion
The SPL gene, which is found only in green plants, is a plant-specific gene. Typically, the coding region of SPL contains an SBP structural domain consisting of approximately 76 amino acids, comprising a nuclear localization signal and two zinc finger structures [10]. The conserved structural domain is the core of the gene and has an important function. The ofSPL11 amino acid sequence also contains the same nuclear localization signal and zinc finger structure (C-C-C-H and C-C-H-C) (Figure 2A). In addition, OfSPL11 protein is hydrophilic. The OfSPL11 gene is most closely related to Olea europaea OeSPL12 (Figure 2B). Furthermore, we confirmed that OfSPL11 is a transcriptional activator that functions in the nucleus. Bioinformatic analysis of the OfSPL11 sequence revealed similarities to SPL genes reported in other species.
Promoters, as important cis-elements for regulating gene expression, play an important role in the regulation of gene expression in plants. At present, the constitutive expression promoter is mostly used to drive the expression of target genes in plant stress resistance gene engineering. The expression pattern of a gene is determined by various cis-acting elements in the promoter region [20]. The presence of cis-acting elements in the OfSPL11 promoter region suggests that OfSPL11 is likely implicated in the response to salt stress (Figure 2D). OfSPL11 promoter transient transformation in the leaves of tobacco and callus of O. fragrans revealed that salt treatment could activate the expression of OfSPL11 directly through its promoter (Figure 5). Furthermore, the presence of many light-responsive elements in the OfSPL11 promoter, such as the Box4, ATCT-motif, I-box, and GATA-motif (Figure 2D), implies that OfSPL11 in O. fragrans may be involved in photoperiod and circadian rhythm responses. The identification of cis-acting elements in the promoter region of OfSPL11 allows us to further understand the molecular mechanism of function.
Various abiotic stresses can have irreversible effects on plants during their life cycle, including salinity, cold, and drought. One of the most prevalent challenges that plants face is salinity. It has the potential to reduce crop quality and yield, resulting in significant losses in agricultural production. Salt stress can also adversely affect the expression levels of many genes in plants [21]. In this study, after NaCl treatment of O. fragrans leaves, the expression level of OfSPL11 in leaves was significantly increased by salt stress, according to qRT-PCR results (Figure 4B), implying that OfSPL11 potentially participates in salt stress response. Transgenic Arabidopsis lines overexpressing the OfSPL11 gene were generated to examine the function of OfSPL11 in salt tolerance (Figure S2). Under high salinity stress, the transgenic lines showed improved phenotypic morphology (Figure 6), demonstrating that the OfSPL11 gene enhances salt tolerance in A. thaliana. The distribution and size of a plant’s root system have an impact on its ability to withstand environmental pressures. In this study, under salt stress, the root system of the OfSPL11 transgenic lines was longer and larger compared to that of WT plants, which likely contributed to salt tolerance by increasing water uptake.
In a previous study, AtSPL9 overexpression lines showed early flowering and sensitivity to abiotic stresses in the form of anthocyanin metabolism regulation in Arabidopsis [22]. Notably, the expression pattern observed in this study indicates that the expression level of the OfSPL11 gene was higher in the young leaves and flower buds of O. fragrans than in other tissues (Figure 4A). In addition, compared with wild-type plants, OE4, OE-8, and OE9 overexpression lines exhibit an early flowering phenotype (Figure S2), implying that OfSPL11 regulates multiple biological processes in O. fragrans, especially in response to salt stress, and regulates the flowering process.
Salt increases oxidative stress by inducing the production of reactive oxygen species (ROSs), such as O2−, singlet oxygen, and H2O2 [23]. The dynamic balance of the synthesis and removal of ROSs in plant cells is disrupted when plants are exposed to environmental stress. Excessive ROSs cause severe oxidative damage to plants and have a negative impact on their growth [24,25,26]. Therefore, maintaining ROS homeostasis and removal of excess ROSs are crucial in improving plant stress resistance [27]. Activated oxygen scavenging is controlled by a number of transcription factors. For instance, NtJERF3 from Nicotiana tabacum overexpression reduced ROS content [28]. Additionally, the Oryza sativa OsMYB2 gene responds to stress conditions by enhancing ROS scavenging ability [29]. In this study, MDA, H2O2, and O2− content in transgenic lines and wild-type plants was almost the same without a significant difference before salt treatment. In contrast, transgenic Arabidopsis lines overexpressing OfSPL11 accumulated much lower levels of H2O2, MDA, and O2− than the wild-type plants under salt treatment (Figure 7). Some abiotic stress response genes, such as COR6.6, CBL1, SOS2, RD29A, COR15A, and SOS3, were expressed at significantly higher levels in OfSPL11 transgenic lines than in wild-type plants (Figure 8), further indicating the importance of OfSPL11 in salt stress resistance. Previous research found that Arabidopsis AtZAT12 (AtRHL41) is a key transcriptional regulator in the abiotic stress expression network and is involved in multiple stress response pathways [30]. In this study, the expression level of OfZAT12 (homologous to Arabidopsis AtRHL41 gene), which was significantly increased by salt stress, and OfZAT12 can specifically bind to the OfSPL11 promoter and positively regulates its expression (Figure 9). It is thus reasonable to speculate that OfSPL11 is an important regulator in the salt stress expression network and is involved in several salt stress response pathways. Overall, these findings suggest that the OfSPL11 gene participates in salt stress through the regulation of several genes involved in lowering oxidative damage, scavenging ROSs, signal transduction, and reducing accumulation of MDA.
5. Conclusions
In conclusion, the full-length CDS of the OfSPL11 gene was isolated from O. fragrans. Subcellular localization results reveal that OfSPL11 is located in the nucleus. Moreover, the yeast transactivation assay revealed that OfSPL11 has transcriptional activation activity. Further analysis of transgenic Arabidopsis lines showed that OfSPL11 positively regulates salt stress tolerance by suppressing the accumulation of MDA and ROSs, alleviating membrane damage, mitigating leaf wilting, and enhancing root growth. Abiotic stress response genes were significantly up-regulated in OfSPL11 overexpression lines. OfZAT12 was able to bind directly and specifically to the promoter of OfSPL11 and regulate its expression.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae8050412/s1. Figure S1: Nucleotide sequence and deduced amino acid sequence of SBP domain alignment of OfSPL11; Figure S2: Phenotype of OfSPL11 transgenic plants; Table S1: Primer sequences used in this study; Table S2: Rosette leaf number and shape of Arabidopsis.
Author Contributions
Conceptualization, H.Z. and S.Z. (Shoukuo Zhu); methodology, S.Z. (Shoukuo Zhu); software, S.Z. (Shoukuo Zhu) and Y.W.; validation, B.D., Q.F. and S.Z. (Shiwei Zhong); formal analysis, Y.W. and S.Z. (Shiwei Zhong); investigation, S.Z. (Shoukuo Zhu); data curation, S.Z. (Shoukuo Zhu); writing—original draft preparation, S.Z. (Shoukuo Zhu); writing—review and editing, H.Z., B.D., Q.F. and S.Z. (Shoukuo Zhu); funding acquisition, H.Z. and B.D. All authors have read and agreed to the published version of the manuscript.
Funding
This work was financially supported by the National Natural Science Foundation of China (Grant No. 32072615; 31902057) and Natural Science Foundation of Zhejiang Province (Grant No. LQ19C160012).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data in this study can be found in the manuscript or in the Supplementary Materials.
Acknowledgments
We thank Gang Zheng for caring for the plant materials and managing the maintenance of the lab.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Kong, C.C.; Ren, C.G.; Li, R.Z.; Xie, Z.H.; Wang, J.P. Hydrogen Peroxide and Strigolactones Signaling Are Involved in Alleviation of Salt Stress Induced by Arbuscular Mycorrhizal Fungus in Sesbania cannabina Seedlings. J. Plant Growth Regul. 2017, 36, 734–742. [Google Scholar] [CrossRef]
- Moles, T.M.; Pompeiano, A.; Huarancca, R.T.; Scartazza, A.; Guglielminetti, L. The efficient physiological strategy of a tomato landrace in response to short-term salinity stress. Plant Physiol. Biochem. 2016, 109, 262–272. [Google Scholar] [CrossRef] [PubMed]
- Zhu, J.K. Regulation of ion homeostasis under salt stress. Curr. Opin. Plant Biol. 2003, 6, 441–445. [Google Scholar] [CrossRef]
- Chen, M.; Yang, Z.; Liu, J.; Zhu, T.; Wei, X.; Fan, H.; Wang, B. Adaptation Mechanism of Salt Excluders under Saline Conditions and Its Applications. Int. J. Mol. Sci. 2018, 19, 3688. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Liu, J.; Zhu, T.; Zhao, C.; Li, L.; Chen, M. The Role of Melatonin in Salt Stress Responses. Int. J. Mol. Sci. 2019, 20, 1735. [Google Scholar] [CrossRef] [Green Version]
- Zhao, K.F.; Song, J.; Fan, H.; Zhou, S.; Zhao, M. Growth response to ionic and osmotic stress of NaCl in salt-tolerant and salt-sensitive maize. J. Integr. Plant Biol. 2010, 52, 468–475. [Google Scholar] [CrossRef]
- Wu, Y.; Fan, X.; Zhang, Y.; Jiang, J.; Sun, L.; Rahman, F.U.; Liu, C. VvSNAT1 overexpression enhances melatonin production and salt tolerance in transgenic Arabidopsis. Plant Physiol. Biochem. 2021, 166, 485–494. [Google Scholar] [CrossRef]
- Wen, X.; Geng, F.; Cheng, Y.; Wang, J. Ectopic expression of CsMYB30 from Citrus sinensis enhances salt and drought tolerance by regulating wax synthesis in Arabidopsis thaliana. Plant Physiol. Biochem. 2021, 166, 777–788. [Google Scholar] [CrossRef]
- Klein, J.; Saedler, H.; Huijser, P. A new family of DNA binding proteins includes putative transcriptional regulators of the Antirrhinum majus floral meristem identity gene SQUAMOSA. Mol. Gen. Genet. 1996, 250, 7–16. [Google Scholar] [CrossRef]
- Yamasaki, K.; Kigawa, T.; Inoue, M.; Tateno, M.; Yamasaki, T.; Yabuki, T.; Aoki, M.; Seki, E.; Matsuda, T.; Nunokawa, E. A novel zinc-binding motif revealed by solution structures of DNA-binding domains of Arabidopsis SBP-family transcription factors. J. Mol. Biol. 2004, 337, 49–63. [Google Scholar] [CrossRef]
- Moreno, A.M.; Harper, C.L.; Krueger, W.R.; Dellaporta, L.S.; Michael, F. liguleless1 encodes a nuclear-localized protein required.for induction of ligules and auricles during maize leaf organogenesis. Genes Dev. 1997, 11, 616–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Preston, J.C.; Hileman, L. Functional evolution in the plant SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) gene family. Front. Plant Sci. 2013, 4, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chao, L.M.; Liu, Y.Q.; Chen, D.Y.; Xue, X.Y.; Mao, Y.B.; Chen, X.Y. Arabidopsis Transcription Factors SPL1 and SPL12 Confer Plant Thermotolerance at Reproductive Stage. Mol. Plant 2017, 10, 735–748. [Google Scholar] [CrossRef] [PubMed]
- Ning, K.; Chen, S.; Huang, H.; Jiang, J.; Yuan, H.; Li, H. Molecular characterization and expression analysis of the SPL gene family with BpSPL9 transgenic lines found to confer tolerance to abiotic stress in Betula platyphylla Suk. Plant Cell Tissue Organ Cult. 2017, 130, 469–481. [Google Scholar] [CrossRef]
- Clough, J.S.; Bent, F.A. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef] [Green Version]
- Saleki, R.; Young, G.P.; Lefebvre, D.D. Mutants of Arabidopsis thaliana Capable of Germination under Saline Conditions. Plant Physiol. 1993, 101, 839–845. [Google Scholar] [CrossRef] [Green Version]
- Kotchoni, O.S.; Kuhns, C.; Ditzer, A.; Kirch, H.H.; Bartels, D. Over-expression of different aldehyde dehydrogenase genes in Arabidopsis thaliana confers tolerance to abiotic stress and protects plants against lipid peroxidation and oxidative stress. Plant Cell Environ. 2006, 29, 1033–1048. [Google Scholar] [CrossRef]
- Kim, S.H.; Woo, D.H.; Kim, J.M.; Lee, S.Y.; Chung, W.S.; Moon, Y.H. Arabidopsis MKK4 mediates osmotic-stress response via its regulation of MPK3 activity. Biochem. Biophys. Res. Commun. 2011, 412, 150–154. [Google Scholar] [CrossRef]
- Livak, J.K.; Schmittgen, D.T. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Du, Q.; Tomkinson, E.A.; Gardner, D.P. Transcriptional Regulation of Neuronal Nicotinic Acetylcholine Receptor Genes. J. Biol. Chem. 1997, 272, 14990–14995. [Google Scholar] [CrossRef] [Green Version]
- Arzani, A. Improving salinity tolerance in crop plants: A biotechnological view. In Vitro Cell. Dev. Biol. Plant. 2008, 44, 373–383. [Google Scholar] [CrossRef]
- Cui, L.G.; Shan, J.X.; Shi, M.; Gao, J.P.; Lin, H.X. The miR156-SPL9-DFR pathway coordinates the relationship between development and abiotic stress tolerance in plants. Plant J. 2014, 80, 1108–1117. [Google Scholar] [CrossRef] [PubMed]
- Hasegawa, M.P.; Bressan, A.R.; Zhu, J.K.; Bohnert, J.H. Plant cellular and molecular responses to high salinity. Plant Biol. 2000, 51, 463–499. [Google Scholar] [CrossRef] [Green Version]
- Mittova, V.; Tal, M.; Volokita, M.; Guy, M. Salt stress induces up-regulation of an efficient chloroplast antioxidant system in the salt-tolerant wild tomato species Lycopersicon pennellii but not in the cultivated species. Physiol. Plant. 2002, 115, 393–400. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Wen, D.; Yang, W.; Meng, Q.; Shi, Q.; Gong, B. Overexpression of Caffeic Acid O-Methyltransferase 1 (COMT1) Increases Melatonin Level and Salt Stress Tolerance in Tomato Plant. J. Plant Growth Regul. 2019, 39, 1221–1235. [Google Scholar] [CrossRef]
- Zheng, X.; Tan, X.D.; Allan, C.A.; Zuo, B.; Zhao, Y.; Reiter, J.R.; Wang, L.; Wang, Z.; Guo, Y.; Zhou, J. Chloroplastic biosynthesis of melatonin and its involvement in protection of plants from salt stress. Sci. Rep. 2017, 7, 41236. [Google Scholar] [CrossRef]
- He, L.; He, T.; Farrar, S.; Ji, L.; Liu, T.; Ma, X. Antioxidants Maintain Cellular Redox Homeostasis by Elimination of Reactive Oxygen Species. Cell. Physiol. Biochem. 2017, 44, 532–553. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, Z.; Zhang, H.; Wang, X.; Huang, R. Transcriptional modulation of ethylene response factor protein JERF3 in the oxidative stress response enhances tolerance of tobacco seedlings to salt, drought, and freezing. Plant Physiol. 2008, 148, 1953–1963. [Google Scholar] [CrossRef] [Green Version]
- Yang, A.; Dai, X.; Zhang, W. A R2R3-type MYB gene, OsMYB2, is involved in salt, cold, and dehydration tolerance in rice. J. Exp. Bot. 2012, 63, 2541–2556. [Google Scholar] [CrossRef]
- Sholpan, D.; Karen, S.; Jesse, C.; Ron, M. Zinc-Finger Protein Zat12 Plays a Central Role in Reactive Oxygen and Abiotic Stress Signaling in Arabidopsis. Plant Physiol. 2005, 139, 847–856. [Google Scholar] [CrossRef] [Green Version]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).