Abstract
Persimmon (Diospyros kaki) anthracnose, predominantly caused by Colletotrichum horii, is a destructive disease of persimmon. Thus, the evaluation of resistance resources is imperative for persimmon breeding and resistant variety deployment. In this study, the isolate from persimmon branches was identified as C. horii by using molecular and morphological characteristics. A total of 142 varieties were tested for anthracnose disease response by inoculation with the conidial suspension of C. horii. A significant variability was observed among the studied accessions. Only two accessions, which had a hypersensitive reaction with a slight lesion at the infection site at 14 days post inoculation, were highly resistant to anthracnose. A total of 7 and 24 accessions exhibited resistant and susceptible responses, respectively, to anthracnose. A total of 109 varieties with the highest proportion (76.76%) of accessions showed high susceptibility to C. horii. A total of 43 persimmon main cultivars were selected for the re-evaluation of the resistance level by counting the natural disease incidence of branches and fruits that showed similar resistant response. Moreover, nine of highly resistant or resistant accessions possessed desirable agronomic characters, including high fruit weight and strong growth potential. These resistant accessions could be used in the breeding of anthracnose-resistant persimmon varieties.
1. Introduction
Persimmon (Diospyros kaki) is regarded to originate from China [1] and is predominantly cultivated in East Asia, including China, Japan, and Korea. China, which has the largest area and production harvest in the world, produces 3.427 million tons, accounting for 75% among all the countries (The Food and Agriculture Organization of the United Nations, FAO, 2021). The persimmon fruit is rich in vitamins, antioxidants, and trace elements that are vital for human health and has been used in various medicinal and chemical industries and for commercial fruit consumption [2,3]. Currently, the cultivation of the persimmon crop is rapidly expanding in Israel, Italy, and Spain, suggesting that persimmon is becoming one of the most popular fruits worldwide.
The collection and evaluation of persimmon germplasms support the safe preservation of genetic diversity and facilitate the selection and breeding of excellent resistant varieties of persimmon [4]. The National Field Genebank for Persimmon (NFGP) in China began to collect persimmon germplasms in 1962 and was first built in 1987 in Meixian County, Shannxi, China [5]. Currently, the NFGP saves more than 1000 persimmon resources from different regions in China and a relatively high number of varieties with a high genetic diversity from other countries [6,7].
Persimmon anthracnose is a severe fungal disease, causing branches and leaves to wither, fruit to rot, and possibly the loss of the entire plant, gravely jeopardizing the development of the persimmon industry [8,9]. Previously, the pathogen that causes anthracnose was reported as Gloeosporium kaki by Shotaro Hori and Seiya Ito [10,11]. Maffei [12] described the leaf spot pathogen of persimmon from a specimen collected in Italy as Colletotrichum kaki. Von [13] later reported that the fungi previously described are the same and synonymous with C. gloeosporioides. However, in 2010, after examining the molecular and morphological characteristics of specimens from China, Japan, and New Zealand, Weir and Johnston renamed the persimmon pathogen as C. horii [14]. Additionally, the persimmon anthracnose can be caused by C. siamense, C. karstii, C. fructicola, C. nymphaeae, and C. melonis [15,16,17,18]. Persimmon anthracnose is a dangerous disease in China and in other nations across the world [15,16,17,18]. Carraro reported that C. fructicola, C. nymphaeae, and C. melonis cause persimmon anthracnose in Brazil [16]. Moreover, C. siamense and C. nymphaeae cause persimmon anthracnose in Korea [15,18]. With increased rain, the anthracnose in persimmon orchards becomes serious because many main varieties are susceptible to anthracnose. In addition to chemical control and cultivation management, breeding anthracnose-resistant varieties based on resistance evaluation is the most effective method to control anthracnose. Thus, screening resistance resources is imperative for breeding resistant varieties.
Abundant Diospyros germplasm resources are in the NFGP of China, and the evaluation and identification of persimmon germplasm resistance to anthracnose can provide new sources for resistance breeding programs. In the present study, we aim to (a) isolate and identify the pathogen of persimmon anthracnose, (b) evaluate the resistance level of 142 persimmon varieties in the NFGP, and (c) identify some anthracnose-resistant varieties for the future application of persimmon breeding.
2. Materials and Methods
2.1. Sample Collection and Pathogen Fungus Isolation
A total of 142 persimmon germplasm resources without infections and insect pests were preserved in the NFGP, Yangling, Shaanxi, China (34°16′56.24″ N, 108°04′27.95″ E; 420 m altitude).
For candidate pathogen isolation, symptomatic twigs of ‘Fuping Jianshi’ persimmon were randomly collected from NFGP at Yangling, China (34°17′52.55″ N, 108°04′05.58″ E) and brought to the laboratory for the isolation of putative causal agents. Samples were surface disinfected for 90 s with 70% ethanol and 1% sodium hypochlorite, rinsed twice with sterile distilled water. Sterilized small branch pieces (5 mm × 5 mm) were placed on PDA plates and then incubated at 25 °C with a photoperiod of 12 h until fungal growth was observed. Pathogen isolates were purified with single-spore culturing prior to use in subsequent experiments. The isolated pathogen fungus was named ‘FJ 3’.
For the microscopic morphology observation of the incidence of tissue samples after infection, the collected branches and fruit samples with different incidence degrees in different diseased parts were cut into small pieces of tissue (5–8 mm) at the junction of disease and health. After cutting, samples were quickly put into FAA fixative. Paraffin sections were prepared in accordance with the reported method [19].
2.2. Molecular Characterization of the Fungus
DNA was extracted from the isolated pathogen, and the genomic DNA sequences of the ITS region of rDNA, beta-tubulin (TUB2) gene, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), chitin synthase (CHS-1), and a partial sequence of the actin (ACT) were amplified with primers of ITS1F/ITS4, T1/Bt2b, GDF1/GDRl, ACT-512F/ACT-783R, and CHS-79F/CHS-345R, respectively [20] (Table S1). The amplified reaction system contained a total volume of 25 μL, including 2.5 μL of 2 × Reaction Mix, 0.2 μL Golden DNA Polymerase, 1 μL template DNA, 1 μL of 10 μmol/L primers, and 9.3 μL ddH2O. PCR conditions were as follows: pre-denaturation at 95 °C for 3 min, denaturation at 95 °C for 30 s, annealing at 56 °C for 30 s, and extension at 72 °C for 1 min for 42 cycles. The last extension was at 72 °C for 5 min. PCR products were detected by 1% agarose gel electrophoresis. Subsequently, products were cloned into the T-Vector pMD19 (TaKaRa, Dalian, China), and three positive clones for each fragment were sequenced (Aoke Technology Co., Ltd, Beijing, China). A phylogenetic tree was obtained using the MEGA5 software [21].
2.3. Artificial Inoculation and Natural Disease Incidence
After 18 days of culturing for purified pathogen isolation, conidia suspensions were prepared by flooding the culture plates and removing the fungal mycelium. The number of conidia was determined using a hemocytometer, and the conidia suspension was adjusted to a 1 × 106/mL concentration as a backup.
For artificial inoculation, an inoculum concentration of 1.0 × 106 conidia/mL for the ‘FJ 3’ fungus was used to infect the uninjured young branches (n = 3) by 5 μL suspensions with a short interval of 1.5 cm (Figure 1). The growth of diseased spots was recorded using a Vernier caliper for 15 consecutive days. The experiment was conducted twice in 2019 and 2020.
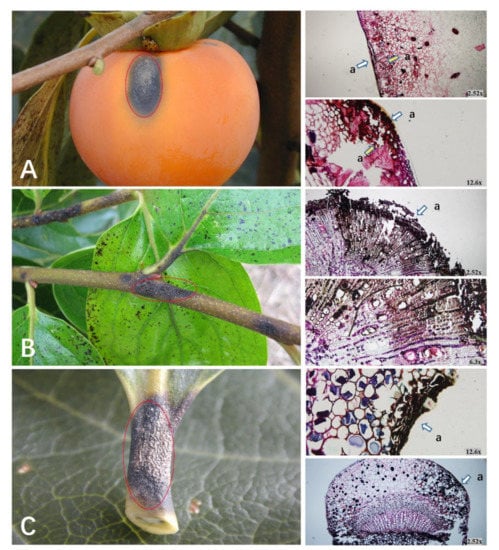
Figure 1.
Anthracnose symptoms and plant tissue sectioning caused by C. horri on persimmon. Disease lesions on (A) fruit, (B) branch, and (C) petiole. The red ellipse indicates the infection site of anthracnose, “a” indicates the location of the disease, and “×” means magnification.
For natural disease incidence in field, diseased fruits (≥20) and branches (≥20) with uniform growth potential were recorded in four different directions of the persimmon tree. The surface or sepals of the fruit exhibiting typical black spots were counted, and the incidence was presented as percent anthracnose infection. Branches showing typical anthracnose lesions were evaluated as infected.
2.4. Disease Resistance Criterion
For the artificial inoculation, the standard of disease classification is shown in Table 1. Disease index (DI) = Σ (number of branches infected in the rank × corresponding disease rank) × 100 / total number of infected branches × highest rank. On the basis of the range of DI, persimmon accessions were classified into highly resistant (0 < DI ≤ 10.0), resistant (10.0 < DI ≤ 30.0), susceptible (30.0 < DI ≤ 50.0), and highly susceptible (50.0 < DI ≤ 100.0, Table 2). If the disease severity varied between two years, a high disease grade of the year should be used as a rating scale of the accession resistance.

Table 1.
Branch lesion standard for persimmon anthracnose.

Table 2.
Levels and types of resistance to persimmon anthracnose.
Under natural field conditions, the severity of fruits and branches was categorized using a 1–4 scale, where highly resistant ≤ 2%, 2% ≤ resistant < 5%, 5% ≤ susceptible < 20%, and highly susceptible ≥ 20%. Fruit and branch severity values were presented as percent infected samples. At least 20 samples were measured for each persimmon variety.
3. Results
3.1. Symptoms of Persimmon Anthracnose Disease
The anthracnose fungus may attack the fruits, twigs, and leaf petioles in susceptible persimmon. When the fruit is infected with anthracnose, black spots appear on the surface of the fruit first and diseased spots slowly expand and dent downwards, finally causing the fruit to soften (Figure 1A). The lesion location had a modest number of stone cells, the peel was softened, and the infection gradually spread throughout the flesh.
Anthracnose symptoms on young twigs first appeared in dark spots, and minute spots developed into dark brown specks, showing a clear dividing line between diseased and symptomless tissues. The anthracnose fungus still attacked the xylem of branches and led to collapse. With the spread of disease spots, adjacent lesions connected until the entire twig was infected (Figure 1B).
The fungus infected petioles and caused similar dark brown spots (Figure 1C). With the development of the disease, leaves gradually fell off.
3.2. Morphological and Molecular Characterization of C. horri
For fungal isolation, symptomatic twigs of persimmon were randomly collected and brought to the laboratory for the isolation of putative causal agents. Samples were surface disinfected for 90 s with 70% ethanol and 1% sodium hypochlorite, then rinsed twice with sterile distilled water. Sterilized small branch pieces (5 mm × 5 mm) were placed on PDA plates. Representative purified isolates of the ‘FJ 3’ fungus colonies resembling Colletotrichum spp. were selected for further phylogenetic and morphological analyses. The ‘FJ 3’ fungal strain grew rapidly on PDA, thereby filling the entire Petri dish (90 mm) after 11 days at 25 °C and 12 h photoperiod. Colonies of ‘FJ 3’ were initially white and gradually became grayish on PDA with dark concentric zonation and regular margins in reverse (Figure 2). Few conidia (no conidial mass) were observed across the colony after 15 days of incubation at 25 °C on PDA. Conidia were hyaline, single-celled, straight, cylindrical apices and measured up to 10.5 to 17.1 μm × 4.0 to 5.56 μm (n = 100, average = 13.6 μm × 4.6 μm, Figure 2). The ‘FJ 3’ isolate showed similar morphological traits with the previously reported C. horri [22].
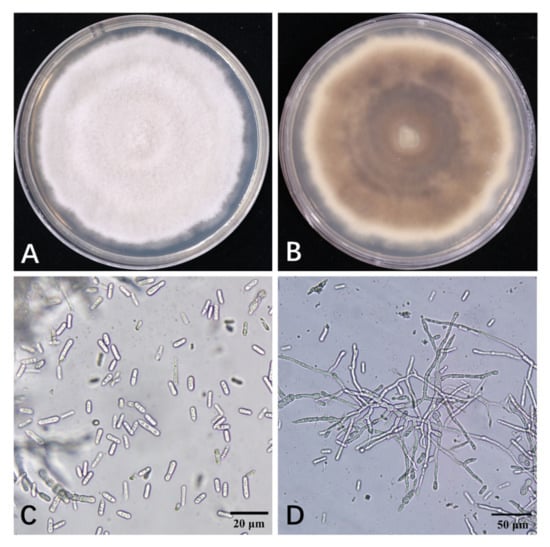
Figure 2.
Cultural and morphological characteristics of the C. horii ‘FJ 3’ isolation growing on PDA: (A) the upper surface, (B) the lower surface, (C) conidia (bar = 20 μm), and (D) mycelial growth (bar = 50 μm) of C. horii.
Five isolates obtained from the ‘Fuping Jianshi’ persimmon were identified as the same fungal species on the basis of multigene phylogenetic and morphological analyses (Figure 3). The persimmon anthracnose ‘FJ 3’ fungus was selected for further phylogenetic analysis. Then, genomic DNA sequences were amplified with ITS1F/ITS4, T1/Bt2b, GDF1/GDRl, ACT-512F/ACT-783R, and CHS-79F/CHS-345R (Weir et al., 2012). These amplicons were submitted to GenBank with accession numbers of OL347726 for ITS, OL364188 for GAPDH, OL364190 for CHS-1, OL364191 for TUB2, and OL364189 for ACT. The BLAST search in GenBank revealed that sequences showed high similarity to those of C. horii. Amplification sequences from the ‘FJ 3’ fungus were identical to JX010450 for CHS-1, 98.34% to GQ329690 for ITS, 98.34% to JX010450 for GAPDH, 99.29% to JX009438 for ACT, and 99.74% to JX010450 for TUB2. The maximum likelihood tree was obtained from a concatenated dataset of ITS, ACT, CHS, GAPDH, and TUB2 gene sequences of the C. gloeosporioides species complex with 1000 bootstrapping, showing that the present ‘FJ 3’ isolate clustered with the reference isolate C. horii (NBRC 7478) with high bootstrap support. Thus, the current ‘FJ 3’ isolate was identified as C. horii on the basis of cultural characteristics and sequence similarity data (Figure 3).

Figure 3.
Phylogenetic analysis using the neighbor-joining method through comparative studies of nucleotide sequences of the internal transcribed spacer (ITS) region of rDNA, glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a partial sequence of the actin (ACT), chitin synthase (CHS-1), and beta-tubulin (TUB2) genes from the present isolates with those of other Colletotrichum spp. in the C. gloeosporioides species complex retrieved from GenBank. The ‘FJ 3’ isolate is emphasized in the red circle. Numbers above the branches represent bootstrap values.
3.3. Evaluation of Anthracnose Resistance among Persimmon Germplasms by Artificial Infection
Persimmon accessions were grown in NFGP at Yangling, China (34°17′52.55″ N, 108°04′05.58″ E). In 2019 and 2020, this area received total rainfall values of 656 and 683 mm, respectively, and average temperatures of 13.0 °C and 13.2 °C, respectively. Rainfall varied between months of the same year, whereas the temperature of the area fluctuated little, especially between different years. The rainfall from July to September accounts for nearly half of the annual rainfall, and this period represents the hottest three months (highest average temperature (26.1–26.3 °C) is observed in July). The temperature and relative humidity from July to September are conducive to the occurrence of the persimmon anthracnose disease.
The resistance level was defined in four categories, including highly resistant, resistant, susceptible, and highly susceptible, on the basis of the DI of inoculated branches in 2019 and 2020 (Figure 4 and Figure 5). A significant variability was observed among the studied accessions. Only two accessions, which had a hypersensitive reaction with a slight lesion at the injection site at 15 days postinoculation, were highly resistant to anthracnose. Seven cultivars exhibited a resistant response to anthracnose and accounted for 4.93% of the 142 accessions (Figure 4). The susceptible response was observed in 24 accessions, which accounted for 16.90% of the total. A total of 109 varieties with the highest proportion (76.76%) of the accessions showed a highly susceptible response to C. horii ‘FJ 3’ (Figure 4 and Table 3).

Figure 4.
Representative varieties of ‘Qianxian Muwashi’ and ‘Hebei Lianhuashi’ exhibiting high susceptibility and susceptibility, respectively, to C. horii after artificial infection in (A) 2019 and (B) 2020.

Figure 5.
Representative varieties of ‘Chengsushi’ and ‘Yuanqu Bayuehong’ exhibiting resistant and highly resistant responses, respectively, to C. horii after artificial infection in (A) 2019 and (B) 2020.

Table 3.
Anthracnose resistance level based on the average lesions of persimmon germplasm inoculated with C. horri isolate.
3.4. Evaluation of the Anthracnose Resistance for Selected Persimmon Accessions by Natural Disease Incidence
A total of 43 persimmon main cultivars were selected for the re-evaluation of anthracnose resistance in the nature field in 2018 and 2019 (Table 4). Four accessions were highly resistant and had no symptomatic anthracnose in branches and fruits, and nine accessions were resistant with average percent disease incidence ≤ 5. In 34 remaining accessions, 5 and 29 were susceptible and highly susceptible, respectively, with percent disease incidence values of 5–20% and 20–100%, respectively. The data of repeated evaluation experiments showed that persimmons performed consistent resistance by artificial infection and natural disease incidence, except for two accessions, i.e., ‘Zaozhuang Ehuangshi’ and ‘Ningbo Tongpenshi’.

Table 4.
Anthracnose resistance level for 43 selected persimmon germplasms based on the severity of fruits and branches in the nature field.
3.5. Agronomic Characteristics of Blast Resistant Finger Millet Genotypes
The agronomic traits of nine anthracnose-resistant germplasms were analyzed, and results showed that highly resistant genotypes, i.e., ‘Kangbing Jianshi’ and ‘Yuanqu Bayuehuang’, had strong growth potential, orange-yellow peel color, glutinous texture, and none had a cross-concave form on the fruit apex (Table S2). ‘Hyakume’, which could naturally remove astringency on the tree during the ripening stage, was classified under the pollination-variant nonastringent type, whereas others were classified under the pollination-constant astringent type. Moreover, ‘Hyakume’ had the highest average fruit weight of 192 g and maximum fruit weight of 386 g. ‘Zhaoan Yuanxiaoshi’ and ‘Baoshan Dafangshi’ had the most days of maintaining fruit crispness, with 27 days. All varieties could be used as female parent materials for resistance breeding. In addition, the ‘Hyakume’ persimmon, with occasional male flowers, could be used as a male parent for the breeding program (Table S2).
4. Discussion
The C. gloeosporioides complex is the most common and polyphagous species complex of the genus. Weir et al. [20] reported that C. gloeosporioides is a complex consisting of 22 species. To date, 57 species have been described [23]. The genus has been voted as one of the top 10 fungal plant pathogens in molecular plant pathology [24]. On the basis of multigene phylogenetic analysis and morphological characteristics, the pathogen causing persimmon anthracnose has been renamed as C. horii, which also belongs to the C. gloeosporioides species complex [14]. To the best of our knowledge, most individual species in the C. gloeosporioides species complex, such as C. horii, C. fructicola, and C. siamense, may cause persimmon anthracnose [15,16,25] (Figure 1). Moreover, some different plants can be infected by the same anthrax species [14,26], C. acutatum from pepper can cause typical persimmon anthracnose symptoms, showing black spot disease in Korea [27,28]. Thus, these species (C. gloeosporioides is a species complex) are extremely difficult to distinguish on the basis of morphological characteristics only. Thus, pathogenic strains from persimmon anthracnose should be identified on the basis of the multigene phylogenetic analysis. In the present study, the ‘FJ 3’ isolate from persimmon was identified as C. horri by the combined analysis of molecular and morphological characteristics. Moreover, the ‘FJ 3’ isolate was tested for pathogenicity on persimmon twigs in the laboratory and nature field to confirm the isolate’s ability to cause the anthracnose disease. Therefore, the identification of the anthracnose pathogen in the national persimmon germplasm resource nursery is helpful to further carry out and deepen the research on the pathogenesis and defense mechanism of this anthracnose and provide a basis for the prevention and control of persimmon anthracnose.
Persimmon anthracnose, a serious disease occurring in many countries where persimmon is cultivated, is predominantly caused by C. gloeosporioides [29]. Persimmon anthracnose symptoms first appear in the spring as spots which then develop into dark lesions. Under high temperature and humidity conditions, adjacent lesions may integrate together, increasing in size until the entire twig is infected [30]. In the present study, the anthracnose pathogen can result in dark lesions in the fruit, calyx, branches, and leaf of persimmon, showing similar results with previous reports [9,15,22,30]. In addition, the C. gloeosporioides species complex consists of important plant pathogens that can cause anthracnose in economic crops, such as chili [31], mango [32], strawberry [33], apples [34], grapes [35], bananas [36,37], and tomato [38]. As a destructive disease of persimmon trees, C. horii can cause leaf defoliation, fruit rot, and even the death of the whole plant [39]. Unfortunately, most persimmon varieties are not resistant to anthracnose. For example, ‘Gongcheng Shuishi’ and ‘Fuping Jianshi’, which have the largest cultivation area in China, are susceptible to persimmon anthracnose. In some persimmon-growing areas in China, anthracnose can reduce the yield of persimmon by more than 50% [40].
Anthracnose-resistant persimmon varieties are important for the breeding of new resistant cultivars and the development of the persimmon industry. However, the evaluation and identification of persimmon germplasms with anthracnose resistance remain unknown. Persimmon is believed to originate in China and has abundant genetic diversity, with at least 1000 varieties [2,7]. Therefore, the identification of excellent persimmon resources resistant to anthracnose is necessary for resistance breeding. On the basis of the artificial infection, two accessions (i.e., ‘Kangbing Jianshi’ and ‘Quyuan Bayuehuang’), which had a hypersensitive reaction with a slight lesion at the injection site at 14 days post inoculation, were highly resistant to anthracnose, and six cultivars (i.e., ‘Yeshi’, ‘Pucheng Fangshi’, ‘Bayuehuang’, ‘Lushi 5’, and ‘ Junqianzi) exhibited a resistant response to anthracnose. Furthermore, 43 main cultivars of persimmon were selected for the re-evaluation of anthracnose resistance in the nature field in 2018 and 2019. The resistance level of germplasm resources was consistent with the previous evaluation results of artificial infection. Moreover, through the evaluation of agronomic characters, the above eight resistance resources showed excellent characters and could be used in resistance breeding in the future. For example, ‘Kangbing Jianshi’ is suitable for making dried persimmon due to the heart-shaped fruit, and ‘Quyuan Bayuehuang’ is early maturing with high yield and strong growth potential.
5. Conclusions
Persimmon anthracnose, a destructive disease for persimmon, is predominantly caused by C. horii pathogen, resulting in shoot wilting, fruit rotting, and even death of the whole plant. In this study, the isolate ‘FJ 3’ from persimmon trees in the NFGP of China was further identified as C. horii by using morphological characteristics and multigene phylogenetic analysis. A total of 142 persimmon varieties were tested for anthracnose disease response by inoculation with the conidial suspension of C. horii in the NFGP. Resistance level was defined in four categories, including highly resistant, resistant, susceptible, and highly susceptible, on the basis of the DI of the inoculated branch. A significant variability was observed among the studied accessions. Only two varieties, which had a hypersensitive reaction with a slight lesion at the injection site at 14 days post inoculation, were highly resistant to anthracnose. Seven cultivars exhibited a resistant response to anthracnose and accounted for 4.93% of the 142 accessions. A susceptible response was observed for 24 accessions, which accounted for 16.90% of the total. A total of 109 varieties with the highest proportion (76.76%) of the accessions showed a highly susceptible response to C. horii. A total of 43 persimmon main cultivars were selected for the re-evaluation of anthracnose resistance by natural disease incidence in 2018 and 2019 and showed similar resistance levels. Moreover, eight highly resistant or resistant accessions possessed desirable agronomic characters. The use of these resistance sources in persimmon breeding should aid in expanding the genetic diversity and in the development of new resistant varieties.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae8020180/s1. These amplicons have been submitted to GenBank with accession numbers of OL347726 for ITS, OL364188 for GAPDH, OL364190 for CHS-1, OL364191 for TUB2, and OL364189 for ACT. Table S1: Primers for pathogen identification; Table S2: The agronomic characteristics of anthracnose resistant persimmon genotypes.
Author Contributions
Conceptualization, C.G. and Y.Y.; methodology, Q.C. and Y.L.; software, Q.C. and J.H.; validation, C.G., Y.Y., and J.H.; writing—original draft preparation, C.G. and J.H.; writing—review and editing, C.G.; visualization, Y.Y.; supervision, Y.Y.; funding acquisition, Y.Y. and C.G. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by National Key Research and Development Program of China (2019YFD1000600) and National Horticulture Germplasm Resources Center (NHGRC2020-NH06).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
Data are contained within the article.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Guan, C.; Liu, S.; Wang, M.; Ji, H.; Ruan, X.; Wang, R.; Yang, Y. Comparative transcriptomic analysis reveals genetic divergence and domestication genes in Diospyros. BMC Plant Biol. 2019, 19, 227. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Luo, Z. Persimmon in China: Domestication and traditional utilizations of genetic resources. Adv. Hortic. Sci. 2008, 22, 239–243. [Google Scholar]
- Woolf, A.B.; Ben-Arie, R. Persimmon (Diospyros kaki L.). In Postharvest Biology and Technology of Tropical and Subtropical Fruits; Yahia, E.M., Ed.; Woodhead Publishing: Cambridge, UK, 2011; Volume 4, pp. 166–193. [Google Scholar]
- Greene, S.; Morris, J. The case for multiple-use plant germplasm collections and a strategy for implementation. Crop Sci. 2001, 41, 886–892. [Google Scholar] [CrossRef]
- Wang, R. Chinese persimmon germplasm resources. Acta Hortic. 1997, 436, 43–50. [Google Scholar]
- Guan, C.; Zhang, P.; Hu, C.; Chachar, S.; Riaz, A.; Wang, R.; Yang, Y. Genetic diversity, germplasm identification and population structure of Diospyros kaki Thunb. from different geographic regions in China using SSR markers. Sci. Hortic. 2019, 251, 233–240. [Google Scholar] [CrossRef]
- Guan, C.; Zhang, Y.; Zhang, P.; Chachar, S.; Wang, R.; Du, X.; Yang, Y. Germplasm conservation, molecular identity and morphological characterization of persimmon (Diospyros kaki Thunb.) in the NFGP of China. Sci. Hortic. 2020, 272, 109490. [Google Scholar] [CrossRef]
- Lee, J.-H.; Han, K.-S.; Lee, S.-C.; Shim, C.-K.; Bae, D.-W.; Kim, D.-K.; Kim, H.-K. Early detection of epiphytic anthracnose inoculum on phyllosphere of Diospyros kaki var. domestica. Plant Pathol. J. 2004, 20, 247–251. [Google Scholar] [CrossRef]
- Zhang, J.-Z. Anthracnose of persimmon caused by Colletotrichum gloeosporioides in China. Asian Australas J. Plant Sci. Biotechnol. 2008, 2, 50–54. [Google Scholar]
- Hori, S. Kaki no Shinbyogai Tansobyo. Engei No Tomo 1910, 6, 58–61. [Google Scholar]
- Ito, S. Gloeosporiose of the Japanese Persimmon. J. Plant Res. 1911, 25, 197–202. [Google Scholar]
- Maffei, L. Una malattia delle foglie del ’Kaki’ dovuta al Colletotrichum Kaki n. sp. Riv. Patol. Veg. 1921, 11, 116–118. [Google Scholar]
- Von, J. A revision of the fungi classified as Gloeosporium. Bibliof. Mycol. 1970, 24, 203. [Google Scholar]
- Weir, B.S.; Johnston, P.R. Characterisation and neotypification of Gloeosporium kaki Hori as Colletotrichum horii nom. nov. Mycotaxon 2010, 111, 209–219. [Google Scholar] [CrossRef]
- Chang, T.; Hassan, O.; Jeon, J.; Shin, J.; Oh, N.; Lim, T. First report of anthracnose of persimmon (Diospyros kaki L. f.) caused by Colletotrichum siamense in Korea. Plant Dis. 2018, 102, 443. [Google Scholar] [CrossRef]
- Carraro, T.; Lichtemberg, P.; Michailides, T.; Pereira, W.; Figueiredo, J.; May-De Mio, L. First report of Colletotrichum fructicola, C. nymphaeae, and C. melonis causing persimmon anthracnose in Brazil. Plant Dis. 2019, 103, 2692. [Google Scholar] [CrossRef]
- Wang, J.; Ai, C.; Yu, X.; An, M.; Sun, S.; Gao, R. First report of Colletotrichum karstii causing anthracnose on persimmon leaves in China. Plant Dis. 2016, 100, 532. [Google Scholar] [CrossRef]
- Hassan, O.; Lee, D.; Chang, T. First report of anthracnose of persimmon caused by Colletotrichum nymphaeae in Korea. Plant Dis. 2019, 103, 1772. [Google Scholar] [CrossRef]
- Fischer, A.H.; Jacobson, K.A.; Rose, J.; Zeller, R. Cutting sections of paraffin-embedded tissues. CSH Protoc. 2008, 3, 4987. [Google Scholar] [CrossRef]
- Weir, B.; Johnston, P.; Damm, U. The Colletotrichum gloeosporioides species complex. Stud. Mycol. 2012, 73, 115–180. [Google Scholar] [CrossRef]
- Tamura, K.; Peterson, D.; Peterson, N.; Stecher, G.; Nei, M.; Kumar, S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 2011, 28, 2731–2739. [Google Scholar] [CrossRef]
- Hassan, O.; Jeon, J.Y.; Chang, T.; Shin, J.S.; Oh, N.K.; Lee, Y.S. Molecular and morphological characterization of Colletotrichum species in the Colletotrichum gloeosporioides complex associated with persimmon anthracnose in South Korea. Plant Dis. 2018, 102, 1015–1024. [Google Scholar] [CrossRef] [PubMed]
- Talhinhas, P.; Baroncelli, R. Colletotrichum species and complexes: Geographic distribution, host range and conservation status. Fungal Divers. 2021, 110, 109–198. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [PubMed]
- Jeon, J.; Hassan, O.; Chang, T.; Lee, D.; Shin, J.; Oh, N. Anthracnose of persimmon (Diospyros kaki) caused by Colletotrichum horii in Sangju, Korea. Plant Dis. 2017, 101, 1035. [Google Scholar] [CrossRef]
- Siddiqui, Y.; Ali, A. Colletotrichum gloeosporioides (Anthracnose). In Postharvest Decay. Control Strategies; Bautista-Baños, S., Ed.; Academic Press: Cambridge, MA, USA; Elsevier Inc.: London, UK, 2014; pp. 337–371. ISBN 978-0-12-411552-1. [Google Scholar]
- Kim, H.R.; Lim, T.H.; Kim, J.-H.; Kim, Y.H.; Kim, H.T. Potential of cross-infection of Colletotrichum species causing anthracnose in persimmon and pepper. Plant Pathol. J. 2009, 25, 13–20. [Google Scholar] [CrossRef]
- Kwon, J.H.; Kim, J.W. First report of fruit black spot of Diospyros kaki caused by Colletotrichum acutatum in Korea. Plant Pathol. J. 2011, 27, 100. [Google Scholar] [CrossRef]
- Tong, Z.J.X. Various stages and amount of Cottetotrichum gloeosporioides on overwintering twigs of persimmon. J. Plant Protect. 2003, 30, 437–438. [Google Scholar]
- Xie, L.; Zhang, J.Z.; Cai, L.; Hyde, K.D. Biology of Colletotrichum horii, the causal agent of persimmon anthracnose. Mycology 2010, 1, 242–253. [Google Scholar] [CrossRef]
- Than, P.P.; Prihastuti, H.; Phoulivong, S.; Taylor, P.W.; Hyde, K.D. Chilli anthracnose disease caused by Colletotrichum species. J. Zhejiang Univ. Sci. 2008, 9, 764–778. [Google Scholar] [CrossRef]
- Vitale, A.; Alfenas, A.C.; Siqueira, D.L.D.; Magistà, D.; Perrone, G.; Polizzi, G. Cultivar resistance against Colletotrichum asianum in the world collection of mango germplasm in southeastern Brazil. Plants 2020, 9, 182. [Google Scholar] [CrossRef]
- Xiao, C.; MacKenzie, S.; Legard, D. Genetic and pathogenic analyses of Colletotrichum gloeosporioides isolates from strawberry and noncultivated hosts. Phytopathology 2004, 94, 446–453. [Google Scholar] [CrossRef]
- González, E.; Sutton, T.B. Population diversity within isolates of Colletotrichum spp. causing Glomerella leaf spot and bitter rot of apples in three orchards in North Carolina. Plant Dis. 2004, 88, 1335–1340. [Google Scholar] [CrossRef] [PubMed]
- Kono, A.; Nakaune, R.; Yamada, M.; Nakano, M.; Mitani, N.; Ueno, T. Effect of culture conditions on conidia formation by Elsinoë ampelina, the causal organism of grapevine anthracnose. Plant Dis. 2009, 93, 481–484. [Google Scholar] [CrossRef] [PubMed]
- Zhimo, V.Y.; Dilip, D.; Sten, J.; Ravat, V.K.; Bhutia, D.D.; Panja, B.; Saha, J. Antagonistic yeasts for biocontrol of the banana postharvest anthracnose pathogen Colletotrichum musae. J. Phytopathol. 2017, 165, 35–43. [Google Scholar] [CrossRef]
- Riera, N.; Ramirez-Villacis, D.; Barriga-Medina, N.; Alvarez-Santana, J.; Herrera, K.; Ruales, C.; Leon-Reyes, A. First report of banana anthracnose caused by Colletotrichum gloeosporioides in Ecuador. Plant Dis. 2019, 103, 763. [Google Scholar] [CrossRef]
- Pardo-De la Hoz, C.; Calderón, C.; Rincón, A.; Cárdenas, M.; Danies, G.; López-Kleine, L.; Restrepo, S.; Jiménez, P. Species from the Colletotrichum acutatum, Colletotrichum boninense and Colletotrichum gloeosporioides species complexes associated with tree tomato and mango crops in Colombia. Plant Pathol. 2016, 65, 227–237. [Google Scholar] [CrossRef]
- Zang, J.; Xu, T. Cytological characteristics of the infection in different species, varieties and organs of persimmon by Colletotrichum gloeosporioides. Mycosystema 2005, 24, 116–122. [Google Scholar]
- Deng, Q.E.; Ding, X.Y.; Li, J.A.; Cui, L.K.; Xu, J.Q. Morphological characteristics and genetic diversity of Colletotrichum horii infecting persimmon tree in China. Eur. J. Plant Pathol. 2020, 156, 437–449. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).