A Preliminary Investigation on the Functional Validation and Interactions of PoWOX Genes in Peony (Paeonia ostii)
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Screening and Cloning of PoWOX Genes
2.3. Analysis of Physicochemical Properties and Phylogenetic Tree of PoWOX Genes
2.4. Multiple Sequence Alignment and Conserved Structural Domain Analysis of PoWOX Genes
2.5. Quantitative Fluorescence Analysis
2.6. Subcellular Localization Assay of PoWOX Proteins in Tobacco
2.7. Yeast Two-Hybrid Assay
2.7.1. Vector Construction
2.7.2. Self-Activation Verification
2.7.3. Yeast Co-Transformation
2.8. Bimolecular Fluorescence Complementation Experiments
3. Results and Analysis
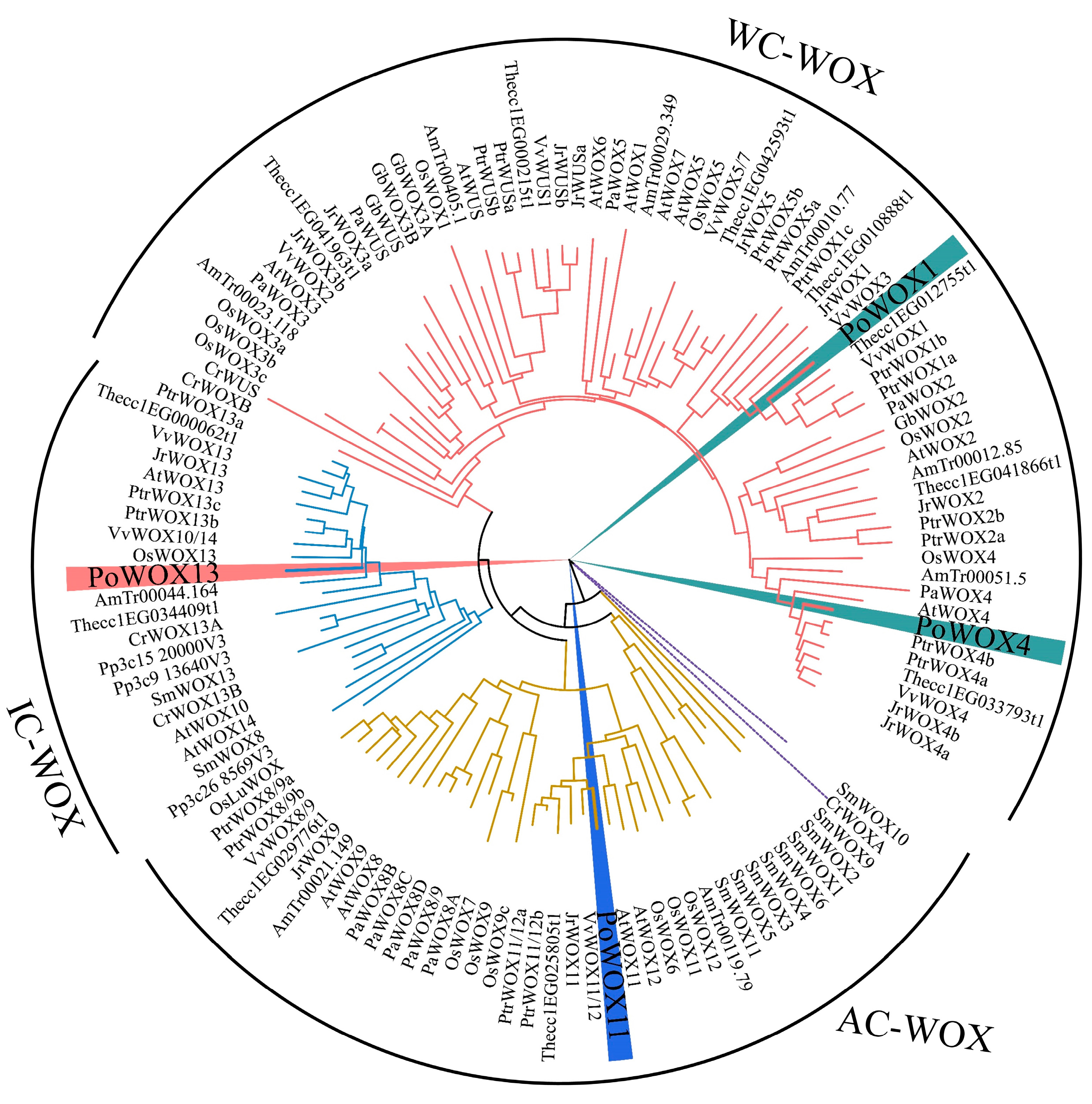
3.1. Cloning and Phylogenetic Analysis of PoWOX Genes in Peony
3.2. Multiple Sequence Alignment and Sequence Characterization Analysis of PoWOX Proteins in Peony
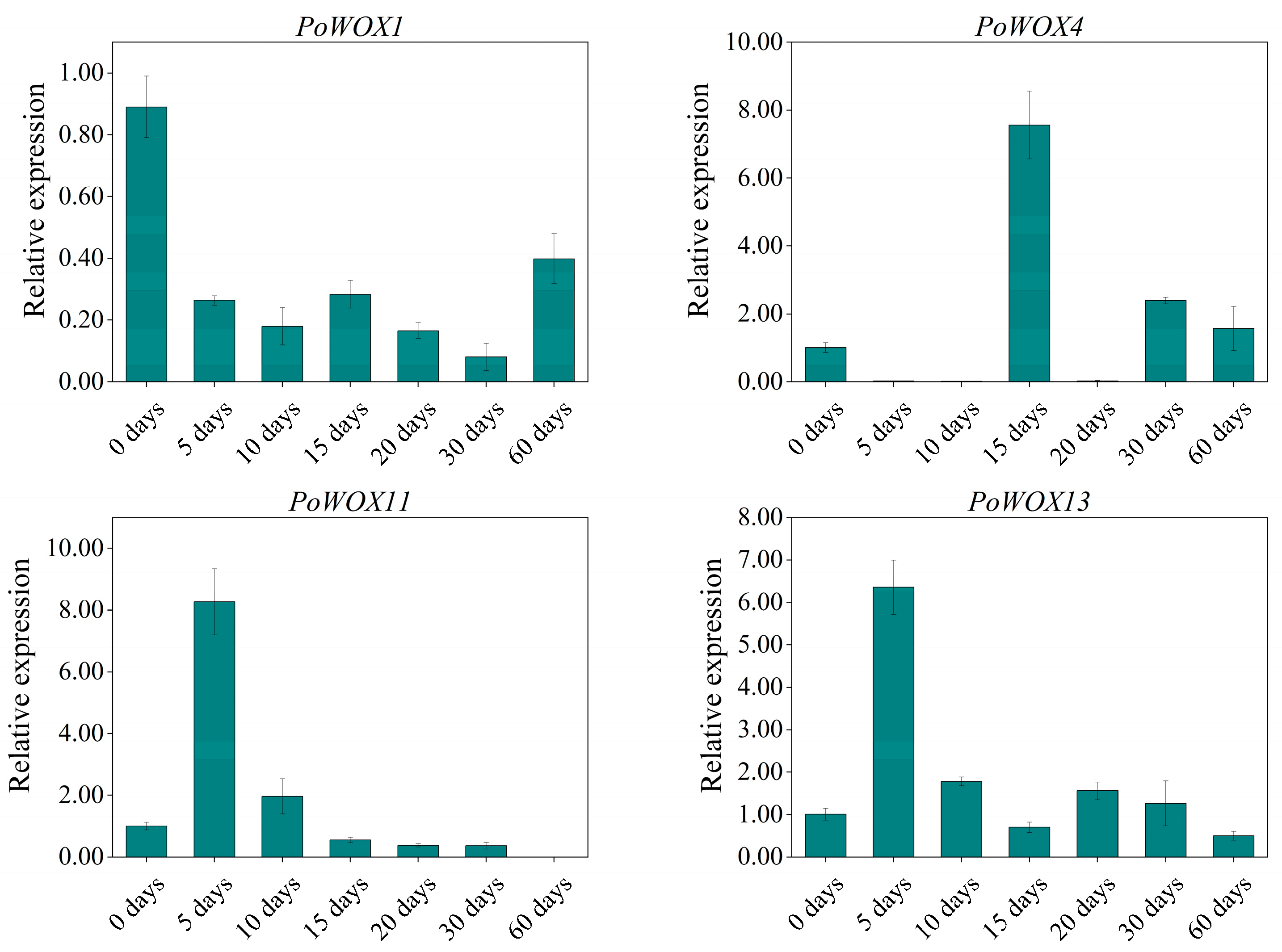
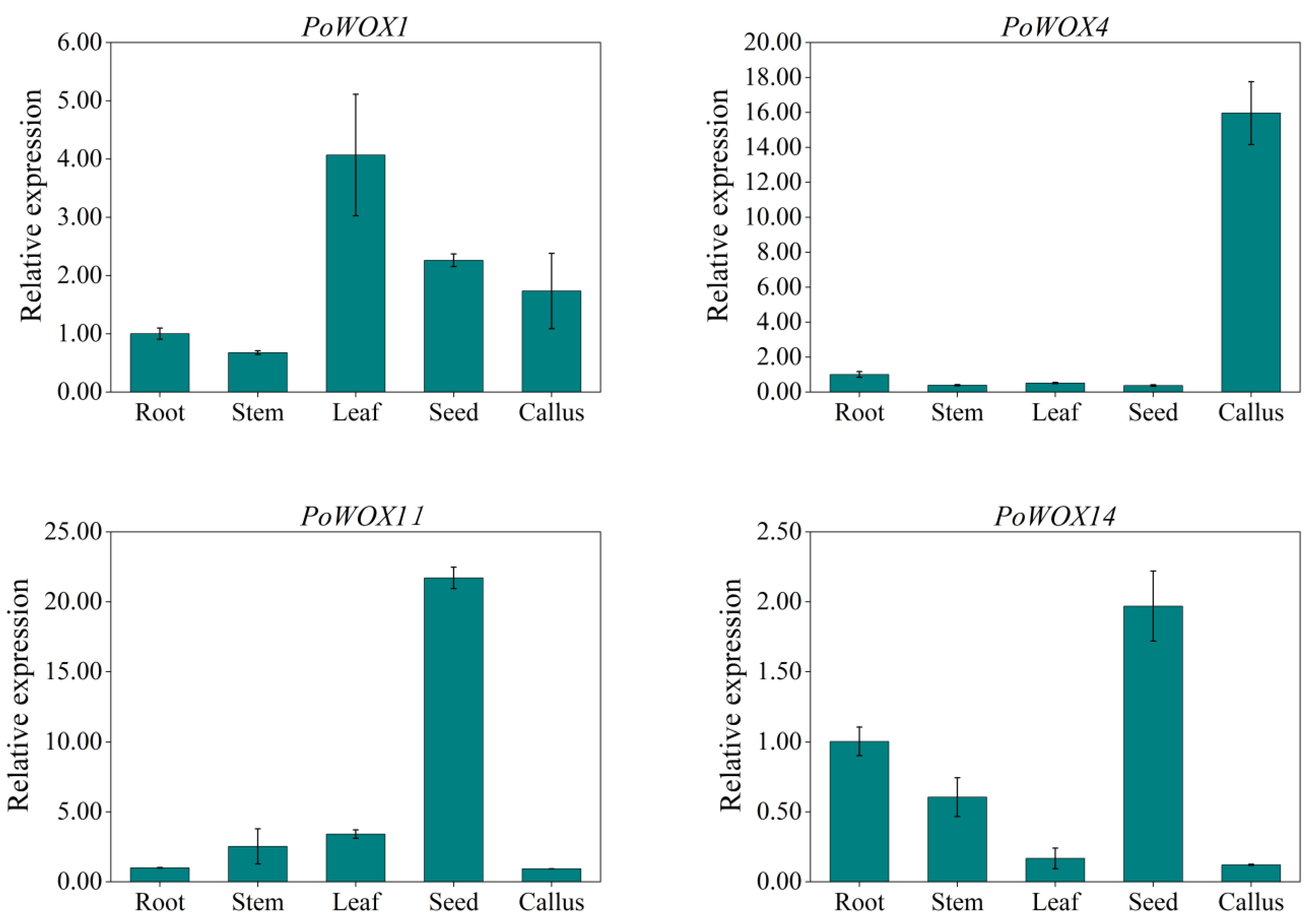
3.3. Analysis of the Expression Pattern of PoWOX Genes in Peony
3.4. Subcellular Localization of PoWOX Proteins
3.5. Results of Interactions between PoWOX Proteins
3.6. BiFC to Verify the Interactions between PoWOX Proteins
4. Discussion
4.1. Evolution Analysis of PoWOX Gene Family
4.2. Expression Pattern Analysis of PoWOX Gene Family
4.3. PoWOX Genes May Form a Dimer to Play a Transcriptional Regulatory Role in Peony Growth and Development
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Gao, J.; Xue, J.; Xue, Y.; Liu, R.; Ren, X.; Wang, S.; Zhang, X. Transcriptome Sequencing and Identification of Key Callus Browning-Related Genes from Petiole Callus of Tree Peony (Paeonia suffruticosa Cv. Kao) Cultured on Media with Three Browning Inhibitors. Plant Physiol. Biochem. 2020, 149, 36–49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Li, X.; Wu, K.; Wang, M.; Liu, P.; Wang, X.; Deng, R. Antioxidant Activities and Chemical Constituents of Flavonoids from the Flower of Paeonia Ostii. Molecules 2016, 22, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Dong, C.; Xue, Z.; Jin, Q.; Xu, Y. De Novo Transcriptome Sequencing and Discovery of Genes Related to Copper Tolerance in Paeonia Ostii. Gene 2016, 576, 126–135. [Google Scholar] [CrossRef]
- Huliang, C.; Fangyun, C.; Liping, P. Determination of the Fatty Acid Composition in Tree Peony Seeds Using Near-Infrared Spectroscopy. J. Am. Oil. Chem. Soc. 2016, 93, 943–952. [Google Scholar] [CrossRef]
- Xie, L.; Niu, L.; Zhang, Y.; Jin, M.; Ji, D.; Zhang, X. Pollen Sources Influence the Traits of Seed and Seed Oil in Paeonia Ostii ‘Feng Dan’. HortScience 2017, 52, 700–705. [Google Scholar] [CrossRef]
- Yu, X.; Zhao, R.; Cheng, F. Seed Germination of Tree and Herbaceous Peonies: A Mini-Review. Seed Sci. Biotechnol. 2007, 1, 11–14. [Google Scholar]
- Zhang, K.; Yao, L.; Zhang, Y.; Baskin, J.M.; Baskin, C.C.; Xiong, Z.; Tao, J. A Review of the Seed Biology of Paeonia Species (Paeoniaceae), with Particular Reference to Dormancy and Germination. Planta 2019, 249, 291–303. [Google Scholar] [CrossRef] [PubMed]
- Zhong, Y.; Cheng, F.; Wu, J. The Study Advances on Functional Genes in Tree Peony (Paeonia sect. Moutan). Mol. Plant Breed. 2016, 14, 2353–2364. [Google Scholar] [CrossRef]
- Yang, Y.; Sun, M.; Li, S.; Chen, Q.; Teixeira da Silva, J.A.; Wang, A.; Yu, X.; Wang, L. Germplasm Resources and Genetic Breeding of Paeonia: A Systematic Review. Hortic. Res. 2020, 7, 107. [Google Scholar] [CrossRef] [PubMed]
- Mayer, K.F.; Schoof, H.; Haecker, A.; Lenhard, M.; Jürgens, G.; Laux, T. Role of WUSCHEL in Regulating Stem Cell Fate in the Arabidopsis Shoot Meristem. Cell 1998, 95, 805–815. [Google Scholar] [CrossRef] [Green Version]
- Kamiya, N.; Nagasaki, H.; Morikami, A.; Sato, Y.; Matsuoka, M. Isolation and Characterization of a Rice WUSCHEL-Type Homeobox Gene That Is Specifically Expressed in the Central Cells of a Quiescent Center in the Root Apical Meristem. Plant J. 2003, 35, 429–441. [Google Scholar] [CrossRef]
- Van der Graaff, E.; Laux, T.; Rensing, S.A. The WUS Homeobox-Containing (WOX) Protein Family. Genome Biol. 2009, 10, 248. [Google Scholar] [CrossRef]
- Haecker, A.; Gross-Hardt, R.; Geiges, B.; Sarkar, A.; Breuninger, H.; Herrmann, M.; Laux, T. Expression Dynamics of WOX Genes Mark Cell Fate Decisions during Early Embryonic Patterning in Arabidopsis Thaliana. Development 2004, 131, 657–668. [Google Scholar] [CrossRef] [Green Version]
- Laux, T.; Mayer, K.F.; Berger, J.; Jürgens, G. The WUSCHEL Gene Is Required for Shoot and Floral Meristem Integrity in Arabidopsis. Development 1996, 122, 87–96. [Google Scholar] [CrossRef]
- Lin, H.; Niu, L.; McHale, N.A.; Ohme-Takagi, M.; Mysore, K.S.; Tadege, M. Evolutionarily Conserved Repressive Activity of WOX Proteins Mediates Leaf Blade Outgrowth and Floral Organ Development in Plants. Proc. Natl. Acad. Sci. USA 2013, 110, 366–371. [Google Scholar] [CrossRef] [Green Version]
- Dolzblasz, A.; Nardmann, J.; Clerici, E.; Causier, B.; van der Graaff, E.; Chen, J.; Davies, B.; Werr, W.; Laux, T. Stem Cell Regulation by Arabidopsis WOX Genes. Mol. Plant 2016, 9, 1028–1039. [Google Scholar] [CrossRef]
- Costanzo, E.; Trehin, C.; Vandenbussche, M. The Role of WOX Genes in Flower Development. Ann. Bot. 2014, 114, 1545–1553. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Zong, J.; Liu, J.; Yin, J.; Zhang, D. Genome-Wide Analysis of WOX Gene Family in Rice, Sorghum, Maize, Arabidopsis and Poplar. J. Integr. Plant Biol. 2010, 52, 1016–1026. [Google Scholar] [CrossRef]
- Jha, P.; Ochatt, S.J.; Kumar, V. WUSCHEL: A Master Regulator in Plant Growth Signaling. Plant Cell Rep. 2020, 39, 431–444. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, R.; Qin, G.; Chen, Z.; Gu, H.; Qu, L.-J. Over-Expression of WOX1 Leads to Defects in Meristem Development and Polyamine Homeostasis in Arabidopsis. J. Integr. Plant. Biol. 2011, 53, 493–506. [Google Scholar] [CrossRef]
- Nakata, M.; Matsumoto, N.; Tsugeki, R.; Rikirsch, E.; Laux, T.; Okada, K. Roles of the Middle Domain-Specific WUSCHEL-RELATED HOMEOBOX Genes in Early Development of Leaves in Arabidopsis. Plant Cell 2012, 24, 519–535. [Google Scholar] [CrossRef] [Green Version]
- Du, F.; Mo, Y.; Israeli, A.; Wang, Q.; Yifhar, T.; Ori, N.; Jiao, Y. Leaflet Initiation and Blade Expansion Are Separable in Compound Leaf Development. Plant J. 2020, 104, 1073–1087. [Google Scholar] [CrossRef]
- Wang, C.; Zhao, B.; He, L.; Zhou, S.; Liu, Y.; Zhao, W.; Guo, S.; Wang, R.; Bai, Q.; Li, Y.; et al. The WOX Family Transcriptional Regulator SlLAM1 Controls Compound Leaf and Floral Organ Development in Solanum Lycopersicum. J. Exp. Bot. 2021, 72, 1822–1835. [Google Scholar] [CrossRef]
- Ji, J.; Strable, J.; Shimizu, R.; Koenig, D.; Sinha, N.; Scanlon, M.J. WOX4 Promotes Procambial Development. Plant Physiol. 2010, 152, 1346–1356. [Google Scholar] [CrossRef] [Green Version]
- Suer, S.; Agusti, J.; Sanchez, P.; Schwarz, M.; Greb, T. WOX4 Imparts Auxin Responsiveness to Cambium Cells in Arabidopsis. Plant Cell 2011, 23, 3247–3259. [Google Scholar] [CrossRef] [Green Version]
- Etchells, J.P.; Provost, C.M.; Mishra, L.; Turner, S.R. WOX4 and WOX14 Act Downstream of the PXY Receptor Kinase to Regulate Plant Vascular Proliferation Independently of Any Role in Vascular Organisation. Development 2013, 140, 2224–2234. [Google Scholar] [CrossRef] [Green Version]
- Wang, L.; Wen, S.; Wang, R.; Wang, C.; Gao, B.; Lu, M. PagWOX11/12a Activates PagCYP736A12 Gene That Facilitates Salt Tolerance in Poplar. Plant Biotechnol. J. 2021, 19, 2249–2260. [Google Scholar] [CrossRef]
- Cheng, S.; Tan, F.; Lu, Y.; Liu, X.; Li, T.; Yuan, W.; Zhao, Y.; Zhou, D.-X. WOX11 Recruits a Histone H3K27me3 Demethylase to Promote Gene Expression during Shoot Development in Rice. Nucleic Acids Res. 2018, 46, 2356–2369. [Google Scholar] [CrossRef] [Green Version]
- Romera-Branchat, M.; Ripoll, J.J.; Yanofsky, M.F.; Pelaz, S. The WOX13 Homeobox Gene Promotes Replum Formation in the Arabidopsis Thaliana Fruit. Plant J. 2013, 73, 37–49. [Google Scholar] [CrossRef]
- Ikeuchi, M.; Iwase, A.; Ito, T.; Tanaka, H.; Favero, D.S.; Kawamura, A.; Sakamoto, S.; Wakazaki, M.; Tameshige, T.; Fujii, H.; et al. Wound-Inducible WUSCHEL RELATED HOMEOBOX 13 Is Required for Callus Growth and Organ Reconnection. Plant Physiol. 2021, 188, 425–441. [Google Scholar] [CrossRef]
- Denis, E.; Kbiri, N.; Mary, V.; Claisse, G.; Conde, E.; Silva, N.; Kreis, M.; Deveaux, Y. WOX14 Promotes Bioactive Gibberellin Synthesis and Vascular Cell Differentiation in Arabidopsis. Plant J. 2017, 90, 560–572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, F.; Liu, F.; Zhao, L. Overexpression of RaWUS Gene of Rosa canina Inducing Shoot Regeneration from Root Tip of Transgenic Tobacco. Sci. Silvae Sin. 2011, 47, 43–52. [Google Scholar]
- Gao, B.; Wen, C.; Fan, L.; Kou, Y.; Ma, N.; Zhao, L. A Rosa Canina WUSCHEL-Related Homeobox Gene, RcWOX1, Is Involved in Auxin-Induced Rhizoid Formation. Plant Mol. Biol. 2014, 86, 671–679. [Google Scholar] [CrossRef] [PubMed]
- Kuang, H. Tissue Culture and Stem Cell Related Gene Cloning and Expression of Rhododendron Equisetum. Master’s Thesis, Fujian Agriculture and Forestry University, Fuzhou City, China, 2014. [Google Scholar]
- Cao, Y.; Han, Y.; Meng, D.; Li, G.; Li, D.; Abdullah, M.; Jin, Q.; Lin, Y.; Cai, Y. Genome-Wide Analysis Suggests the Relaxed Purifying Selection Affect the Evolution of WOX Genes in Pyrus bretschneideri, Prunus persica, Prunus mume, and Fragaria vesca. Front. Genet. 2017, 8, 78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Liu, Z.; Lyu, M.; Yuan, Y.; Wu, B. Characterization of JsWOX1 and JsWOX4 during Callus and Root Induction in the Shrub Species Jasminum Sambac. Plants 2019, 8, 79. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, S.; Cheng, S.; Wang, Z.; Li, S.; Jin, X.; Lan, L.; Yang, B.; Yu, K.; Ni, X.; Li, N.; et al. Draft Genome of the Famous Ornamental Plant Paeonia suffruticosa. Ecol. Evol. 2020, 10, 4518–4530. [Google Scholar] [CrossRef] [PubMed]
- Potter, S.C.; Luciani, A.; Eddy, S.R.; Park, Y.; Lopez, R.; Finn, R.D. HMMER Web Server: 2018 Update. Nucleic Acids Res. 2018, 46, W200–W204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilkins, M.R.; Gasteiger, E.; Bairoch, A.; Sanchez, J.C.; Williams, K.L.; Appel, R.D.; Hochstrasser, D.F. Protein Identification and Analysis Tools in the ExPASy Server. Methods Mol. Biol. 1999, 112, 531–552. [Google Scholar] [CrossRef] [PubMed]
- Chou, K.-C.; Shen, H.-B. Plant-MPLoc: A Top-down Strategy to Augment the Power for Predicting Plant Protein Subcellular Localization. PLoS ONE 2010, 5, e11335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, J.; Tian, F.; Yang, D.-C.; Meng, Y.-Q.; Kong, L.; Luo, J.; Gao, G. PlantTFDB 4.0: Toward a Central Hub for Transcription Factors and Regulatory Interactions in Plants. Nucleic Acids Res. 2017, 45, D1040–D1045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, J.D.; Higgins, D.G.; Gibson, T.J. CLUSTAL W: Improving the Sensitivity of Progressive Multiple Sequence Alignment through Sequence Weighting, Position-Specific Gap Penalties and Weight Matrix Choice. Nucleic Acids Res. 1994, 22, 4673–4680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Johnson, J.; Grant, C.E.; Noble, W.S. The MEME Suite. Nucleic Acids Res. 2015, 43, W39–W49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Crooks, G.E.; Hon, G.; Chandonia, J.-M.; Brenner, S.E. WebLogo: A Sequence Logo Generator. Genome Res. 2004, 14, 1188–1190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Hedman, H.; Zhu, T.; von Arnold, S.; Sohlberg, J.J. Analysis of the WUSCHEL-RELATED HOMEOBOX Gene Family in the Conifer Picea abies Reveals Extensive Conservation as Well as Dynamic Patterns. BMC Plant. Biol. 2013, 13, 89. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakakibara, K.; Reisewitz, P.; Aoyama, T.; Friedrich, T.; Ando, S.; Sato, Y.; Tamada, Y.; Nishiyama, T.; Hiwatashi, Y.; Kurata, T.; et al. WOX13-like Genes Are Required for Reprogramming of Leaf and Protoplast Cells into Stem Cells in the Moss Physcomitrella Patens. Development 2014, 141, 1660–1670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, Y.; Liu, J.; Zeng, M.; He, J.; Qin, P.; Huang, H.; Xu, L. Identification of WOX Family Genes in Selaginella Kraussiana for Studies on Stem Cells and Regeneration in Lycophytes. Front. Plant Sci. 2016, 7, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Xu, L. Recruitment of IC-WOX Genes in Root Evolution. Trends Plant Sci. 2018, 23, 490–496. [Google Scholar] [CrossRef] [PubMed]
- Wójcik, A.M.; Wójcikowska, B.; Gaj, M.D. Current Perspectives on the Auxin-Mediated Genetic Network That Controls the Induction of Somatic Embryogenesis in Plants. Int. J. Mol. Sci. 2020, 21, 1333. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weigel, D.; Jürgens, G. Stem Cells That Make Stems. Nature 2002, 415, 751–754. [Google Scholar] [CrossRef] [PubMed]
- Sablowski, R. The Dynamic Plant Stem Cell Niches. Curr. Opin. Plant Biol. 2007, 10, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Yadav, R.K.; Perales, M.; Gruel, J.; Girke, T.; Jönsson, H.; Reddy, G.V. WUSCHEL Protein Movement Mediates Stem Cell Homeostasis in the Arabidopsis Shoot Apex. Genes Dev. 2011, 25, 2025–2030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daum, G.; Medzihradszky, A.; Suzaki, T.; Lohmann, J.U. A Mechanistic Framework for Noncell Autonomous Stem Cell Induction in Arabidopsis. Proc. Natl. Acad. Sci. USA 2014, 111, 14619–14624. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schlegel, J.; Denay, G.; Wink, R.; Pinto, K.G.; Stahl, Y.; Schmid, J.; Blümke, P.; Simon, R.G. Control of Arabidopsis Shoot Stem Cell Homeostasis by Two Antagonistic CLE Peptide Signalling Pathways. Elife 2021, 10, e70934. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, K.; Perales, M.; Snipes, S.; Yadav, R.K.; Diaz-Mendoza, M.; Reddy, G.V. DNA-Dependent Homodimerization, Sub-Cellular Partitioning, and Protein Destabilization Control WUSCHEL Levels and Spatial Patterning. Proc. Natl. Acad. Sci. USA 2016, 113, E6307–E6315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sloan, J.; Hakenjos, J.P.; Gebert, M.; Ermakova, O.; Gumiero, A.; Stier, G.; Wild, K.; Sinning, I.; Lohmann, J.U. Structural Basis for the Complex DNA Binding Behavior of the Plant Stem Cell Regulator WUSCHEL. Nat. Commun. 2020, 11, 2223. [Google Scholar] [CrossRef] [PubMed]
- Su, Y.H.; Zhou, C.; Li, Y.J.; Yu, Y.; Tang, L.P.; Zhang, W.J.; Yao, W.J.; Huang, R.; Laux, T.; Zhang, X.S. Integration of Pluripotency Pathways Regulates Stem Cell Maintenance in the Arabidopsis Shoot Meristem. Proc. Natl. Acad. Sci. USA 2020, 117, 22561–22571. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liu, X.; Engstrom, E.M.; Nimchuk, Z.L.; Pruneda-Paz, J.L.; Tarr, P.T.; Yan, A.; Kay, S.A.; Meyerowitz, E.M. Control of Plant Stem Cell Function by Conserved Interacting Transcriptional Regulators. Nature 2015, 517, 377–380. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.; Yan, A.; Han, H.; Li, T.; Geng, Y.; Liu, X.; Meyerowitz, E.M. HAIRY MERISTEM with WUSCHEL Confines CLAVATA3 Expression to the Outer Apical Meristem Layers. Science 2018, 361, 502–506. [Google Scholar] [CrossRef] [Green Version]
- Sun, W.; Gao, D.; Xiong, Y.; Tang, X.; Xiao, X.; Wang, C.; Yu, S. Hairy Leaf 6, an AP2/ERF Transcription Factor, Interacts with OsWOX3B and Regulates Trichome Formation in Rice. Mol. Plant 2017, 10, 1417–1433. [Google Scholar] [CrossRef] [Green Version]






| Genes | Primer Sequence F (5’-3’) | Primer Sequence R (5’-3’) |
|---|---|---|
| PoWOX1 | GCTCTAGAATGTATATGATGGGTTATAATGATGGCGGAG | TCCCCCCGGGATTCCTCAACGGAAGGAACTCAAAATACT |
| PoWOX4 | CGTCTAGAATGGGAAACATGAAGGTTCATCAGTTC | TCCCCCCGGGTCTTCCTTCCGGGTGTAATGGAA |
| PoWOX11 | GCTCTAGAATTCATGTGTTTTATCTTTTTTCTCTCAACTCT | TCCCCCCGGGAGTGGTTCTTGAAACTAGGAAATAGCTTTC |
| PoWOX13 | GCTCTAGAATGGGGTTAGCAAAAAAGATTTTAGAAAGT | TCCCCCCGGGCGAAGAATGTCAAATTCTTCCTCCTG |
| Primers Name | Primers Sequence (5′-3′) |
|---|---|
| Q-PoWOX1-F | CGTTGGCGGCAATGAAGAAGAATC |
| Q-PoWOX1-R | GGCAATTAGGAGGACTCAAGTTGGTAT |
| Q-PoWOX4-F | CCGCAACAGTCTTGGTCTTAGCC |
| Q-PoWOX4-R | TTCCTCATCTCTACATCTCACCTCTTCC |
| Q-PoWOX11-F | GCAACGCCAGATTCAAGCAAGTC |
| Q-PoWOX11-R | AAGAGGAACCAGCAAGACAAGAAGATG |
| Q-PoWOX13-F | ATGACGGACGAGCAAATAGAGGAACTT |
| Q-PoWOX13-R | CCGCTGCCTGGTAGTGATCTTCTG |
| Q-Poubiquitin-F | TCCTCCACCTCCTACCTTCCGACTC |
| Q-Poubiquitin-R | CGATCCTCCTGAGCCAAGCGTCAT |
| Primers Name | Primers Sequence (5′-3′) |
|---|---|
| PHG-PoWOX1-F | AGTCTCTCTCTCAAGCTTGATGTATATGATGGGTTATAATGATGGC |
| PHG-PoWOX1-R | CGGGTCATGAGCTCCTGCAATTCCTCAACGGAAGGAACT |
| PHG-PoWOX4-F | AGTCTCTCTCTCAAGCTTGATGGGAAACATGAAGGTTCATCA |
| PHG-PoWOX4-R | CGGGTCATGAGCTCCTGCATCTTCCTTCCGGGTGTAATG |
| PHG-PoWOX11-F | AGTCTCTCTCTCAAGCTTGATGGAAGATCATGACCCTAACA |
| PHG-PoWOX11-R | CGGGTCATGAGCTCCTGCAAGTGGTTCTTGAAACTAGGAAATAG |
| PHG-PoWOX13-F | AGTCTCTCTCTCAAGCTTGATGGGGTTAGCAAAAAAGATTTTAG |
| PHG-PoWOX13-R | CGGGTCATGAGCTCCTGCACCGAAGAATGTCAAATTCTTCCT |
| Primer Name | Upstream Primer F (5′-3′) | Downstream Primer R (5′-3′) | ||
|---|---|---|---|---|
| BK-PoWOX1 | ATCTCAGAGGAGGACCTGCAATGTATATGATGGGTTATAA | NdeI | AGGGGTTATGCTAGTTATGCATTCCTCAACGGAAGGAACT | NotI |
| BK-PoWOX4 | ATCTCAGAGGAGGACCTGCAATGGGAAACATGAAGGTTCA | NdeI | AGGGGTTATGCTAGTTATGCTCTTCCTTCCGGGTGTAATG | NotI |
| BK-PoWOX13 | ATCTCAGAGGAGGACCTGCAATGGGGTTAGCAAAAAAGAT | NdeI | AGGGGTTATGCTAGTTATGCCCGAAGAATGTCAAATTCTT | NotI |
| BK-PoWOX11 | ATCTCAGAGGAGGACCTGCAATGGAAGATCATGACCCTAA | NdeI | AGGGGTTATGCTAGTTATGCAGTGGTTCTTGAAACTAGGA | NotI |
| AD-PoWOX1 | GACGTACCAGATTACGCTCAATGTATATGATGGGTTATAA | NdeI | TATTAAGGGTTCCGGATCGCATTCCTCAACGGAAGGAACT | NotI |
| AD-PoWOX4 | GACGTACCAGATTACGCTCAATGGGAAACATGAAGGTTCA | NdeI | TATTAAGGGTTCCGGATCGCTCTTCCTTCCGGGTGTAATG | NotI |
| AD-PoWOX13 | GACGTACCAGATTACGCTCAATGGGGTTAGCAAAAAAGAT | NdeI | TATTAAGGGTTCCGGATCGCCCGAAGAATGTCAAATTCTT | NotI |
| AD-PoWOX11 | GACGTACCAGATTACGCTCAATGGAAGATCATGACCCTAA | NdeI | TATTAAGGGTTCCGGATCGCAGTGGTTCTTGAAACTAGGA | NotI |
| Gene Name | Upstream Primer F (5′-3′) | Downstream Primer R (5′-3′) | ||
|---|---|---|---|---|
| YCE-PoWOX1 | TTACGCTGGGCCCAGGCCTAATGTATATGATGGGTTATAA | SpeI | CGGTACCCTCGAGGTCGACGATTCCTCAACGGAAGGAACT | BamHI |
| YCE-PoWOX4 | TTACGCTGGGCCCAGGCCTAATGGGAAACATGAAGGTTCA | SpeI | CGGTACCCTCGAGGTCGACGTCTTCCTTCCGGGTGTAATG | BamHI |
| YCE-PoWOX13 | TTACGCTGGGCCCAGGCCTAATGGGGTTAGCAAAAAAGAT | SpeI | CGGTACCCTCGAGGTCGACGCCGAAGAATGTCAAATTCTT | BamHI |
| YCE-PoWOX11 | TTACGCTGGGCCCAGGCCTAATGGAAGATCATGACCCTAA | SpeI | CGGTACCCTCGAGGTCGACGAGTGGTTCTTGAAACTAGGA | BamHI |
| YNE-PoWOX1 | GGATCTTGGGCCCAGGCCTAATGTATATGATGGGTTATAA | SpeI | CGGTACCCTCGAGGTCGACGATTCCTCAACGGAAGGAACT | BamHI |
| YNE-PoWOX4 | GGATCTTGGGCCCAGGCCTAATGGGAAACATGAAGGTTCA | SpeI | CGGTACCCTCGAGGTCGACGTCTTCCTTCCGGGTGTAATG | BamHI |
| YNE-PoWOX13 | GGATCTTGGGCCCAGGCCTAATGGGGTTAGCAAAAAAGAT | SpeI | CGGTACCCTCGAGGTCGACGCCGAAGAATGTCAAATTCTT | BamHI |
| YNE-PoWOX11 | GGATCTTGGGCCCAGGCCTAATGGAAGATCATGACCCTAA | SpeI | CGGTACCCTCGAGGTCGACGAGTGGTTCTTGAAACTAGGA | BamHI |
| Gene | GenBank Accession | ORF (bp) | Protein Length (aa) | MW (Da) | pI | Instability Coefficient | GRAVY | Aliphatic Index | Subcellular Localization |
|---|---|---|---|---|---|---|---|---|---|
| PoWOX1 | OM457047 | 951 | 317 | 36562.68 | 8.71 | 55.93 | −0.93 | 60.63 | Nucleus |
| PoWOX4 | OM457050 | 633 | 211 | 24300.55 | 9.36 | 47.58 | −0.91 | 60.52 | Nucleus |
| PoWOX11 | OM457049 | 765 | 255 | 28165.35 | 6.13 | 67.96 | −0.46 | 61.88 | Nucleus |
| PoWOX13 | OM457048 | 858 | 286 | 32543.31 | 4.92 | 64.70 | −0.80 | 69.27 | Nucleus |
| Protein | α-Helix | β-Sheet | Extended Chain | Random Coil |
|---|---|---|---|---|
| PoWOX1 | 29.65% | 12.62% | 3.15% | 54.57% |
| PoWOX4 | 25.41% | 6.63% | 4.42% | 63.54% |
| PoWOX11 | 20.11% | 15.76% | 9.24% | 54.89% |
| PoWOX13 | 39.51% | 8.39% | 3.15% | 48.95% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, M.; Zhang, W.; Chang, Y.; Ma, Y.; Deng, Y.; Fan, K.; Zhang, X.; Jiang, Z.; Hu, T. A Preliminary Investigation on the Functional Validation and Interactions of PoWOX Genes in Peony (Paeonia ostii). Horticulturae 2022, 8, 266. https://doi.org/10.3390/horticulturae8030266
Xia M, Zhang W, Chang Y, Ma Y, Deng Y, Fan K, Zhang X, Jiang Z, Hu T. A Preliminary Investigation on the Functional Validation and Interactions of PoWOX Genes in Peony (Paeonia ostii). Horticulturae. 2022; 8(3):266. https://doi.org/10.3390/horticulturae8030266
Chicago/Turabian StyleXia, Mengsi, Wenbo Zhang, Yanting Chang, Yanjun Ma, Yayun Deng, Keke Fan, Xue Zhang, Zehui Jiang, and Tao Hu. 2022. "A Preliminary Investigation on the Functional Validation and Interactions of PoWOX Genes in Peony (Paeonia ostii)" Horticulturae 8, no. 3: 266. https://doi.org/10.3390/horticulturae8030266
APA StyleXia, M., Zhang, W., Chang, Y., Ma, Y., Deng, Y., Fan, K., Zhang, X., Jiang, Z., & Hu, T. (2022). A Preliminary Investigation on the Functional Validation and Interactions of PoWOX Genes in Peony (Paeonia ostii). Horticulturae, 8(3), 266. https://doi.org/10.3390/horticulturae8030266






