miR398 Attenuates Heat-Induced Leaf Cell Death via Its Target CSD1 in Chinese Cabbage
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials and Growth Condition
2.2. Cloning and Generation of Transgenic Plants
2.3. Heat Treatment and Measurement of Leaf Cell Death
2.4. miRNA Isolation and Northern Blot Analysis
2.5. 5′ RACE (Rapid Amplification of cDNA Ends)
2.6. Real-Time qRT-PCR
2.7. Sequence Alignment and Phylogenetic Analysis
2.8. Degradome Analysis
2.9. Statistical Analysis
3. Results
3.1. Characterization of the miR398 and Its Targets Genes in B. rapa
3.2. Response of Bra-miR398a and Its Target Genes to Heat Stress in B. rapa
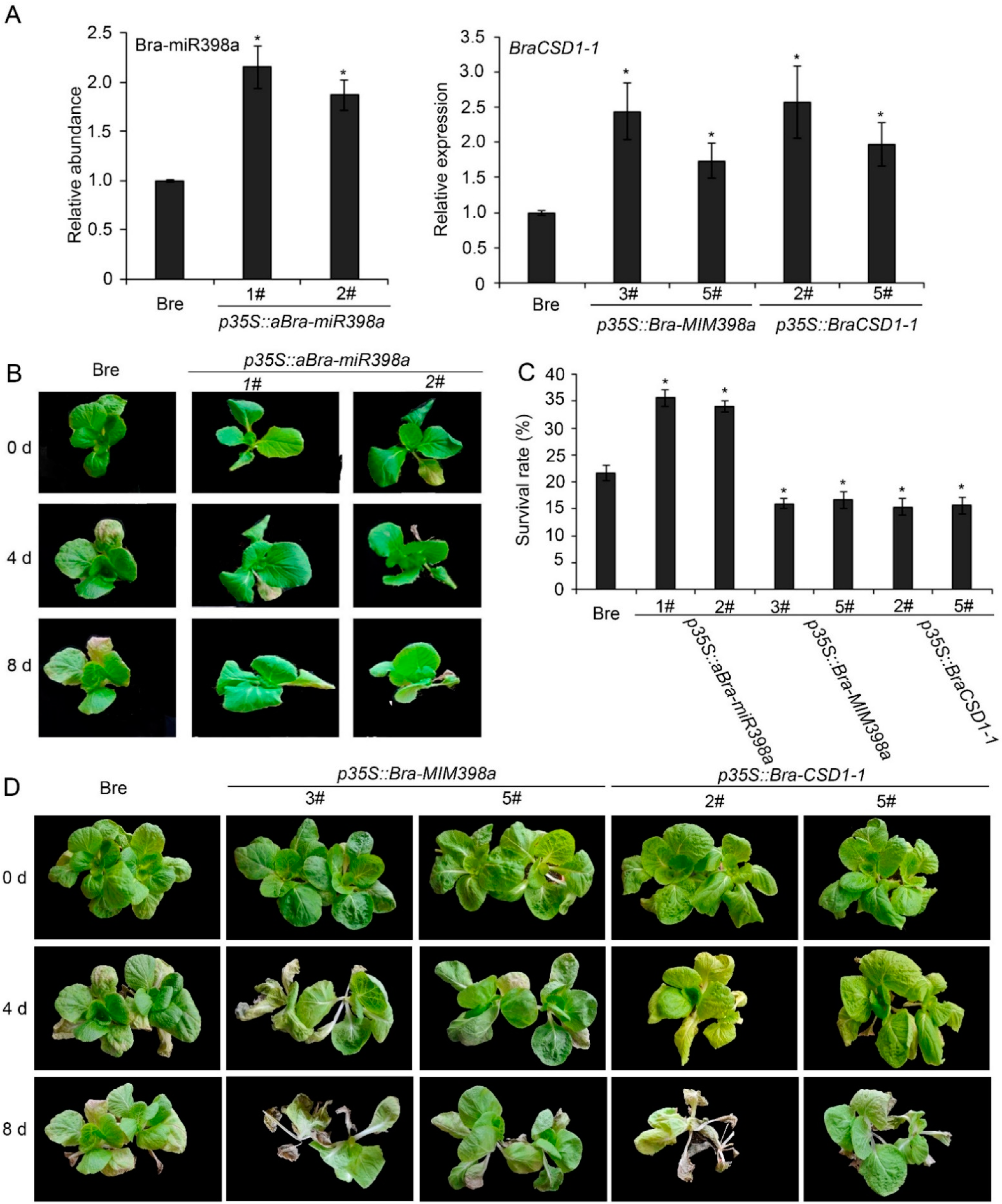
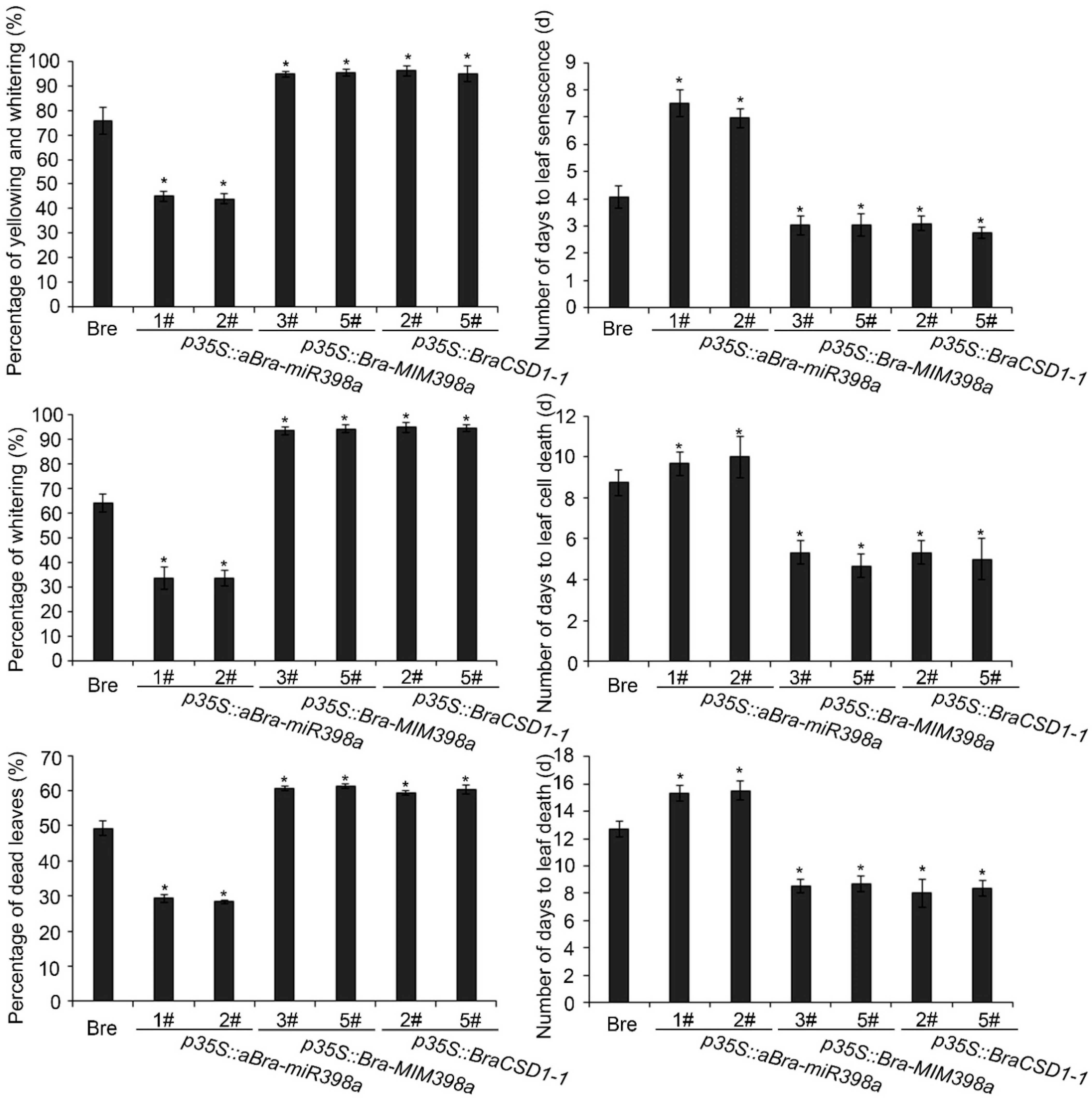
3.3. Bra-miR398a Aids in the Prevention of Leaf Death and Plant Death
3.4. Bra-miR398a Regulated Heat-Induced Leaf Cell Death Independent with Cu2+-Mediated Pathway
3.5. Stress-Related Marker Genes Were Upregulated by Bra-miR398a
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cui, Y.; Peng, Y.; Zhang, Q.; Xia, S.; Ruan, B.; Xu, Q.; Yu, X.; Zhou, T.; Liu, H.; Zeng, D.; et al. Disruption of EARLY LESION LEAF 1, encoding a cytochrome P450 monooxygenase, induces ROS accumulation and cell death in rice. Plant J. 2021, 105, 942–956. [Google Scholar] [CrossRef] [PubMed]
- Ramiro, D.A.; Melotto-Passarin, D.M.; Barbosa, M.; Santos, F.D.; Gomez, S.; Júnior, N.M.; Lam, E.; Carrer, H. Expression of Arabidopsis Bax Inhibitor-1 in transgenic sugarcane confers drought tolerance. Plant Biotechnol. J. 2016, 14, 1826–1837. [Google Scholar] [CrossRef] [PubMed]
- Lincoln, J.E.; Sanchez, J.P.; Zumstein, K.; Gilchrist, D.G. Plant and animal PR1 family members inhibit programmed cell death and suppress bacterial pathogens in plant tissues. Mol. Plant Pathol. 2018, 19, 2111–2123. [Google Scholar] [CrossRef] [PubMed]
- Fraser, M.S.; Dauphinee, A.N.; Gunawardena, A.H.L.A.N. Determining the effect of calcium on cell death rate and perforation formation during leaf development in the novel model system, the lace plant (Aponogeton madagascariensis). J. Microsc. 2020, 278, 132–144. [Google Scholar] [CrossRef] [PubMed]
- Woo, H.R.; Kim, H.J.; Lim, P.O.; Nam, H.G. Leaf Senescence: Systems and Dynamics Aspects. Annu. Rev. Plant Biol. 2019, 70, 347–376. [Google Scholar] [CrossRef] [PubMed]
- Hensel, L.L.; Grbic, V.; Baumgarten, D.A.; Bleecker, A.B. Developmental and Age-Related Processes That Influence the Longevity and Senescence of Photosynthetic Tissues in Arabidopsis. Plant Cell 1993, 5, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Kim, J.H.; Lyu, J.I.; Woo, H.R.; Lim, P.O. New insights into the regulation of leaf senescence in Arabidopsis. J. Exp. Bot. 2018, 69, 787–799. [Google Scholar] [CrossRef]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef]
- Zhu, C.; Ding, Y.; Liu, H. MiR398 and plant stress responses. Physiol. Plant 2011, 143, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Song, X.W.; Li, Y.; Cao, X.F.; Qi, Y.J. MicroRNAs and their regulatory roles in plant-environment interactions. Annu. Rev. Plant Biol. 2019, 70, 489–525. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.W.; Bartel, D.P. Computational identification of plant microRNAs and their targets, including a stress-induced miRNA. Mol. Cell 2004, 14, 787–799. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Zhu, J.K. Novel and stress-regulated microRNAs and other small RNAs from Arabidopsis. Plant Cell 2004, 16, 2001–2019. [Google Scholar] [CrossRef]
- Li, Y.; Li, X.; Yang, J.; He, Y. Natural antisense transcripts of MIR398 genes suppress microR398 processing and attenuate plant thermotolerance. Nat. Commun. 2020, 11, 5351. [Google Scholar] [CrossRef] [PubMed]
- Bouché, N. New insights into miR398 functions in Arabidopsis. Plant Signal. Behav. 2010, 6, 684–686. [Google Scholar] [CrossRef] [PubMed]
- Bonnet, E.; Wuyts, J.; Rouze, P.; Van, D.P.Y. Detection of 91 potential conserved plant microRNAs in Arabidopsis thaliana and Oryza sativa identifies important target genes. Proc. Natl. Acad. Sci. USA 2004, 101, 11511–11516. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Lu, X.; Zeng, H.; Zhang, Y.; Zhu, J. Heat stress induction of miR398 triggers a regulatory loop that is critical for thermotolerance in Arabidopsis. Plant J. 2013, 74, 840–851. [Google Scholar] [CrossRef]
- Jagadeeswaran, G.; Saini, A.; Sunkar, R. Biotic and abiotic stress down-regulate miR398 expression in Arabidopsis. Planta 2009, 229, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Kapoor, A.; Zhu, J.K. Posttranscriptional induction of two Cu/Zn superoxide dismutase genes in Arabidopsis is mediated by downregulation of miR398 and important for oxidative stress tolerance. Plant Cell 2006, 18, 2051–2065. [Google Scholar] [CrossRef]
- Abdel-Ghany, S.E.; Pilon, M. MicroRNA-mediated systemic down-regulation of copper protein expression in response to low copper availability in Arabidopsis. J. Biol. Chem. 2008, 283, 15932–15945. [Google Scholar] [CrossRef]
- Jia, X.; Wang, W.X.; Ren, L.; Chen, Q.J.; Mendu, V.; Willcut, B.; Dinkins, R.; Tang, X.; Tang, G. Differential and dynamic regulation of miR398 in response to ABA and salt stress in Populus tremula and Arabidopsis thaliana. Plant Mol. Biol. 2009, 71, 51–59. [Google Scholar] [CrossRef] [PubMed]
- Yamasaki, H.; Abdel-Ghany, S.E.; Cohu, C.M.; Kobayashi, Y.; Shikanai, T.; Pilon, M. Regulation of copper homeostasis by micro-RNA in Arabidopsis. J. Biol. Chem. 2007, 282, 16369–16378. [Google Scholar] [CrossRef]
- Larkindale, J.; Mishkind, M.; Vierling, E. Plant Abiotic Stress; Jenks, M.A., Hasegawa, P.M., Eds.; Blackwell: Burlington, NC, USA, 2005; pp. 100–144. [Google Scholar]
- Tian, X.; Wang, F.; Zhao, Y.; Lan, T.; Yu, K.; Zhang, L.; Qin, Z.; Hu, Z.; Yao, Y.; Ni, Z.; et al. Heat shock transcription factor A1b regulates heat tolerance in wheat and Arabidopsis through OPR3 and jasmonate signalling pathway. Plant Biotechnol. J. 2020, 18, 1109–1111. [Google Scholar] [CrossRef] [PubMed]
- Battisti, D.S.; Naylor, R.L. Historical warnings of future food insecurity with unprecedented seasonal heat. Science 2009, 323, 240–244. [Google Scholar] [CrossRef] [PubMed]
- Sunkar, R.; Li, Y.F.; Jagadeeswaran, G. Functions of microRNAs in plant stress responses. Trends Plant Sci. 2012, 17, 196–203. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Z.H.; Lin, Z.G.; Chen, H.H.; Chen, Z.H.; Center, G.C. Temporal and spatial variation of temperature suitability index for Brassica parachinesis in Guangdong. Guangdong Agric. Sci. 2016, 3, 66–71. [Google Scholar]
- Jiang, J.; Bai, J.; Li, S.; Li, X.; Yang, L.; He, Y. HTT2 promotes plant thermotolerance in Brassica rapa. BMC Plant Biol. 2018, 18, 127. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zhang, S.; Bai, J.; He, Y. Tuning growth cycles of Brassica crops via natural antisense transcripts of BrFLC. Plant Biotechnol. J. 2016, 14, 905–914. [Google Scholar] [CrossRef] [PubMed]
- Ren, W.; Wu, F.; Bai, J.; Li, X.; Yang, X.; Xue, W.; Liu, H.; He, Y. BcpLH organizes a specific subset of microRNAs to form a leafy head in Chinese cabbage (Brassica rapa ssp. pekinensis). Hortic. Res. 2020, 7, 1. [Google Scholar] [CrossRef]
- Schwab, R.; Ossowski, S.; Riester, M.; Warthmann, N.; Weigel, D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 2006, 18, 1121–1133. [Google Scholar] [CrossRef]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef]
- Franco-Zorrilla, J.M.; Valli, A.; Todesco, M.; Mateos, I.; Puga, M.I.; Rubio-Somoza, I.; Leyva, A.; Weigel, D.; Garcia, J.A.; Paz-Ares, J. Target mimicry provides a new mechanism for regulation of microRNA activity. Nat. Genet 2007, 39, 1033–1037. [Google Scholar] [CrossRef] [PubMed]
- Nooden, L.D.; Hillsberg, J.W.; Schneider, M.J. Induction of leaf senescence in Arabidopsis thaliana by long days through a light-dosage effect. Physiol. Plant. 1996, 96, 491–495. [Google Scholar] [CrossRef]
- He, Y.; Bai, J.; Wu, F.; Mao, Y. In planta transformation of Brassica rapa and B. napus via vernalization-infiltration methods. Protoc. Exch. 2013. [Google Scholar] [CrossRef]
- Liu, Z.; Jia, L.; Wang, H.; He, Y. HYL1 regulates the balance between adaxial and abaxial identity for leaf flattening via miRNA-mediated pathways. J. Exp. Bot. 2011, 62, 4367–4381. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef] [PubMed]
- Varkonyi-Gasic, E.; Wu, R.; Wood, M.; Walton, E.F.; Hellens, R.P. Protocol: A highly sensitive RT-PCR method for detection and quantification of microRNAs. Plant Methods 2007, 3, 12. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔ CT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Piast, M.; Kustrzeba-Wojcicka, I.; Matusiewicz, M.; Banas, T. Molecular evolution of enolase. Acta Biochim. Pol. 2005, 52, 507–513. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chen, L.; Zhang, L.; Li, X.; Liu, Y.; Wu, Z.; Dong, F.; Wan, L.; Liu, K.; Hong, D.; et al. BnaC9.SMG7b Functions as a Positive Regulator of the Number of Seeds per Silique in Brassica napus by Regulating the Formation of Functional Female Gametophytes. Plant Physiol. 2015, 169, 2744–2760. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zhang, X.; Yao, Q.; Yuan, Y.; Li, X.; Wei, F.; Zhao, Y.; Zhang, Q.; Wang, Z.; Jiang, W.; et al. The miRNAs and their regulatory networks responsible for pollen abortion in Ogura-CMS Chinese cabbage revealed by high-throughput sequencing of miRNAs, degradomes, and transcriptomes. Front. Plant Sci. 2015, 6, 894. [Google Scholar] [CrossRef][Green Version]
- Yu, X.; Willmann, M.R.; Anderson, S.J.; Gregory, B.D. Genome-Wide Mapping of Uncapped and Cleaved Transcripts Reveals a Role for the Nuclear mRNA Cap-Binding Complex in Cotranslational RNA Decay in Arabidopsis. Plant Cell 2016, 28, 2385–2397. [Google Scholar] [CrossRef] [PubMed]
- Martin, M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet. J. 2011, 17, 10. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Lu, X.; Guan, Q.; Zhu, J. Downregulation of CSD2 by a heat-inducible miR398 is required for thermotolerance in Arabidopsis. Plant Signal Behav. 2013, 8, e54952. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yu, X.; Wang, H.; Lu, Y.; de Ruiter, M.; Cariaso, M.; Prins, M.; van Tunen, A.; He, Y. Identification of conserved and novel microRNAs that are responsive to heat stress in Brassica rapa. J. Exp. Bot. 2012, 63, 1025–1038. [Google Scholar] [CrossRef]
- He, Y.H.; Tang, W.N.; Swain, J.D.; Green, A.L.; Jack, T.P.; Gan, S.S. Networking senescence-regulating pathways by using Arabidopsis enhancer trap lines. Plant Physiol. 2001, 126, 707–716. [Google Scholar] [CrossRef]
- Kishor, P.; Hong, Z.; Miao, G.H.; Hu, C.; Verma, D. Overexpression of [delta]-Pyrroline-5-Carboxylate Synthetase Increases Proline Production and Confers Osmotolerance in Transgenic Plants. Plant Physiol. 1995, 108, 1387–1394. [Google Scholar] [CrossRef] [PubMed]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot. 2007, 58, 221–227. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef]
- Xiong, L.; Zhu, J.K. Molecular and genetic aspects of plant responses to osmotic stress. Plant Cell Env. 2002, 25, 131–139. [Google Scholar] [CrossRef]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef]
- Ding, Y.F.; Wang, G.Y.; Fu, Y.P.; Zhu, C. The role of miR398 in plant stress responses. Yi Chuan 2010, 32, 129–134. [Google Scholar] [CrossRef] [PubMed]
- Huo, X.; Wang, C.; Teng, Y.; Liu, X. Identification of miRNAs associated with dark-induced senescence in Arabidopsis. BMC Plant Biol. 2015, 15, 266. [Google Scholar] [CrossRef]
- Zhu, T.; De Lima, C.F.F.; De Smet, I. The Heat is On: How Crop Growth, Development and Yield Respond to High Temperature. J. Exp. Bot. 2021, 72, 7359–7373. [Google Scholar] [CrossRef]
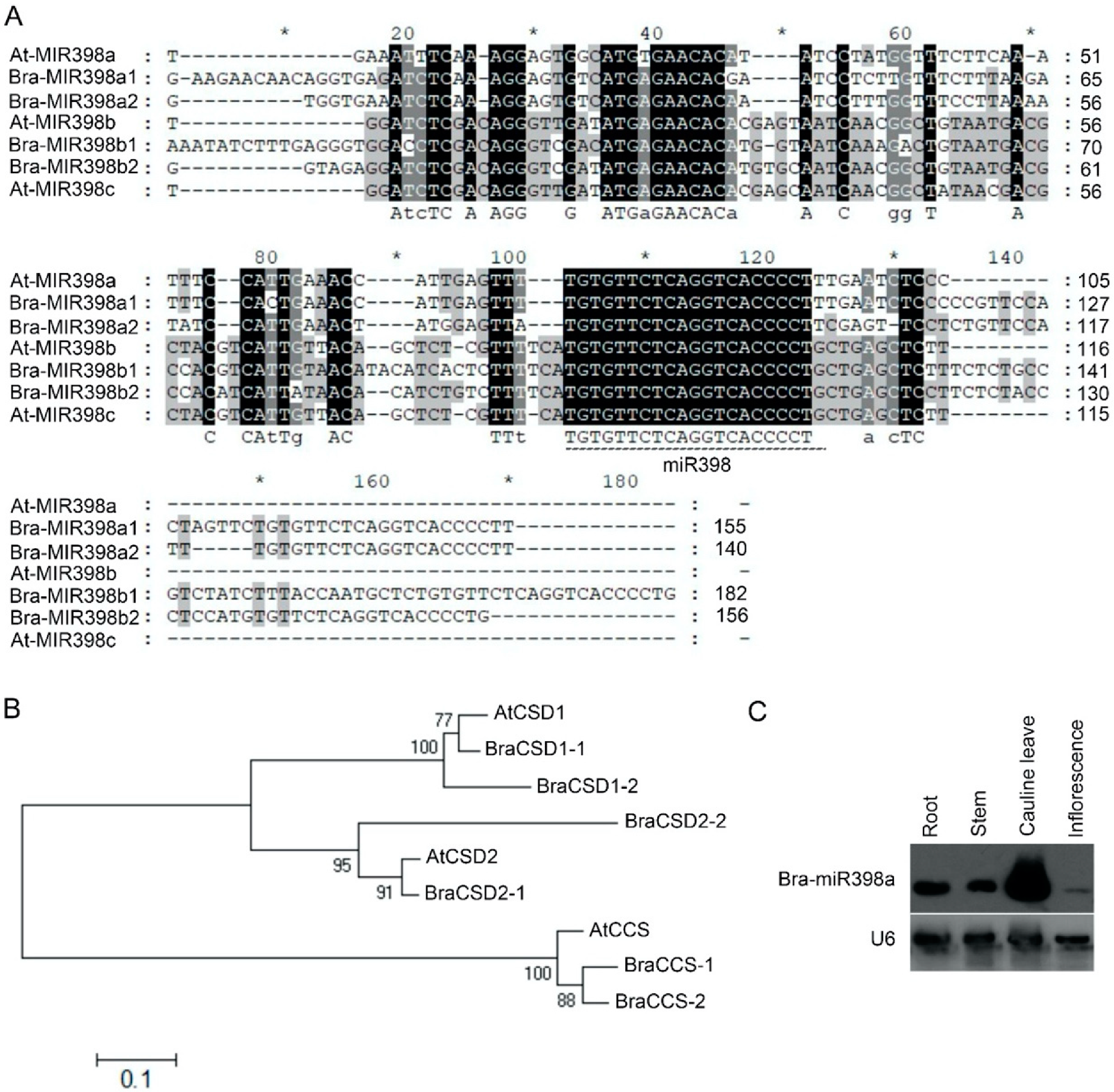



| miR398 and Targets in Arabidopsis thaliana | Homologous Genes in B. rapa |
|---|---|
| ath-miR398a | Bra-MIR398a1/a2 |
| ath-miR398b | Bra-MIR398b1/b2 |
| ath-miR398c | |
| AtCSD1 (At1g08830) | BraCSD1-1 (Bra031642) BraCSD1-2 (Bra018596) |
| AtCSD2 (At2g28190) | BraCSD2-1 (Bra034394) |
| BraCSD2-2 (Bra011971) | |
| AtCCS (At1g12520) | BraCCS-1 (Bra016768) |
| BraCCS-2 (Bra026968) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, B.; Jiang, J.; Bai, J.; Wang, X.; Li, Y.; Shao, W.; Hu, S.; He, Y.; Yu, X. miR398 Attenuates Heat-Induced Leaf Cell Death via Its Target CSD1 in Chinese Cabbage. Horticulturae 2022, 8, 299. https://doi.org/10.3390/horticulturae8040299
Cao B, Jiang J, Bai J, Wang X, Li Y, Shao W, Hu S, He Y, Yu X. miR398 Attenuates Heat-Induced Leaf Cell Death via Its Target CSD1 in Chinese Cabbage. Horticulturae. 2022; 8(4):299. https://doi.org/10.3390/horticulturae8040299
Chicago/Turabian StyleCao, Biting, Jianxia Jiang, Jinjuan Bai, Xuan Wang, Yajie Li, Wenna Shao, Shengwu Hu, Yuke He, and Xiang Yu. 2022. "miR398 Attenuates Heat-Induced Leaf Cell Death via Its Target CSD1 in Chinese Cabbage" Horticulturae 8, no. 4: 299. https://doi.org/10.3390/horticulturae8040299
APA StyleCao, B., Jiang, J., Bai, J., Wang, X., Li, Y., Shao, W., Hu, S., He, Y., & Yu, X. (2022). miR398 Attenuates Heat-Induced Leaf Cell Death via Its Target CSD1 in Chinese Cabbage. Horticulturae, 8(4), 299. https://doi.org/10.3390/horticulturae8040299







