Systematic Characterization of Brassica napus HIR Gene Family Reveals a Positive Role of BnHIR2.7 in Sclerotinia sclerotiorum Resistance
Abstract
1. Introduction
2. Materials and Methods
2.1. Plants, Strain Materials, and Growth Conditions
2.2. Identification of BnHIR Gene Family in B. napus
2.3. Phylogenetic Analysis, Gene Structure, and Protein Motif Analysis
2.4. Analysis of Cis-Acting Elements and Expression Patterns of BnHIR Genes
2.5. Overexpression Vector Construction and Plant Transformation
2.6. Phenotypic Observation of A. thaliana Transgenic and Mutant Plants
2.7. Expression Analysis of BnHIR2.7 under the Infection of S. sclerotiorum
2.8. Statistical Analysis
3. Results
3.1. Identification and Characterization of the B. napus HIR Family
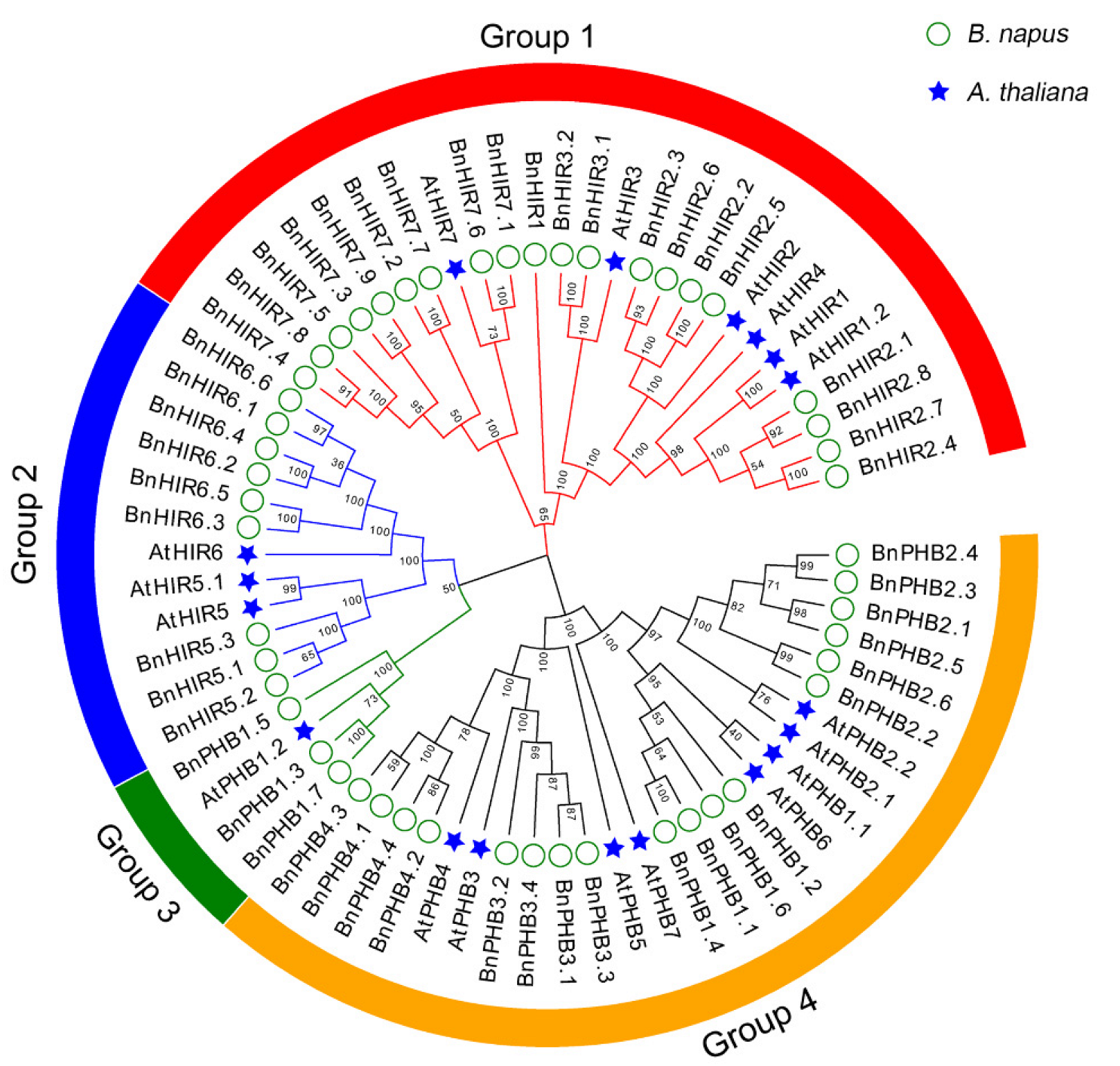
3.2. Phylogenetic Analysis of HIR Proteins in B. napus
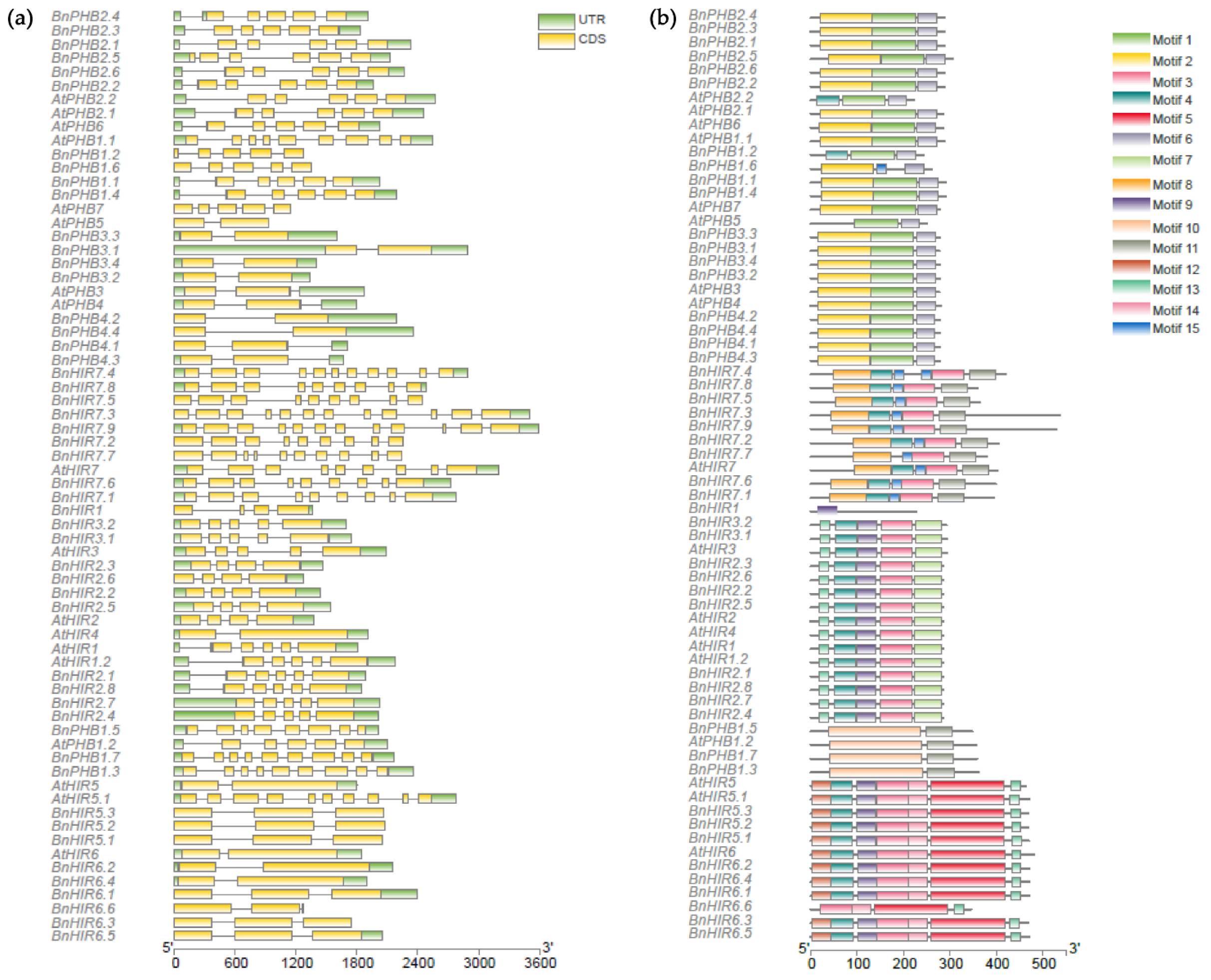
3.3. Gene Structure and Conserved Motif Analysis
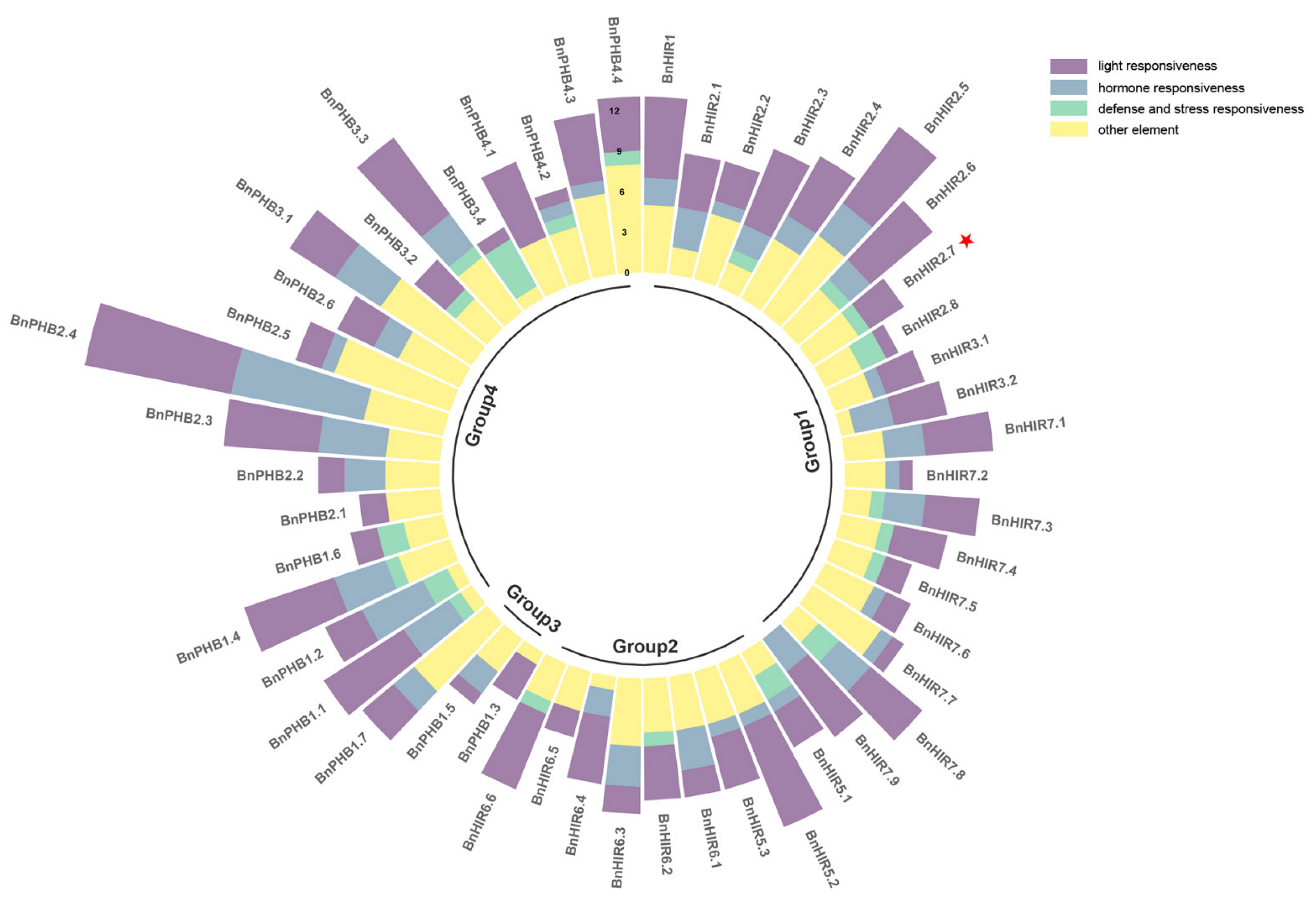
3.4. Cis-Acting Elements in the Promoter Regions of BnHIR Genes
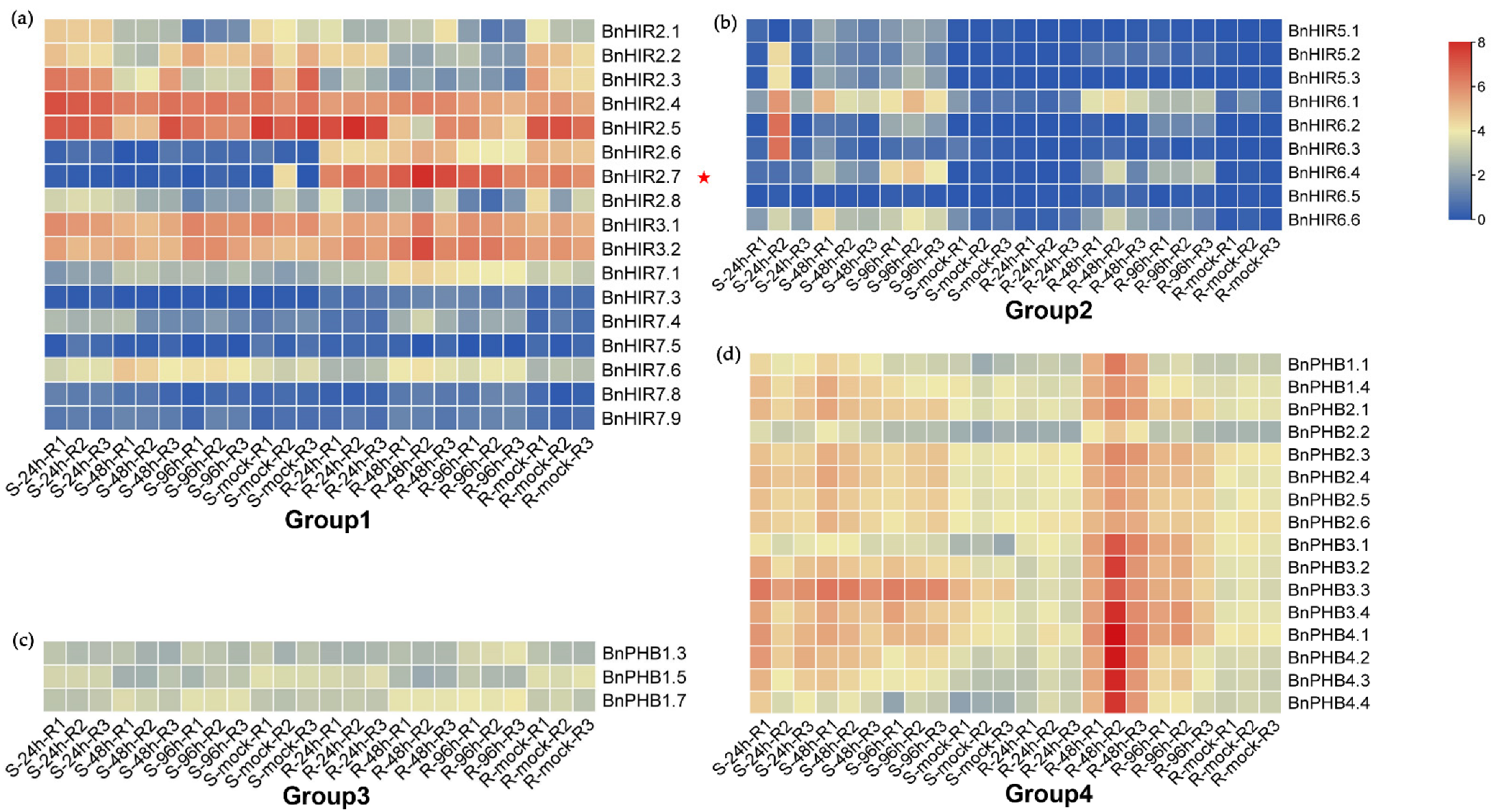
3.5. Expression Patterns of BnHIR Genes at Different Developmental Stages
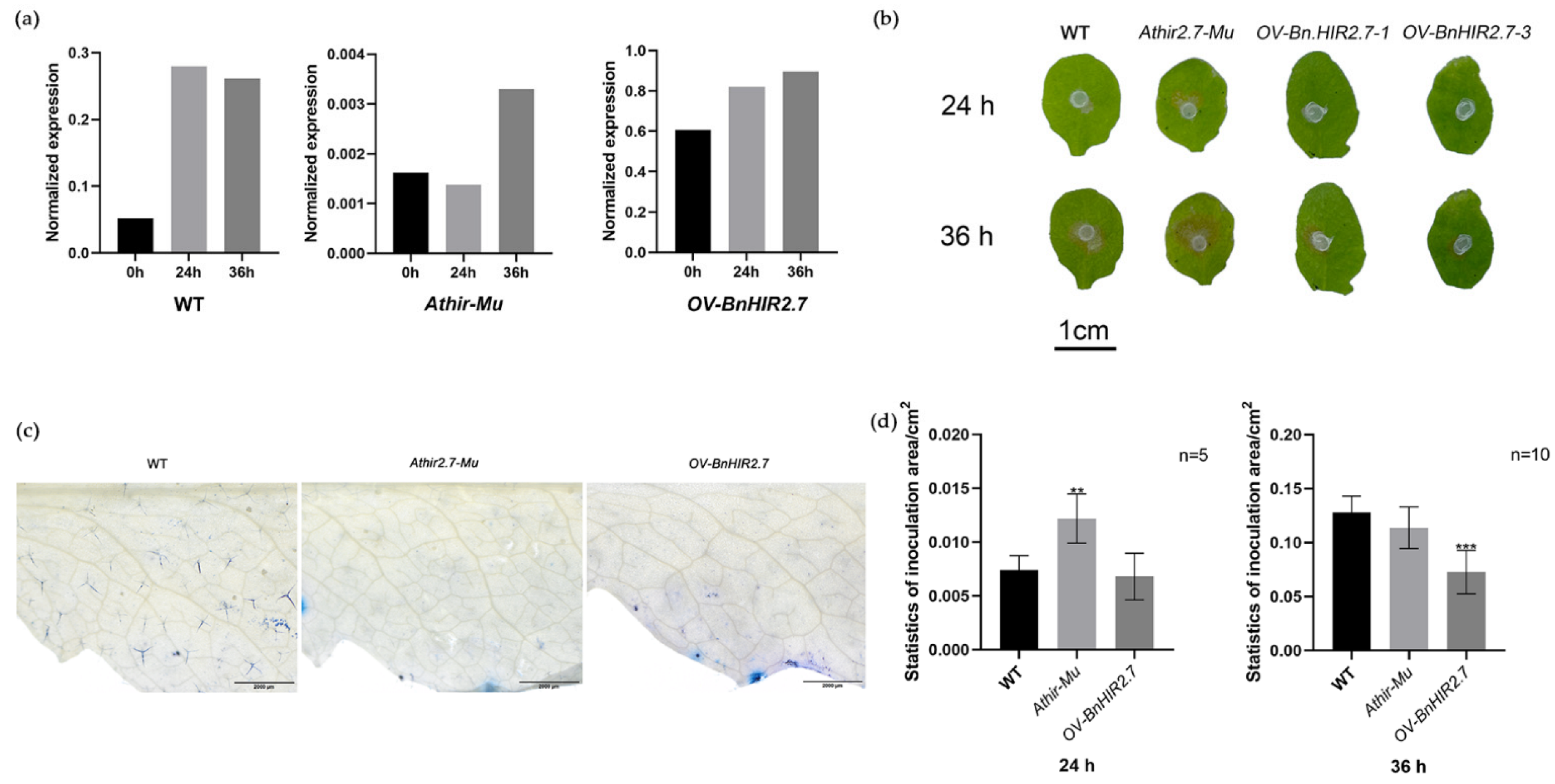
3.6. Expression Analysis of BnHIRs under S. sclerotiorum Induction
3.7. Functional Characterization of BnHIR2.7 in Transgenic Arabidopsis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Abbreviations
References
- Zuo, R.; Xie, M.; Gao, F.; Sumbal, W.; Cheng, X.; Liu, Y.; Bai, Z.; Liu, S. The Characterization of the Phloem Protein 2 Gene Family Associated with Resistance to Sclerotinia sclerotiorum in Brassica napus. Int. J. Mol. Sci. 2022, 23, 3934. [Google Scholar] [CrossRef]
- Ding, L.; Li, T.; Guo, X.; Li, M.; Liu, X.; Cao, J.; Tan, X. Sclerotinia Stem Rot Resistance in Rapeseed: Recent Progress and Future Prospects. J. Agric. Food Chem. 2021, 69, 2965–2978. [Google Scholar] [CrossRef]
- Nadimpalli, R.; Yalpani, N.; Johal, G.S.; Simmons, C.R. Prohibitins, Stomatins, and Plant Disease Response Genes Compose a Protein Superfamily That Controls Cell Proliferation, Ion Channel Regulation, and Death. J. Biol. Chem. 2000, 275, 29579–29586. [Google Scholar] [CrossRef] [PubMed]
- Atkinson, M.M.; Keppler, L.D.; Orlandi, E.W.; Baker, C.J.; Mischke, C.F. Involvement of Plasma-Membrane Calcium Influx in Bacterial Induction of the K+/H+ And Hypersensitive Responses in Tobacco. Plant Physiol. 1990, 92, 215–221. [Google Scholar] [CrossRef]
- Beers, E.P.; McDowell, J.M. Regulation and Execution of Programmed Cell Death in Response to Pathogens, Stress and Developmental Cues. Curr. Opin. Plant Biol. 2001, 4, 561–567. [Google Scholar] [CrossRef]
- Lam, E.; Kato, N.; Lawton, M. Programmed cell death, mitochondria and the plant hypersensitive response. Nature 2001, 411, 848–853. [Google Scholar] [CrossRef]
- Di, C.; Xu, W.; Su, Z.; Yuan, J.S. Comparative Genome Analysis of PHB Gene Family Reveals Deep Evolutionary Origins and Diverse Gene Function. BMC Bioinform. 2010, 11, S22. [Google Scholar] [CrossRef]
- Qi, Y.; Tsuda, K.; Nguyen, L.V.; Wang, X.; Lin, J.; Murphy, A.S.; Glazebrook, J.; Thordal-Christensen, H.; Katagiri, F. Physical Association of Arabidopsis Hypersensitive Induced Reaction Proteins (Hirs) with the Immune Receptor RPS2. J. Biol. Chem. 2011, 286, 31297–31307. [Google Scholar] [CrossRef]
- Zhou, L.; Cheung, M.Y.; Li, M.W.; Fu, Y.; Sun, Z.; Sun, S.M.; Lam, H.M. Rice Hypersensitive Induced Reaction Protein 1 (OsHIR1) Associates with Plasma Membrane and Triggers Hypersensitive Cell Death. BMC Plant Biol. 2010, 10, 290. [Google Scholar] [CrossRef]
- Duan, Y.; Guo, J.; Shi, X.; Guan, X.; Liu, F.; Bai, P.; Huang, L.; Kang, Z. Wheat Hypersensitive-Induced Reaction Genes Tahir1 and Tahir3 Are Involved in Response to Stripe Rust Fungus Infection and Abiotic Stresses. Plant Cell Rep. 2012, 32, 273–283. [Google Scholar] [CrossRef]
- Karrer, E.E.; Beachy, R.N.; Holt, C.A. Cloning of Tobacco Genes That Elicit the Hypersensitive Response. Plant Mol. Biol. 1998, 36, 681–690. [Google Scholar] [CrossRef] [PubMed]
- Wen, X.; Niu, T.; Kong, X. In Silico Analysis of PHB Gene Family in Maize. Plant Growth Regul. 2013, 73, 181–191. [Google Scholar] [CrossRef]
- Rostoks, N.; Schmierer, D.; Kudrna, D.; Kleinhofs, A. Barley Putative Hypersensitive Induced Reaction Genes: Genetic Mapping, Sequence Analyses and Differential Expression in Disease Lesion Mimic Mutants. Theor. Appl. Genet. 2003, 107, 1094–1101. [Google Scholar] [CrossRef]
- Jung, H.W.; Hwang, B.K. The Leucine-Rich Repeat (LRR) Protein, Calrr1, Interacts with the Hypersensitive Induced Reaction (HIR) Protein, Cahir1, and Suppresses Cell Death Induced by the Cahir1 Protein. Mol. Plant Pathol. 2007, 8, 503–514. [Google Scholar] [CrossRef]
- Chalhoub, B.; Denoeud, F.; Liu, S.; Parkin, I.A.P.; Tang, H.; Wang, X.; Chiquet, J.; Belcram, H.; Tong, C.; Samans, B.; et al. Early Allopolyploid Evolution in the Post-Neolithic Brassica napus Oilseed Genome. Science 2014, 345, 950–953. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Altschul, S.F.; Madden, T.L.; Schaffer, A.A.; Zhang, J.H.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A New Generation of Protein Database Search Programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef]
- Fan, Y.; Yu, M.; Liu, M.; Zhang, R.; Sun, W.; Qian, M.; Duan, H.; Chang, W.; Ma, J.; Qu, C.; et al. Genome-Wide Identification, Evolutionary and Expression Analyses of the GALACTINOL SYNTHASE Gene Family in Rapeseed and Tobacco. Int. J. Mol. Sci. 2017, 18, 2768. [Google Scholar] [CrossRef]
- Chou, K.C.; Shen, H.B. Plant-mPLoc: A Top-Down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: Multiple Sequence Alignment with High Accuracy and High Throughput. Nucleic Acids Res. 2004, 32, 1792–1797. [Google Scholar] [CrossRef]
- Guindon, S.; Dufayard, J.F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Lefort, V.; Longueville, J.E.; Gascuel, O. SMS: Smart Model Selection in PhyML. Mol. Biol. Evol. 2017, 34, 2422–2424. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.H. Evolview v3: A Webserver for Visualization, Annotation, and Management of Phylogenetic Trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and Analyzing DNA and Protein Sequence Motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant. 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a Database of Plant Cis-Acting Regulatory Elements and a Portal to Tools for In Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Chao, H.; Li, T.; Luo, C.; Huang, H.; Ruan, Y.; Li, X.; Niu, Y.; Fan, Y.; Sun, W.; Zhang, K.; et al. BrassicaEDB: A Gene Expression Database for Brassica Crops. Int. J. Mol. Sci. 2020, 21, 5831. [Google Scholar] [CrossRef]
- Abedi, A.; Hajiahmadi, Z.; Kordrostami, M.; Esmaeel, Q.; Jacquard, C. Analyses of Lysin-motif Receptor-like Kinase (LysM-RLK) Gene Family in Allotetraploid Brassica napus L. and Its Progenitor Species: An In Silico Study. Cells 2021, 11, 37. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA Extraction Protocol for Plants Containing High Polysaccharide and Polyphenol Components. Plant Mol. Biol. Rep. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Schmidt, R.; Willmitzer, L. High Efficiency Agrobacterium tumefaciens-Mediated-Transformation of Arabidopsis thaliana Leaf and Cotyledon Explants. Plant Cell Rep. 1988, 7, 583–586. [Google Scholar] [CrossRef]
- Lu, K.; Li, T.; He, J.; Chang, W.; Zhang, R.; Liu, M.; Yu, M.; Fan, Y.; Ma, J.; Sun, W.; et al. qPrimerDB: A Thermodynamics-Based Gene-Specific Qrt-PCR Primer Database for 147 Organisms. Nucleic Acids Res. 2018, 46, D1229–D1236. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE Guidelines: Minimum Information for Publication of Quantitative Real-Time PCR Experiments. Clin. Chem. 2009, 55, 611–622. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Zhang, L.; Wu, Y.; Cao, Y.; Lu, C. Comparison of Five Endogenous Reference Genes for Specific PCR Detection and Quantification of Brassica napus. J. Agric. Food Chem. 2010, 58, 2812–2817. [Google Scholar] [CrossRef] [PubMed]
- Swift, M.L. GraphPad Prism, Data Analysis, and Scientific Graphing. J. Chem. Inf. Comput. Sci. 1997, 37, 411–412. [Google Scholar] [CrossRef]
- Jung, H.W.; Lim, C.W.; Lee, S.C.; Choi, H.W.; Hwang, C.H.; Hwang, B.K. Distinct Roles of the Pepper Hypersensitive Induced Reaction Protein Gene Cahir1 in Disease and Osmotic Stress, as Determined by Comparative Transcriptome and Proteome Analyses. Planta 2008, 227, 409–425. [Google Scholar] [CrossRef]
- Morrow, I.C.; Parton, R.G. Flotillins and the PHB Domain Protein Family: Rafts, Worms and Anaesthetics. Traffic 2005, 6, 725–740. [Google Scholar] [CrossRef]
- Huang, F.; Ye, X.; Wang, Z.; Ding, Y.; Cai, X.; Yu, L.; Waseem, M.; Abbas, F.; Ashraf, U.; Chen, X.; et al. The prohibitins (PHB) Gene Family in Tomato: Bioinformatic Identification and Expression Analysis Under Abiotic and Phytohormone Stresses. GM Crop. Food 2021, 12, 535–550. [Google Scholar] [CrossRef]
- Song, M.; Peng, X.; Du, C.; Lei, L.; Zhang, T.; Xiang, Y. Genome-Wide Analysis of the PHB Gene Family in Glycine max (L.). Merr. Genes Genom. 2017, 39, 1095–1106. [Google Scholar] [CrossRef]
- Van Aken, O.; Pečenková, T.; van de Cotte, B.; De Rycke, R.; Eeckhout, D.; Fromm, H.; De Jaeger, G.; Witters, E.; Beemster, G.T.; Inzé, D.; et al. Mitochondrial Type-I Prohibitins of Arabidopsis thaliana are required for Supporting Proficient Meristem Development. Plant J. 2007, 52, 850–864. [Google Scholar] [CrossRef]
- Wang, Y.; Ries, A.; Wu, K.; Yang, A.; Crawford, N.M. The Arabidopsis prohibitin Gene PHB3 Functions in Nitric Oxide-Mediated Responses and in Hydrogen Peroxide-Induced Nitric Oxide Accumulation. Plant Cell 2010, 22, 249–259. [Google Scholar] [CrossRef]
- Artal-Sanz, M.; Tsang, W.Y.; Willems, E.M.; Grivell, L.A.; Lemire, B.D.; van der Spek, H.; Nijtmans, L.G. The Mitochondrial Prohibitin Complex Is Essential for Embryonic Viability and Germline Function in Caenorhabditis elegans. J. Biol. Chem. 2003, 278, 32091–32099. [Google Scholar] [CrossRef]
- Chen, J.C.; Jiang, C.Z.; Reid, M.S. Silencing a Prohibitin Alters Plant Development and Senescence. Plant J. 2005, 44, 16–24. [Google Scholar] [CrossRef]
- Ahn, C.S.; Lee, J.H.; Reum Hwang, A.; Kim, W.T.; Pai, H.S. Prohibitin is Involved in Mitochondrial Biogenesis in Plants. Plant J. 2006, 46, 658–667. [Google Scholar] [CrossRef]
- Artal-Sanz, M.; Tavernarakis, N. Prohibitin Couples Diapause Signalling to Mitochondrial Metabolism During Ageing in C. elegans. Nature 2009, 461, 793–797. [Google Scholar] [CrossRef]
- Merkwirth, C.; Langer, T. Prohibitin Function Within Mitochondria: Essential Roles for Cell Proliferation and Cristae Morphogenesis. Biochim. Biophys. Acta 2009, 1793, 27–32. [Google Scholar] [CrossRef]
- Merkwirth, C.; Dargazanli, S.; Tatsuta, T.; Geimer, S.; Löwer, B.; Wunderlich, F.T.; von Kleist-Retzow, J.C.; Waisman, A.; Westermann, B.; Langer, T. Prohibitins Control Cell Proliferation and Apoptosis by Regulating OPA1-Dependent Cristae Morphogenesis in Mitochondria. Genes 2008, 22, 476–488. [Google Scholar] [CrossRef]
- Wang, Z.; Fang, H.; Chen, Y.; Chen, K.; Li, G.; Gu, S.; Tan, X. Overexpression of BnWRKY33 in Oilseed Rape Enhances Resistance to Sclerotinia sclerotiorum. Mol. Plant Pathol. 2015, 15, 677–689. [Google Scholar] [CrossRef]
- Hu, H.; Tang, Y.; Wu, J.; Chen, F.; Yang, Y.; Pan, X.; Dong, X.; Jin, X.; Liu, S.; Du, X. Brassica napus Mediator Subunit16 Induces BnMED25- and BnWRKY33-Activated Defense Signaling to Confer Sclerotinia sclerotiorum Resistance. Front. Plant Sci. 2021, 12, 663536. [Google Scholar] [CrossRef]
- Sun, Q.; Lin, L.; Liu, D.; Wu, D.; Fang, Y.; Wu, J.; Wang, Y. CRISPR/Cas9-Mediated Multiplex Genome Editing of the BnWRKY11 and BnWRKY70 Genes in Brassica napus L. Int. J. Mol. Sci. 2018, 19, 2716. [Google Scholar] [CrossRef]
- Wang, Z.; Bao, L.; Zhao, F.; Tang, M.; Chen, T.; Li, Y.; Wang, B.; Fu, B.; Fang, H.; Li, G.; et al. BnaMPK3 Is a Key Regulator of Defense Responses to the Devastating Plant Pathogen Sclerotinia sclerotiorum in Oilseed Rape. Front. Plant Sci. 2019, 10, 91. [Google Scholar] [CrossRef]
- He, Y.; Zhang, Z.; Xu, Y.; Chen, S.; Cai, X. Genome-Wide Identification of Rapid Alkalinization Factor Family in Brassica napus and Functional Analysis of BnRALF10 in Immunity to Sclerotinia sclerotiorum. Front. Plant Sci. 2022, 3, 877404. [Google Scholar] [CrossRef]

| Gene Name | Gene ID | Homologs in A. thaliana | Amino Acids | MW (kDa) a | pI b | Subcellular Localization |
|---|---|---|---|---|---|---|
| BnHIR5.1 | BnaA02g32070D | AT5G25260 | 469 | 51,982.35 | 5.57 | Nucleus |
| BnHIR5.2 | BnaA02g32090D | AT5G25260 | 468 | 51,756.07 | 5.42 | Nucleus |
| BnHIR5.3 | BnaC02g40750D | AT5G25260 | 467 | 51,708.93 | 5.40 | Nucleus |
| BnHIR6.1 | BnaA02g34440D | AT5G64870 | 469 | 51,663.18 | 6.12 | Nucleus |
| BnHIR6.2 | BnaA06g23810D | AT5G64870 | 470 | 51,848.36 | 5.95 | Nucleus |
| BnHIR6.3 | BnaA07g33580D | AT5G64870 | 467 | 51,499.11 | 6.60 | Nucleus |
| BnHIR6.4 | BnaC03g49410D | AT5G64870 | 470 | 51,853.33 | 5.88 | Nucleus |
| BnHIR6.5 | BnaC06g38190D | AT5G64870 | 469 | 51,551.08 | 6.39 | Nucleus |
| BnHIR6.6 | BnaCnng58920D | AT5G64870 | 345 | 37,805.16 | 6.13 | Nucleus |
| BnHIR1 | BnaC09g53100D | AT5G62740 | 227 | 25,972.59 | 4.99 | Chloroplast, Nucleus |
| BnHIR2.1 | BnaA02g35710D | AT1G69840 | 285 | 31,218.54 | 5.49 | Mitochondrion |
| BnHIR2.2 | BnaA03g27520D | AT3G01290 | 286 | 31,610.32 | 5.89 | Mitochondrion |
| BnHIR2.3 | BnaA05g34000D | AT3G01290 | 286 | 31,529.24 | 6.01 | Mitochondrion |
| BnHIR2.4 | BnaA07g28360D | AT1G69840 | 286 | 31,316.54 | 5.27 | Mitochondrion |
| BnHIR2.5 | BnaC03g32530D | AT3G01290 | 286 | 31,610.32 | 5.89 | Mitochondrion |
| BnHIR2.6 | BnaC05g48690D | AT3G01290 | 286 | 31,529.24 | 6.01 | Mitochondrion |
| BnHIR2.7 | BnaC06g30890D | AT1G69840 | 286 | 31,316.54 | 5.27 | Mitochondrion |
| BnHIR2.8 | BnaCnng44030D | AT1G69840 | 285 | 31,087.34 | 5.35 | Mitochondrion |
| BnHIR3.1 | BnaA03g13070D | AT5G51570 | 292 | 32,465.11 | 5.28 | Mitochondrion |
| BnHIR3.2 | BnaC03g15940D | AT5G51570 | 292 | 32,366.02 | 5.38 | Mitochondrion |
| BnPHB1.1 | BnaA01g08360D | AT4G28510 | 290 | 32,048.98 | 9.45 | Mitochondrion |
| BnPHB1.2 | BnaA08g13690D | AT4G28510 | 242 | 27,374.50 | 8.69 | Mitochondrion |
| BnPHB1.3 | BnaA09g19330D | AT2G03510 | 361 | 41,160.82 | 5.44 | Mitochondrion |
| BnPHB1.4 | BnaC01g09910D | AT4G28510 | 290 | 32,048.98 | 9.45 | Mitochondrion |
| BnPHB1.5 | BnaC02g47990D | AT2G03510 | 347 | 39,719.46 | 6.10 | Mitochondrion |
| BnPHB1.6 | BnaC08g13090D | AT4G28510 | 260 | 28,591.94 | 9.06 | Mitochondrion |
| BnPHB1.7 | BnaC09g21530D | AT2G03510 | 358 | 40,821.52 | 5.56 | Mitochondrion |
| BnPHB2.1 | BnaA08g27620D | AT1G03860 | 288 | 31,909.77 | 9.37 | Mitochondrion |
| BnPHB2.2 | BnaA09g50790D | AT1G03860 | 288 | 32,033.87 | 9.39 | Mitochondrion |
| BnPHB2.3 | BnaA10g02220D | AT1G03860 | 288 | 31,923.74 | 9.35 | Mitochondrion |
| BnPHB2.4 | BnaC05g02140D | AT1G03860 | 288 | 31,908.68 | 9.37 | Mitochondrion |
| BnPHB2.5 | BnaC08g00370D | AT1G03860 | 306 | 34,181.58 | 9.55 | Mitochondrion |
| BnPHB2.6 | BnaC08g43560D | AT1G03860 | 288 | 31,991.83 | 9.37 | Mitochondrion |
| BnPHB3.1 | BnaA04g10580D | AT5G40770 | 277 | 30,481.01 | 7.94 | Mitochondrion |
| BnPHB3.2 | BnaA07g15170D | AT5G40770 | 277 | 30,446.99 | 7.94 | Mitochondrion |
| BnPHB3.3 | BnaC04g32860D | AT5G40770 | 277 | 30,481.01 | 7.94 | Mitochondrion |
| BnPHB3.4 | BnaC06g13200D | AT5G40770 | 277 | 30,481.01 | 7.94 | Mitochondrion |
| BnPHB4.1 | BnaA06g32230D | AT3G27280 | 278 | 30,372.77 | 6.94 | Mitochondrion |
| BnPHB4.2 | BnaA09g01960D | AT3G27280 | 278 | 30,330.69 | 6.94 | Mitochondrion |
| BnPHB4.3 | BnaC07g24140D | AT3G27280 | 278 | 30,367.75 | 6.99 | Mitochondrion |
| BnPHB4.4 | BnaC09g01400D | AT3G27280 | 278 | 30,330.69 | 6.94 | Mitochondrion |
| BnHIR7.1 | BnaA01g16560D | AT4G27585 | 395 | 43,279.16 | 6.30 | Mitochondrion |
| BnHIR7.2 | BnaA02g09860D | AT4G27585 | 404 | 43,570.06 | 9.05 | Mitochondrion |
| BnHIR7.3 | BnaA02g09870D | AT4G27585 | 535 | 58,018.04 | 5.71 | Nucleus |
| BnHIR7.4 | BnaA03g12010D | AT4G27585 | 420 | 45,807.70 | 8.50 | Mitochondrion |
| BnHIR7.5 | BnaA10g07200D | AT4G27585 | 364 | 39,713.44 | 8.89 | Mitochondrion |
| BnHIR7.6 | BnaC01g20150D | AT4G27585 | 398 | 43,625.71 | 6.74 | Mitochondrion |
| BnHIR7.7 | BnaC02g13810D | AT4G27585 | 378 | 40,677.62 | 9.13 | Nucleus |
| BnHIR7.8 | BnaC03g14740D | AT4G27585 | 359 | 39,006.84 | 9.02 | Mitochondrion |
| BnHIR7.9 | BnaC02g13820D | AT4G27585 | 528 | 57,171.01 | 5.67 | Mitochondrion |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, M.; Tang, Y.; Yu, M.; Fan, Y.; Khan, S.U.; Chang, W.; Li, X.; Wei, S.; Wei, L.; Qu, C.; et al. Systematic Characterization of Brassica napus HIR Gene Family Reveals a Positive Role of BnHIR2.7 in Sclerotinia sclerotiorum Resistance. Horticulturae 2022, 8, 874. https://doi.org/10.3390/horticulturae8100874
Li M, Tang Y, Yu M, Fan Y, Khan SU, Chang W, Li X, Wei S, Wei L, Qu C, et al. Systematic Characterization of Brassica napus HIR Gene Family Reveals a Positive Role of BnHIR2.7 in Sclerotinia sclerotiorum Resistance. Horticulturae. 2022; 8(10):874. https://doi.org/10.3390/horticulturae8100874
Chicago/Turabian StyleLi, Mengqi, Yuqiao Tang, Mengna Yu, Yonghai Fan, Shahid Ullah Khan, Wei Chang, Xiaodong Li, Siyu Wei, Lijuan Wei, Cunmin Qu, and et al. 2022. "Systematic Characterization of Brassica napus HIR Gene Family Reveals a Positive Role of BnHIR2.7 in Sclerotinia sclerotiorum Resistance" Horticulturae 8, no. 10: 874. https://doi.org/10.3390/horticulturae8100874
APA StyleLi, M., Tang, Y., Yu, M., Fan, Y., Khan, S. U., Chang, W., Li, X., Wei, S., Wei, L., Qu, C., Li, J., & Lu, K. (2022). Systematic Characterization of Brassica napus HIR Gene Family Reveals a Positive Role of BnHIR2.7 in Sclerotinia sclerotiorum Resistance. Horticulturae, 8(10), 874. https://doi.org/10.3390/horticulturae8100874







