Metabolic Profiling of Organic Acids Reveals the Involvement of HuIPMS2 in Citramalic Acid Synthesis in Pitaya
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Determination of Organic Acids
2.3. RNA Extraction and cDNA Synthesis
2.4. Gene Expression Analyses
2.5. Gene Isolation and Sequence Analyses
2.6. Subcellular Localization
2.7. Statistical Analysis
3. Results
3.1. Changes of Organic Acid Contents during Fruit Development of Pitayas
3.2. Changes of Total Organic and Composition Ratios
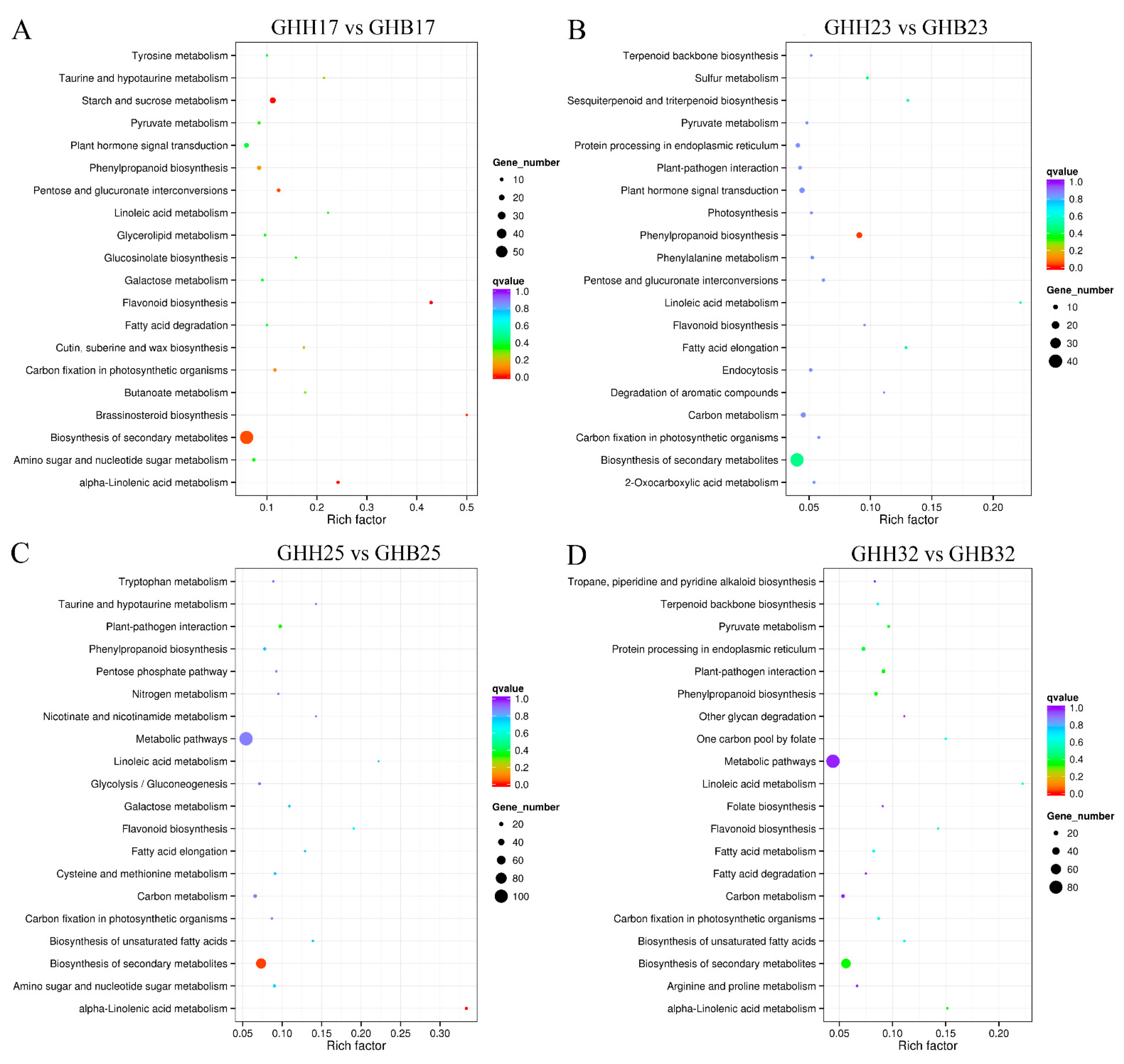
3.3. Analyses of Transcriptome Data of ‘GHB’ and ‘GHH’ Pitayas
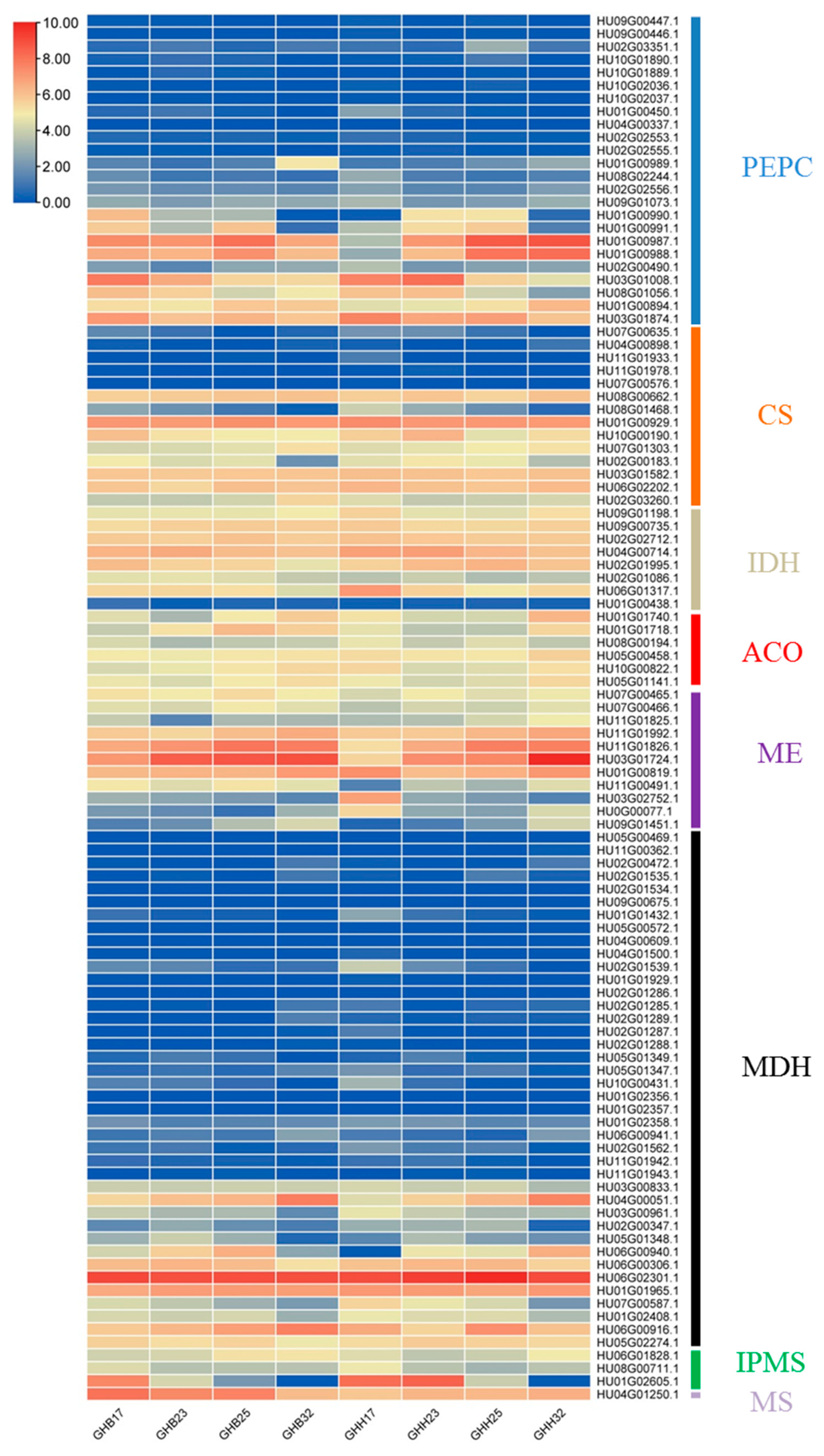
3.4. Expression Analyses of Organic Acid Metabolism-Related Genes
3.5. Cloning, Sequence and Evolutionary Analyses of HuIPMS
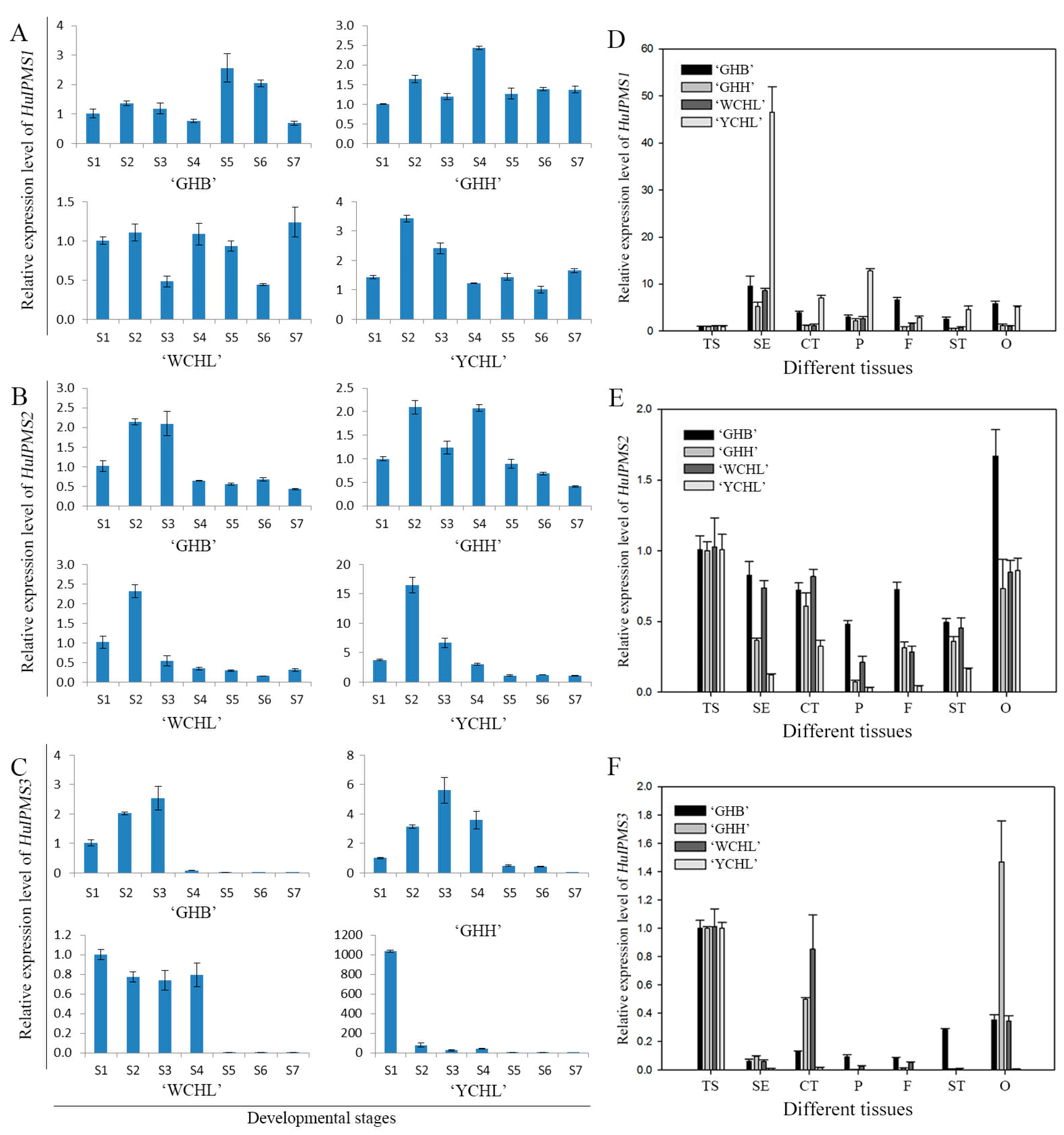
3.6. Expression Analyses of HuIPMSs
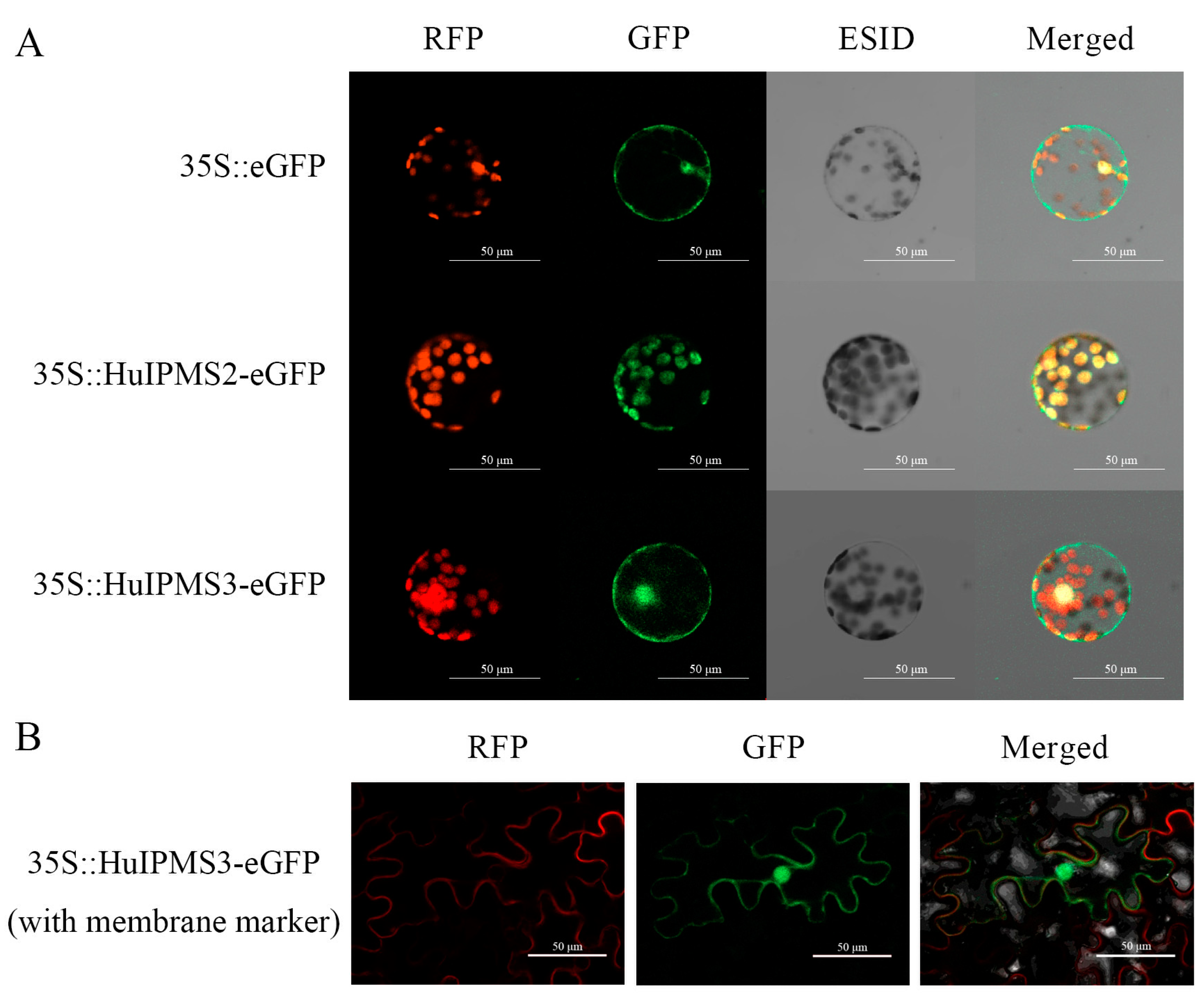
3.7. Subcellular Localization of HuIPMSs Protein
4. Discussion
4.1. Accumulation of Organic Acids in the Four Pitaya Cultivars
4.2. The Relationship between Organic Acid Contents and Metabolism-Associated Genes
4.3. Role of HuIPMS2 in Citramalic Acid Synthesis of Pitaya
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yin, X.; Li, J.H.; Shin, H.D.; Du, G.C.; Liu, L.; Chen, J. Metabolic engineering in the biotechnological production of organic acids in the tricarboxylic acid cycle of microorganisms: Advances and prospects. Biotechnol. Adv. 2015, 33, 830–841. [Google Scholar] [CrossRef] [PubMed]
- Batista-Silva, W.; Nascimento, V.L.; Medeiros, D.B.; Nunes-Nesi, A.; Ribeiro, D.M.; Zsögön, A.; Araújo, W.L. Modifications in organic acid profiles during fruit development and ripening: Correlation or causation? Front Plant Sci. 2018, 9, 1689. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.X.; Liu, X.H.; Chen, L.S. Developmental changes in pulp organic acid concentration and activities of acid-metabolising enzymes during the fruit development of two loquat (Eriobotrya japonica Lindl.) cultivars differing in fruit acidity. Food Chem. 2009, 114, 657–664. [Google Scholar] [CrossRef]
- Ma, B.Q.; Yuan, Y.Y.; Gao, M.; Li, C.Y.; Ogutu, C.; Li, M.J.; Ma, F.W. Determination of predominant organic acid components in Malus species: Correlation with apple domestication. Metabolites 2018, 8, 74. [Google Scholar] [CrossRef] [Green Version]
- Sha, S.F.; Li, J.C.; Wu, J.; Zhang, S.L. Changes in the organic acid content and related metabolic enzyme activities in developing ‘Xinping’ pear fruit. Afr. J. Agric. Res. 2011, 6, 3560–3567. [Google Scholar]
- Hulme, A.C. A New Hydroxy-acid in the Peel of Apple Fruits. Nature 1953, 172, 346. [Google Scholar] [CrossRef]
- Sugimoto, N.; Engelgau, P.; Jones, A.D.; Song, J.; Beaudry, R. Citramalate synthase yields a biosynthetic pathway for isoleucine and straight- and branched-chain ester formation in ripening apple fruit. Proc. Natl. Acad. Sci USA 2021, 118, e2009988118. [Google Scholar] [CrossRef]
- Frantom, P.A. Structural and functional characterization of alpha-isopropylmalate synthase and citramalate synthase, members of the LeuA dimer superfamily. Arch. Biochem. Biophys. 2012, 519, 202–209. [Google Scholar] [CrossRef]
- Hulme, A.C.; Wooltorton, L.S.C. Determination and isolation of the non-volatile acids of pome fruits and a study of acid changes in apples during storage. J. Sci. Food Agric. 1958, 9, 150–158. [Google Scholar] [CrossRef]
- Howell, D.M.; Xu, H.; White, R.H. (R)-citramalate synthase in methanogenic archaea. J. Bacteriol. 1999, 181, 331–333. [Google Scholar] [CrossRef] [Green Version]
- Johnson, D.W.; Eastham, G.R.; Poliakoff, M.; Huddle, T.A. Method of Producing Arcylic and Methacrylic Acid. U.S. Patent 8,933,179, 13 January 2015. [Google Scholar]
- Webb, J.; Springthorpe, V.; Rossoni, L.; Minde, D.P.; Langer, S.; Walker, H.; Alstrom-Moore, A.; Larson, T.; Lilley, K.; Eastham, G.; et al. Systems analyses reveal the resilience of Escherichia coli physiology during accumulation and export of the nonnative organic acid citramalate. Msystems 2019, 4, e00187-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Webb, J.P.; Arnold, S.A.; Baxter, S.; Hall, S.J.; Eastham, G.; Stephens, G. Efficient bio-production of citramalate using an engineered Escherichia coli strain. Microbiology 2018, 164, 133–141. [Google Scholar] [CrossRef]
- Zhang, K.C.; Woodruff, A.P.; Xiong, M.; Zhou, J.; Dhande, Y.K. A synthetic metabolic pathway for production of the platform chemical isobutyric acid. ChemSusChem 2011, 4, 1068–1070. [Google Scholar] [CrossRef] [PubMed]
- Hossain, A.H.; Hendrikx, A.; Punt, P.J. Identification of novel citramalate biosynthesis pathways in Aspergillus niger. Fungal Biol. Biotechnol. 2019, 6, 19. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, Y.; Yadav, J.S.; Sih, C.J. (S)-citramalic acid, a useful chiral synthon for the synthesis of 15-deoxy-16(S)-hydroxy-16-methylprostaglandins. Tetrahedron Lett. 1980, 21, 2481–2482. [Google Scholar] [CrossRef]
- Wilkes, J.B.; Wall, R.G. Citric Acid and Citramalic Acid Preparation. U.S. Patent 4,022,823, 10 May 1977. [Google Scholar]
- Takao, Y.; Takahashi, T.; Yamada, T.; Goshima, T.; Isogai, A.; Sueno, K.; Fujii, T.; Akao, T. Characteristic features of the unique house sake yeast strain Saccharomyces cerevisiae Km67 used for industrial sake brewing. J. Biosci. Bioeng. 2018, 126, 617–623. [Google Scholar] [CrossRef] [PubMed]
- Le Bellec, F.; Vaillant, F. Pitahaya (pitaya) (Hylocereus spp.). In Postharvest Biology and Technology of Tropical and Subtropical Fruits; Woodhead Publishing: Cambridge, UK, 2011; pp. 247–273e. ISBN 978-0-85709-090-4. [Google Scholar]
- Zhang, Z.K.; Xing, Y.M.; Ramakrishnan, M.; Chen, C.B.; Xie, F.F.; Hua, Q.Z.; Chen, J.Y.; Zhang, R.; Zhao, J.T.; Hu, G.B.; et al. Transcriptomics-based identification and characterization of genes related to sugar metabolism in ‘Hongshuijing’ pitaya. Hortic. Plant J. 2021; in press. [Google Scholar] [CrossRef]
- Hua, Q.Z.; Chen, C.B.; Tel Zur, N.; Wang, H.C.; Wu, J.Y.; Chen, J.Y.; Zhang, Z.K.; Zhao, J.T.; Hu, G.B.; Qin, Y.H. Metabolomic characterization of pitaya fruit from three red-skinned cultivars with different pulp colors. Plant Physiol. Biochem. 2018, 126, 117–125. [Google Scholar] [CrossRef]
- Wu, Y.W.; Xu, J.; Shi, M.Y.; Han, X.M.; Li, W.Y.; Zhang, X.W.; Wen, X.P. Pitaya: A potential plant resource of citramalic acid. CyTA-J. Food 2020, 18, 249–256. [Google Scholar] [CrossRef] [Green Version]
- Hu, Z.; Wang, H.; Hu, G.B. Measurement of sugars, organic acids and vitamin C in litchi fruit by performance liquid chromatography. J. Fruit Sci. 2005, 22, 582–585. [Google Scholar]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Chen, C.B.; Wu, J.Y.; Hua, Q.Z.; Tel-Zur, N.; Xie, F.F.; Zhang, Z.K.; Chen, J.Y.; Zhang, R.; Hu, G.B.; Zhao, J.T.; et al. Identification of reliable reference genes for quantitative real-time PCR normalization in pitaya. Plant Methods 2019, 15, 70. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Fan, X.G.; Zhao, H.D.; Wang, X.M.; Cao, J.K.; Jiang, W.B. Sugar and organic acid composition of apricot and their contribution to sensory quality and consumer satisfaction. Sci. Hortic. 2017, 225, 553–560. [Google Scholar] [CrossRef]
- Minamikawa, M.F.; Nonaka, K.; Kaminuma, E.; Kajiya-Kanegae, H.; Onogi, A.; Goto, S.; Yoshioka, T.; Imai, A.; Hamada, H.; Hayashi, T.; et al. Genome-wide association study and genomic prediction in citrus: Potential of genomics-assisted breeding for fruit quality traits. Sci. Rep. 2017, 7, 4721. [Google Scholar] [CrossRef]
- Ramadan, N.S.; Wessjohann, L.A.; Mocan, A.; Vodnar, D.C.; El-Sayed, N.H.; El-Toumy, S.A.; Mohamed, D.A.; Aziz, Z.A.; Ehrlich, A.; Farag, M.A. Nutrient and sensory metabolites profiling of Averrhoa carambola L. (Starfruit) in the context of its origin and ripening stage by GC/MS and chemometric analysis. Molecules 2020, 25, 2423. [Google Scholar] [CrossRef]
- Xu, J.D.; Yan, J.J.; Li, W.J.; Wang, Q.Y.; Wang, C.X.; Guo, J.X.; Geng, D.L.; Guan, Q.M.; Ma, F.W. Integrative analyses of widely targeted metabolic profiling and transcriptome data reveals molecular insight into metabolomic variations during apple (Malus domestica) fruit development and ripening. Int. J. Mol. Sci. 2020, 21, 4797. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.M.; Oh, T.J.; Kim, S.H.; Kim, H.Y.; Chung, H.; Min, D.S.; Auh, J.H.; Lee, H.J.; Lee, J.; Choi, H.K. Comprehensive metabolic profiles of mulberry fruit (Morus alba Linnaeus) according to maturation stage. Food Sci. Biotechnol. 2016, 25, 1035–1041. [Google Scholar] [CrossRef] [PubMed]
- Qiu, G.L.; Zhuang, Q.G.; Li, Y.F.; Li, S.Y.; Chen, C.; Li, Z.H.; Zhao, Y.Y.; Yang, Y.; Liu, Z.B. Correlation between fruit weight and nutritional metabolism during development in CPPU-treated Actinidia chinensis ‘Hongyang’. PeerJ 2020, 8, e9724. [Google Scholar] [CrossRef]
- Wu, X.H.; Tovilla-Coutino, D.B.; Eiteman, M.A. Engineered citrate synthase improves citramalic acid generation in Escherichia coli. Biotechnol. Bioeng. 2020, 117, 2781–2790. [Google Scholar] [CrossRef]
- Walker, R.P.; Paoletti, A.; Leegood, R.C.; Famiani, F. Phosphorylation of phosphoenolpyruvate carboxykinase (PEPCK) and phosphoenolpyruvate carboxylase (PEPC) in the flesh of fruits. Plant Physiol. Biochem. 2016, 108, 323–327. [Google Scholar] [CrossRef]
- Yao, Y.X.; Li, M.; Zhai, H.; You, C.X.; Hao, Y.J. Isolation and characterization of an apple Cytosolic malate dehydrogenase gene reveal its function in malate synthesis. J. Plant Physiol. 2011, 168, 474–480. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.X.; Dong, Q.L.; Zhai, H.; You, C.X.; Hao, Y.J. The functions of an apple Cytosolic malate dehydrogenase gene in growth and tolerance to cold and salt stresses. Plant Physiol. Biochem. 2011, 49, 257–264. [Google Scholar] [CrossRef]
- Chen, Q.Q.; Wang, B.P.; Ding, H.Y.; Zhang, J.; Li, S.C. Review: The role of NADP-malic enzyme in plants under stress. Plant Sci. 2019, 281, 206–212. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.H.; Wang, S.; Zhang, Y.; Guo, R.Z.; Wu, M.S.; Chen, Q.; Xue, Z.H. Effects of chitosan and calcium chloride treatments on malic acid-metabolizing enzymes and the related gene expression in post-harvest pear cv. ‘Huang guan’. Sci. Hortic. 2014, 165, 252–259. [Google Scholar] [CrossRef]
- Gao, L.; Zhao, S.J.; Lu, X.Q.; He, N.; Zhu, H.J.; Dou, J.L.; Liu, W.G. Comparative transcriptome analysis reveals key genes potentially related to soluble sugar and organic acid accumulation in watermelon. PLoS ONE 2018, 13, e0190096. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Wei, X.X.; Ali, M.M.; Rizwan, H.M.; Li, B.Q.; Li, H.; Jia, K.J.; Yang, X.L.; Ma, S.F.; Li, S.J.; et al. Changes in the content of organic acids and expression analysis of citric acid accumulation-related genes during fruit development of yellow (Passiflora edulis F. Flavicarpa) and purple (Passiflora edulis F. Edulis) passion fruits. Int. J. Mol. Sci. 2021, 22, 5765. [Google Scholar] [CrossRef] [PubMed]
- Leroy, B.; De Meur, Q.; Moulin, C.; Wegria, G.; Ruddy, W. New insight into the photoheterotrophic growth of the isocytrate lyase-lacking purple bacterium Rhodospirillum rubrum on acetate. Microbiology 2015, 161, 1061–1072. [Google Scholar] [CrossRef]
- Atsumi, S.; Liao, J.C. Directed evolution of Methanococcus jannaschii citramalate synthase for biosynthesis of 1-propanol and 1-butanol by Escherichia coli. Appl. Environ. Microbiol. 2008, 74, 7802–7808. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.H.; Eiteman, M.A. Synthesis of citramalic acid from glycerol by metabolically engineered Escherichia coli. J. Ind. Microbiol. Biotechnol. 2017, 44, 1483–1490. [Google Scholar] [CrossRef]
- Textor, S.; de Kraker, J.W.; Hause, B.; Gershenzon, J.; Tokuhisa, J. MAM3 catalyzes the formation of all aliphatic glucosinolate chain lengths in Arabidopsis. Plant Physiol. 2007, 144, 60–71. [Google Scholar] [CrossRef] [Green Version]


| Cultivars (n = 3) | Stages | Contents (mg/g) | ||||
|---|---|---|---|---|---|---|
| Malic Acid | Citric Acid | Citramalic Acid | Oxalic Acid | Ascorbic Acid | ||
| ‘GHH’ | S1 | 2.177 ± 0.064 d | 0.481 ± 0.018 b | 3.136 ± 0.609 bc | 0.515 ± 0.077 b | 0.123 ± 0.004 a |
| S2 | 1.753 ± 0.326 d | 0.377 ± 0.063 b | 3.997 ± 0.309 ab | 0.713 ± 0.087 a | 0.105 ± 0.015 bc | |
| S3 | 1.638 ± 0.460 d | 0.385 ± 0.106 b | 3.542 ± 0.856 abc | 0.514 ± 0.040 b | 0.067 ± 0.024 b | |
| S4 | 5.487 ± 0.984 c | 0.543 ± 0.100 b | 4.380 ± 0.306 a | 0.434 ± 0.036 bc | 0.065 ± 0.021 b | |
| S5 | 11.133 ± 0.729 b | 0.849 ± 0.13 ab | 3.662 ± 0.28 abc | 0.451 ± 0.131 bc | 0.066 ± 0.018 b | |
| S6 | 13.493 ± 1.430 a | 1.218 ± 0.375 a | 2.609 ± 0.192 c | 0.413 ± 0.040 bc | 0.066 ± 0.007 b | |
| S7 | 7.228 ± 0.523 c | 1.055 ± 0.090 a | 0.209 ± 0.061 d | 0.283 ± 0.056 c | 0.022 ± 0.011 c | |
| ‘GHB’ | S1 | 2.922 ± 0.247 d | 0.000 ± 0.000 b | 4.062 ± 0.699 b | 0.054 ± 0.018 c | 0.179 ± 0.016 a |
| S2 | 3.990 ± 0.646 cd | 0.000 ± 0.000 b | 5.597 ± 0.000 a | 0.095 ± 0.021 b | 0.203 ± 0.013 a | |
| S3 | 4.449 ± 0.062 cd | 0.000 ± 0.000 b | 5.931 ± 0.880 a | 0.139 ± 0.009 a | 0.178 ± 0.030 a | |
| S4 | 15.196 ± 1.500 a | 0.165 ± 0.030 a | 4.295 ± 0.187 b | 0.073 ± 0.024 bc | 0.090 ± 0.006 b | |
| S5 | 18.116 ± 2.735 a | 0.122 ± 0.030 a | 3.160 ± 0.228 bc | 0.072 ± 0.014 bc | 0.048 ± 0.006 c | |
| S6 | 7.159 ± 0.204 c | 0.202 ± 0.023 a | 2.693 ± 0.283 c | 0.036 ± 0.011 c | 0.034 ± 0.002 c | |
| S7 | 11.072 ± 1.226 b | 0.138 ± 0.018 a | 1.241 ± 0.071 d | 0.041 ± 0.003 c | 0.052 ± 0.003 c | |
| ‘WCHL’ | S1 | 0.848 ± 0.0140 e | 0.000 ± 0.000 d | 4.045 ± 0.059 b | 0.582 ± 0.012 a | 0.280 ± 0.007 a |
| S2 | 1.489 ± 0.152 e | 0.000 ± 0.000 d | 4.499 ± 0.406 b | 0.526 ± 0.155 a | 0.185 ± 0.034 b | |
| S3 | 3.379 ± 1.076 d | 0.433 ± 0.210 c | 5.815 ± 0.310 a | 0.323 ± 0.067 b | 0.154 ± 0.003 b | |
| S4 | 13.997 ± 1.095 a | 1.704 ± 0.219 a | 4.021 ± 0.205 b | 0.256 ± 0.071 bc | 0.185 ± 0.004 b | |
| S5 | 10.183 ± 0.507 b | 0.952 ± 0.083 b | 1.437 ± 0.078 c | 0.248 ± 0.067 bc | 0.093 ± 0.003 c | |
| S6 | 5.624 ± 0.313 c | 0.451 ± 0.018 c | 0.910 ± 0.000 d | 0.154 ± 0.033 c | 0.058 ± 0.001 d | |
| S7 | 5.945 ± 0.950 c | 0.569 ± 0.110 c | 0.293 ± 0.010 e | 0.189 ± 0.029 bc | 0.047 ± 0.009 d | |
| ‘YCHL’ | S1 | 2.422 ± 0.140 de | 1.818 ± 0.237 f | 1.561 ± 0.237 b | 0.692 ± 0.022 a | 0.220 ± 0.012 a |
| S2 | 2.698 ± 0.140 cd | 2.503 ± 0.135 e | 1.935 ± 0.260 a | 0.653 ± 0.021 a | 0.177 ± 0.015 a | |
| S3 | 3.058 ± 0.071 c | 3.060 ± 0.126 de | 1.498 ± 0.092 b | 0.690 ± 0.005 a | 0.196 ± 0.014 a | |
| S4 | 5.122 ± 0.275 a | 6.736 ± 0.418 a | 0.632 ± 0.054 c | 0.617 ± 0.036 a | 0.120 ± 0.039 b | |
| S5 | 3.837 ± 0.400 b | 5.125 ± 0.224 b | 0.000 ± 0.000 d | 0.458 ± 0.003 b | 0.006 ± 0.001 c | |
| S6 | 2.138 ± 0.162 e | 4.185 ± 0.261 c | 0.000 ± 0.000 d | 0.331 ± 0.040 c | 0.003 ± 0.000 c | |
| S7 | 1.576 ± 0.202 f | 3.620 ± 0.298 cd | 0.000 ± 0.000 d | 0.263 ± 0.075 c | 0.015 ± 0.001 c | |
| Cultivars | HuIPMS1 | HuIPMS2 | HuIPMS3 |
|---|---|---|---|
| ‘GHB’ | −0.083 | 0.871 * | 0.848 * |
| ‘GHH’ | 0.350 | 0.786 * | 0.590 |
| ‘WCHL’ | −0.072 | 0.473 | 0.886 ** |
| ‘YCHL’ | 0.782 | 0.811 * | 0.453 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, J.; Yuan, Y.; Xie, F.; Zhang, Z.; Chen, J.; Zhang, R.; Zhao, J.; Hu, G.; Qin, Y. Metabolic Profiling of Organic Acids Reveals the Involvement of HuIPMS2 in Citramalic Acid Synthesis in Pitaya. Horticulturae 2022, 8, 167. https://doi.org/10.3390/horticulturae8020167
Chen J, Yuan Y, Xie F, Zhang Z, Chen J, Zhang R, Zhao J, Hu G, Qin Y. Metabolic Profiling of Organic Acids Reveals the Involvement of HuIPMS2 in Citramalic Acid Synthesis in Pitaya. Horticulturae. 2022; 8(2):167. https://doi.org/10.3390/horticulturae8020167
Chicago/Turabian StyleChen, Jiaxuan, Yuanju Yuan, Fangfang Xie, Zhike Zhang, Jianye Chen, Rong Zhang, Jietang Zhao, Guibing Hu, and Yonghua Qin. 2022. "Metabolic Profiling of Organic Acids Reveals the Involvement of HuIPMS2 in Citramalic Acid Synthesis in Pitaya" Horticulturae 8, no. 2: 167. https://doi.org/10.3390/horticulturae8020167
APA StyleChen, J., Yuan, Y., Xie, F., Zhang, Z., Chen, J., Zhang, R., Zhao, J., Hu, G., & Qin, Y. (2022). Metabolic Profiling of Organic Acids Reveals the Involvement of HuIPMS2 in Citramalic Acid Synthesis in Pitaya. Horticulturae, 8(2), 167. https://doi.org/10.3390/horticulturae8020167








