Abstract
A plant microbiome is an important factor in plant growth, stress resistance, health status, and consumer quality and safety. The rhizosphere microbiome evolves in a negotiation between microbial communities that inhabit soil and plant root tissue. In this study, the rhizosphere and root internal tissue microbiome of six varieties of lettuce were analyzed in normal conditions and under salinity stress. The metabarcoding analysis used 16S rRNA gene and ITS2 region sequencing. The microbiomes of root samples were significantly less diverse with different members of the community compared to those of the rhizosphere. A significant effect of lettuce variety was found on the diversity index for bacteria and fungi. Varieties formed very different communities of bacteria in roots. Pseudomonas, Herbaspirillum, Mycobacterium, potentially pathogenic Enterobacter, and other genera were more prevalent in certain varieties. Salinity stress had a significant negative impact on bacterial diversity and community composition, whereas the diversity of fungi has not changed significantly, and the fungal community has changed less than the bacterial one. Changes were more evident in varieties that were more resistant to salinity stress than in sensitive varieties.
1. Introduction
Lettuce (Lactuca sativa L.) is one of the most freshly consumed vegetables [1]. It is an important source of fiber, vitamins, antioxidants, and other nutrients that positively impact human health and well-being [2,3]. Lettuce comprises many varieties with diverse phenotypes [4]. Globally, lettuces are grown in an area of about 1.8 million hectares in field or greenhouse conditions [5]. They are grown in many soil or substrate types, including degraded saline soil fields.
Plants and the environment are inhabited by various microbial species that form complex microbial communities called microbiomes. Plant microbiomes are currently understood as the sum of all microbial genomes that inhabit plant leaves, fruits, and rhizospheres in interconnected microbial network complexes [6,7,8,9]. Members of these communities affect plants during their growth and can act as neutral, beneficial, or harmful. Plant-associated microorganisms have been proven to affect the entire plant’s growth process, nutrition, productivity, and disease resistance [10,11].
Research of plant-associated microbiomes is currently focused mainly on the rhizosphere [12]. The rhizosphere is an interaction zone between the soil and the plants, rich in microbial diversity. The decomposition of organic matter, nutrient cycling and other ecologically important microbe-driven processes are very intensive here. Available data suggest that plants are involved in the selection process when the microbiome of the rhizosphere is distinguished from soil. Plant products such as carbohydrates, amino acids or other secondary root-secreted metabolites (rhizodeposits) create specific conditions for microbial growth [13]. It is assumed that plants could modulate the rhizosphere microbiota by selective stimulation of microbiome members beneficial for their health and growth [14]. Rhizodeposition leads to development of specific rhizosphere microbiomes, and it is also the first step in selection of endophytic microbial assemblage within root tissues. Some pilot studies showed that the plant genotype is responsible for the second step of root microbiome shaping [15]. The effect of plant genotypes on root-associated microbiomes was found primarily on longer living plants such as trees and perennial crops [16,17] but also in some annual crops [18]. It is mainly connected to the different physiology of plant genotypes [19].
Plant microbiomes not only affect the plant itself, but some microbiome members can directly or indirectly affect human health [14]. Plant tissues may be successfully colonized by pathogenic bacteria such as Salmonella, Escherichia coli, Campylobacter, Listeria, or Enterobacteriaceae [20,21]. Members of other bacterial genera (Pseudomonas, Burkholderia, Pantoea and others) may act as potential pathogens, although they are common members of the rhizosphere and plant microbiomes [22,23,24]. Even though some contamination may occur during handling [25], it is usually associated with agricultural practices such as organic fertilizers [26] or untreated irrigation water [27]. As lettuce is consumed almost solely in its fresh state without any heat treatment, any contamination by human pathogenic bacteria may be dangerous [28]. The rhizosphere is considered to be a playground in which complex microbial interactions may allow or suppress the development of plant or human pathogens [29].
Salinity of soil is one of the key factors that limits agricultural use of the soil in certain areas [30]. Salinity stress affects plant growth, productivity, and health [31]. Selection of saline-tolerant varieties can open a way to use even degraded saline soil. Some lettuce varieties have been already selected to tolerate high salinity, and future research is desirable [32]. Recent studies showed distinctive composition of microbiomes in saline soils [33]. It is also associated with the specific development of plant microbiomes under salinity stress [34,35]. Supposedly, such microbiomes help the plant to withstand the harmful conditions of saline soil [36]. Plants participate in this selection primarily through their products such as exopolysaccharides, playing a role in root biofilm formation [37]. Research of microbiomes in saline-stressed plants can be useful in future selections of microbiome members that can increase plant tolerance to salinity stress [38,39]. Several strains of plant growth-promoting rhizobacteria with such ability were already described [40,41].
Understanding the biological processes that shape the structure and dynamics of a microbiome of the rhizosphere is a fundamental step to ensure plant productivity and in producing safe food [10]. Although some studies examined the interaction between plant genotype and salinity, differences in tolerance were not reported for used genotypes. Salinity-tolerant varieties should maintain the development of microbiomes in salt soils due to the ability to support bacteria with root exudates.
The aim of the study was to characterize and compare the development of microbiomes in the rhizosphere and internal root tissue of six different varieties of lettuce and to assess the reaction of these microbiomes to salinity stress. We hypothesize that the lettuce microbiome depends on the variety, and the reaction to salinity is different in varieties that are naturally tolerant of high salinity.
2. Materials and Methods
Six varieties of lettuce—Bibb, SM09PA, Romana Larga Blanca, Dark Green Romaine, Pavane, and Sentry were used in the research. The first three have a higher salt tolerance according to Xu and Mou [32] and Adhikari et al. [42]. The lettuce seeds were surface sterilized for 2 min in 0.5% sodium hypochlorite and then washed three times with sterile distilled water. Seeds were pre-germinated on filter paper in a Petri dish moistened by sterile water. Pre-germination took seven days in a climate chamber under a 16 h light and 8 h dark regime, at 20 and 10 °C, respectively. The germinated seeds were then transplanted into 90 × 80 × 80 mm pots (single seed per pot) filled with commercially available growing substrate (gardening substrate with vitality complex, AgroCS, Říkov, Czech Republic).
Lettuce was grown in six pots for each variety. Growing conditions were based on the modified protocol from Wei et al. [43], with light cycles of a 16 hour’s day and 8 hour’s night, a temperature of 18–20 °C, and 60% humidity. The air was purified from pathogens. The plants were watered every two days with sterile distilled water to maintain the desired humidity. After two weeks, watering in an experimental group (three pots per variety) with a solution of 100 mM NaCl induced osmotic stress whereas the control group (3 pots) was watered with distilled water. According to the protocol [43], samples were watered on the 1st, 4th, and 7th day of the stress period by 30 mL and then by 40 mL on days 10 and 13. Samples were collected two weeks after the stress induction. The total conductivity of EC1:5 measured in soil water extract (ratio 1:5 w/v) reached 4.64 ± 0.31 dS/m in the treated samples compared to 1.36 ± 0.16 dS/m in the control soil after the stress period.
Each variety provided 2 types of samples—the internal root microbiome and the root surface soil microbiome (the rhizosphere). Roots were cleaned from the soil softly by a brush, and rhizosphere samples were prepared as the leachate of the root surface into 0.9% saline solution in 50 mL falcon tubes. The suspension of soil particles containing the microbiome of the rhizosphere was centrifuged for 20 min at 6000× g, and the resulting pellets were used for DNA extraction. The roots were then rinsed several times with sterile distilled water, the surface was sterilized with 2% sodium hypochlorite solution for 5 min, and was again rinsed with water several times. The prepared roots were used for DNA extraction.
2.1. DNA Extraction, Amplification, and Sequencing
DNA was extracted using the MOBIO Powersoil DNA extraction kit (Qiagen, Hilden, Germany). Garnet particles in the kit were replaced by 2 mm diameter zirconium oxide beads to better disintegrate the root tissue. In total, 250 mg of sample (root tissue or rhizosphere pellet) was homogenized with the BeadBug homogenizer (Benchmark scientific, Sayreville, NJ, USA).
General bacterial primers 515F and 806R [44] enhanced by 8 bp identification sequence (tag) were used for amplification of V4 region of 16S rRNA gene. For analysis of fungal community, primers g ITS7 and ITS4 [45] were used in the amplification of the ITS2 region (Supplement Table S1). The composition of the PCR mixture was as follows: 15 µL KAPA HIFI HS MIX 2X (Roche, Indianapolis, IN, USA), 4 µL of each primer with a concentration of 2.5 µM, and 1 µL of extracted DNA. The amplification was carried out in the Stratagene mx3005p thermal cycler (Agilent, Santa Clara, CA, USA) with the following configuration. Initial denaturation for 90 s at 98 °C was followed by 35 cycles of denaturation for 15 s at 98 °C, annealing for 15 s at 62 °C, and by extension for 15 s at 72 °C. The final extension was 2 min at 72 °C. The PCR products were purified using a PCR purification kit (Jena Bioscience, Jena, Germany). Then, the PCR products were quantified by qubit (Thermo scientifics, Waltham, MA, USA), diluted to the same concentration and pooled together. Illumina adapters were attached by TruSeq LT PCR free kit (Illumina, San Diego, CA, USA) with modification involving the skip of DNA fragmentation and size selection. The library was quantified by qPCR using NebNext Quantification kit (New England BioLabs, Ipswich, MA, USA), diluted to 4 nM concentration, and denatured. The MiSeq Reagent Kit v3 (600-cycle) was used for sequencing and a 20 pM library with 1% PhiX spike was loaded into the cartridge.
2.2. Sequences Analysis
Acquired data was processed in the SEED2 environment (version 2.12) [46]. Forward and reverse readings were joined with join2fastqc and sequences with overall quality less than Q30 were removed from further analysis. The primers were removed, and the sequences were processed by the Vsearch [47] algorithm to detect chimeras, which were also removed from further analysis. Chimera-free sequences were clustered to operational taxonomic units (OTUs) using Vsearch set at 97% similarity level. The most abundant sequence was found in each cluster (OTU), and such sequences for each OTU were identified using the RDP classifier [48]. Sequences of chloroplasts, mitochondria, and the ITS region of plants were removed from further analysis. The most abundant sequences in each OTU were aligned with MAAFT [49], and a phylogenetic tree was constructed using PhyML [50]. Using the phylogenetic tree and OTU table, a weighted Unifrac [51] distance matrix was calculated in the R statistical environment [52]. Non-metric multidimensional scaling (NMDS) analysis and permutational multivariate analysis of variance (PERMANOVA) statistics based on the Unifrac matrix were obtained with the package Vegan [53]. Heatmaps were made using the Heatmap3 package [54] in R. Linear discriminant analysis effect size (LefSe) [55] was used to compare the abundance of taxa between varieties and the discovery of biomarkers. EdgeR [56] was used to analyze changes in tax abundance due to the salinity stress in each variety.
Tables of OTUs were rarefied to the lowest sequence count among samples for alpha diversity assessment. Alpha diversity was described by OTU Richness, Shannon’s index, and Pielou’s corrected evenness. The indices were statistically evaluated using the multifactor ANOVA in R.
3. Results
There were 1,104,413 and 1,137,914 high-quality chimera-free sequences acquired for the ITS2 region and the 16S rRNA gene, respectively. All sequence data was submitted to GenBank databases as part of BioProject PRJNA893639. The mean number of sequences per sample was 15,339 for ITS2 and 15,804 for the 16S rRNA gene. However, samples taken from the plants’ roots contained 12–30% of sequences identified as lettuce ITS2, which were removed before analysis. Similarly, 16S rRNA gene sequences from root samples showed a considerably high portion of chloroplast (21–29%) and mitochondria (16–23%) sequences that needed to be removed prior to the analysis. Rhizosphere samples contained less than 10% of such sequences. In total, 4429 OTUs were generated from 16 rRNA gene sequences, and 1830 OTUs were found for ITS sequences.
3.1. Diversity and Structure of Bacterial Community
Using 3-factor ANOVA, the sample type was identified as the most important factor affecting diversity indices (p < 0.001) (Table 1). Furthermore, the effect of osmotic stress was highly significant (p < 0.001). Variety did not affect the richness significantly (p = 0.183) but evenness and Shannon’s index varied significantly (p < 0.001) among lettuce varieties. Moreover, highly significant interactions between variety and sample type, variety and stress, as well as between sample type and stress (all p < 0.001) were detected. These interactions indicate a mixed response of microbiomes to salinity stress among lettuce varieties. Moreover, this response of microbiomes in the rhizosphere zone and root tissue is clearly not the same. The lowest values of indices were found in the root samples of salinity tolerant varieties in saline conditions (Supplement Table S2).

Table 1.
Alpha diversity indices of bacterial microbiomes in root and rhizosphere of six varieties of lettuce under normal conditions and salinity stress.
The primary driver of the differences in the bacterial community composition was the variety (PERMANOVA p < 0.001; R2 = 0.15). In addition, the sample type and salinity stress significantly affected the community, and all interactions were significant. Multivariate dispersion of root samples was significantly wider than dispersion of rhizosphere samples (BETADISPER p < 0.001). The difference in dispersion can affect PERMANOVA results, so root tissue and rhizosphere samples were analyzed separately (Figure 1). Variety was the most significant factor in both sample types, and differences were confirmed by pairwise comparison (Supplement Table S3). For certain varieties (Bibb, Romana Larga Blanca, Dark Green Romaine, and SM09PA), salinity stressed samples of lettuce roots were clearly separated from control root samples according to PERMANOVA results (p < 0.001). The clustering of the varieties was also observed in the rhizosphere samples. The effect of salinity stress was not clear, despite the significant interaction between stress and variety (p = 0.007).
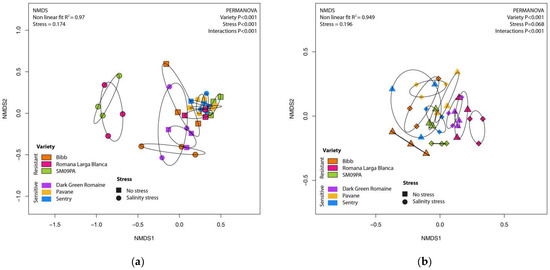
Figure 1.
NMDS scatterplot of bacterial community in roots (a) and rhizosphere (b) of six lettuce varieties in normal conditions and under salinity stress.
Rhizosphere microbiome samples contained slightly less Proteobacteria and Firmicutes but more Actinobacteria, Chloroflexi, and Planctomyces phyla (Figure 2). Analysis at the genera level led to clear clustering depending on the sample type and variety (Figure 3). The most common genera were Acidobacteria group GP6, Pseudomonas, Herbaspirillum, and Flavobacterium. In the rhizosphere of non-stressed plants, LefSe analysis showed Mycobacterium and Sedimenticola to be biomarkers for the Pavane variety, and Herbaspirillum and Conexibacter for the Sentry variety. Romana Larga Blanca’s variety biomarkers were Ktenobacter and Streptomyces. Limnobacter’s and Aquabacterium’s were selected for the Bibb variety. Dokdonella and Salinibacterium were biomarkers for the Dark Green Romaine variety, and Blastocladella for the SM09PA variety. It was not fully consistent with the analysis of non-stressed plant root samples where Pseudomonas, Mycobacterium, and Actinomadura have been found as biomarkers for the Pavane variety. Conexibacter, Burkholderia, Sacharibacteria, Halomonas, and others were biomarkers for the Sentry variety whereas Ktedonobacter, Otitutus, and Aridibacter were biomarkers for the Dark Green Romaine variety. Ornatilinea and Thermogutta were selected for the Bibb variety, and Cellvibrio, Staphylococcus, and Methylophilus for the SM09PA variety. Bar charts (Figure 4) show the first 20 biomarker OTUs for varieties in rhizospheres and root samples, which reflect the biomarker genera.
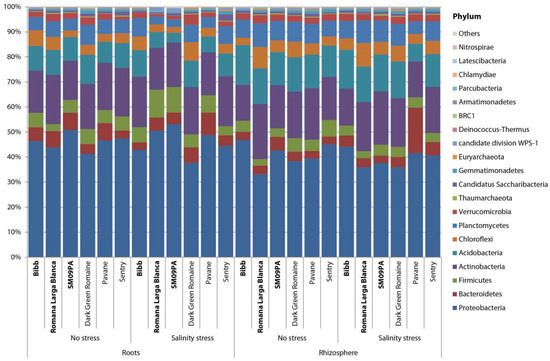
Figure 2.
Bar chart of bacterial phyla composition in roots and rhizosphere of six lettuce varieties in normal conditions and under salinity stress.
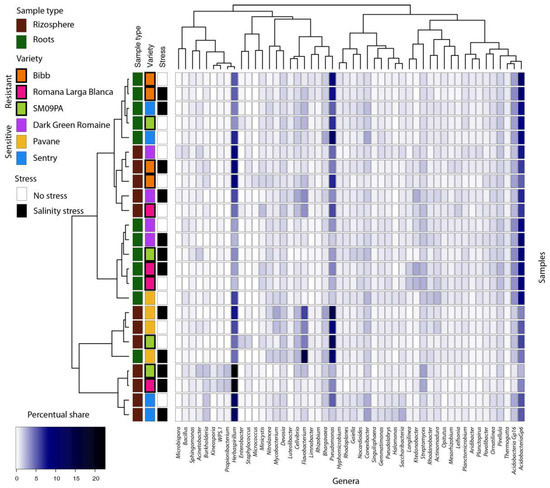
Figure 3.
Heatmap of bacterial genera composition in roots and rhizosphere of six lettuce varieties in normal conditions and under salinity stress. Only genera with min. 2% occurrences in any samples are listed. Dendrograms are based on occurrence of genera in samples (Bray–Curtis distance, complete clustering).
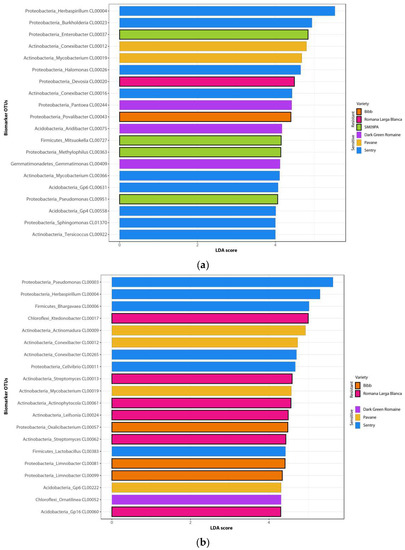
Figure 4.
Bar charts of the first 20 LefSe biomarkers among bacterial OTUs in roots (a) and rhizosphere (b) of six lettuce varieties.
Due to the significant interactions of factors, the microbiome response to salinity stress was analyzed separately for each sample type (Supplement Tables S4 and S5).
The most significant changes were found in the varieties Romana Larga Blanca and SM09PA, where some genera were changed by up to five two-fold logs.
In these varieties, Herbaspirillum was positively affected by salinity whereas the occurrence of Pseudomonas, Minicystis, or Enterobacter was lower. Among the less prevalent genera, Kineosporia, Mogibacterium, Cupriavidus, Methyloceanibacter, Pseudobacteroides, and Gelria were positively affected by salinity in the roots. In rhizosphere samples, salinity affected the microbiomes of Pavane and SM09PA varieties. Microbiome reactions to salinity were often opposite in those varieties. Pseudomonas, Flavobacterium, Cellvibrio, and Luteolibacter increased in Pavane but decreased in SM09PA. There was also a very significant increase of known halotolerant bacteria Jeotgalicoccus found in some samples. Other halotolerant genera such as Halomonas, Halobacillus, or Thiohalobacter showed an increase, but also decreased depending on the variety and sample type.
3.2. Diversity and Structure of Fungal Community
Similar to the bacteria, most of the difference between the diversity estimates of fungi (Table 2) was attributed to sample type, where all indices were significantly lower in the root samples than in the rhizospheres (p < 0.001 for all indices). The effect of the variety was lower (p < 0.001, p = 0.036, and p = 0.029 for richness, evenness, and Shannon’s index respectively). Contrary to bacterial diversity, salinity stress was not a significant factor for any of the indices (p = 0.565, p = 0.890, and p = 0.984). However, certain interactions of stress and variety were found (p = 0.002, p = 0.003, and p < 0.001) and there was great variance among samples (Supplement Table S6).

Table 2.
Alpha diversity indices of fungal microbiomes in root and rhizosphere of six varieties of lettuce under normal conditions and salinity stress.
The composition of the fungal microbiome was quite different between the sample types (PERMANOVA p < 0.001; R2 = 0.25). In the NMDS plot (not shown), samples created distinct groups and again, their dispersions were significantly different (BETADISPER p < 0.001). In the rhizosphere, lettuce varieties formed clearly separated clusters (PERMANOVA p < 0.001; R2 = 0.48) but any grouping of root samples according to the variety in the NMDS plot was not obvious (PERMANOVA p = 0.086) (Figure 5). The pairwise comparison between the varieties demonstrated significant results in the rhizosphere only (Supplement Table S1). Salinity stress did not cause significant changes of fungal microbiome in the root samples although differences caused by salinity stress in the rhizosphere were significant (PERMANOVA p < 0.001; R2 = 0.15). Interactions between the variety and salinity stress (PERMANOVA p < 0.001) indicated different reactions of the root and the rhizosphere microbiomes to stress in particular varieties.
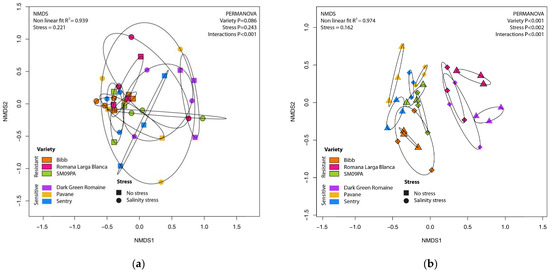
Figure 5.
NMDS scatterplot of fungal community in roots (a) and rhizosphere (b) of six lettuce varieties in normal conditions and under salinity stress.
Significantly less Zygomycota but more Basidiomycota were observed in the root samples (Figure 6). Furthermore, most prevalent genera were different in the sample types (Figure 7). Actinomucor and Arthrobotrys were the most prevalent in the roots whereas Candida and Malassezia were the most prevalent in the rhizosphere.
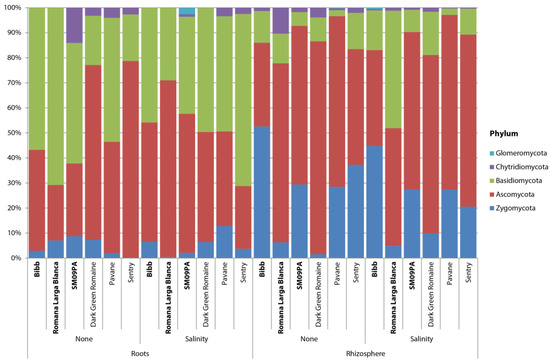
Figure 6.
Bar chart of fungal phyla composition in roots and rhizosphere of six lettuce varieties in normal conditions and under salinity stress.
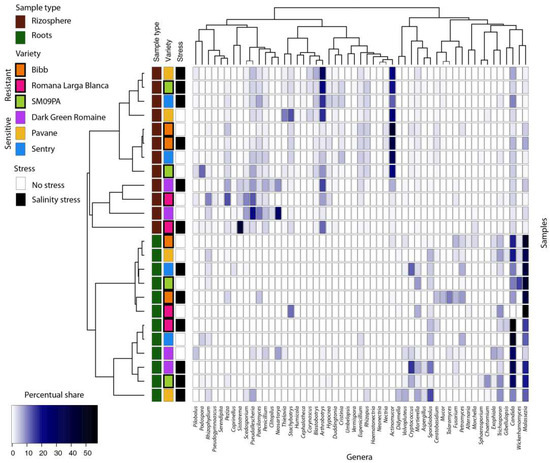
Figure 7.
Heatmap of the most common fungal genera in samples from lettuce roots and rhizosphere. Only genera with min. 3% occurrences in any samples are listed. Dendrograms are based on occurrence of genera in samples (Bray–Curtis distance, complete clustering).
Actinomucor, Malassezia, and Rhizopus were identified as biomarker taxa for the Bibb variety whereas Sistotrema and Scedosporium were identified for the Romana Larga Blanca variety. Arthrobortys, Thielavia, and Blastobotrys were biomarkers for the Pavane variety. Neosartorya, Pseudaellescherichia, Paecilomyces, and Penicillium were found for the Dark green variety, and Cristinia for the Sentry variety. The biomarkers for SM09PA were Podosphora and Pilobolus. Despite different biomarker genera, some OTUs from the same fungal species were found as biomarkers for certain varieties (Figure 8). LefSe analysis did not show any statistically significant results for any variety in the root samples.
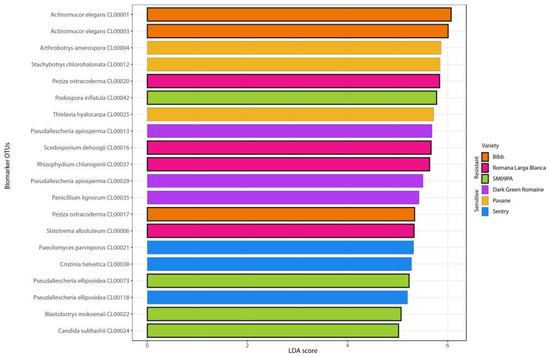
Figure 8.
Bar chart of the first 20 LefSe biomarkers among fungal OTUs in rhizosphere of six lettuce varieties.
In the root samples, there was no differently abundant taxon in response to salinity stress. However, several differentially abundant taxa were found for samples of the rhizosphere (Supplement Table S7). Among them, Vermispora was the only one that was solely positively affected, whereas others increased and decreased in certain varieties.
4. Discussion
Plant microbiomes are currently considered an integral part of the plant holobiont system, where the synergy between plants and related microbiota provides important system services [57,58]. However, plant pathogenic or human pathogenic members of microbiome can cause loss of yields or possess a threat for consumer health [59]. This study examined microbiome changes in the roots and rhizosphere of six varieties of lettuce. One of the main characteristics of varieties was their resistance to salinity, thus microbiomes were also examined in salinity stress conditions, and both factors affected the microbial diversity and community.
The microbiome of lettuce has been targeted by several studies that analyzed the impact of soil conditions and agricultural practices on the composition of microbiomes. In the study by Schreite, et al. [60], the bacterial community in the rhizosphere of field-grown lettuce was analyzed by denaturing gradient gel electrophoresis and pyrosequencing of the 16S rRNA gene. The microbiome was mainly shaped by the type of soil and the stage of development. In all three studied soil types, Proteobacteria phyla was enhanced in lettuce rhizosphere. Sphingomonas, Rhizobium, and Pseudomonas were among the most abundant genera. In the study of Iliev et al. [61], 98% of sequences came from nine phyla (Proteobacteria, Actinobacteria, Firmicutes, Acidobacteria, Chloroflexi, Bacteroidetes, Gemmatimonadetes, Verrucomicrobia, and Nitrospira). Cardinale et al. [62] reported that Proteobacteria, Bacteriodetes, Chloroflexi, and Actinobacteria dominated in the lettuce root microbiome. The results from those authors are in concordance with the findings of this study, where the most common phyla were Proteobacteria, Actinobacteria, Acidobacteria, Planctomyces, Firmicutes, Chloroflexi, and Bacteroidetes.
Iliev et al. [61] also showed that conventional fertilization reduced the diversity of bacteria in lettuce-related rhizospheres and recommended bio-organic fertilizers because they can increase the occurrence of bacteria previously known to suppress plant pathogens. Fertilization of lettuce by feather-based compost significantly changed the composition of the rhizosphere microbiome. [63].
Sun et al. [64] analyzed the effect of manure fertilization of lettuce. They also showed a significant shift in the bacterial community and altered resistome in the soil end episphere of lettuce. However, the endosphere of lettuce remains almost unchanged, which is important in terms of consumer safety. Erlacher et al. [65] pointed out some potentially human pathogenic bacteria within the rhizosphere and phyllosphere of lettuce. Later, the effect of biotic stress on the abundance and structure of Enterobacteriaceae was analyzed [66].
Enterobacteriaceae and other potential human pathogenic bacteria in lettuce microbiomes has gained great attention as lettuce is consumed in its fresh state, and any contamination can result in health issues. Several microbiome studies assessed the microbial community structure in lettuce leaves to examine potential human pathogens. Yeon-Cheol, Su-Jin, and Da-Young [23] used 16S rRNA gene-based sequencing to identify foodborne pathogens in lettuce during different seasons and the potentially pathogenic bacteria such as Bacillus spp., Enterococcus casseliflavus, Klebsiella pneumonia, and Pseudomonas aeruginosa were identified. Within this study, the presence of the genus Enterobacter achieved up to 1.5% of the total community in the roots of the SM09PA variety. It was also detected in the roots of other varieties as well. Furthermore, members of the bacterial genera Pseudomonas, Pantoea, or Burkholderia that were very common in the root samples can be potentially pathogenic in specific cases [22]. On the other hand, certain strains of Pantoea, Pseudomonas, or Acinetobacter may provide some growth promotion and disease resistance for plants [67]. Some human pathogenic bacteria live internalized in plant tissue and transmission between the internal tissue of the root and the consumed parts of the plant (leaves) is presumed. Metagenomic sequencing of lettuce leaves indicated that the pre-storage bacterial community is variable, usually dominated by the species Erwiniaceae and Pseudomonadaceace, and after cold storage, differences based on varieties emerge [68].
The association of microbiomes to the plant genotypse is less commonly studied, even in other plant species. The performed studies of lettuce microbiota showed that the structure of the phyllosphere microbiota is more influenced by the morphological difference of lettuce phenotypes than the lettuce genotype [69,70]. However, the root-associated microbiota needs to be studied and defined more precisely. Comparison of root microbiota in ancient and modern lettuce varieties and its wild ancestor Lactuca serriola showed that the domestication of lettuce led to the diversification of bacteria in the root system [62]. This study shows that different microbiomes could develop in the root area of certain lettuce varieties. Significantly different diversity and community composition were found among varieties. Herbaspirillum, Enterobacter, Burkholderia, Conexibacter, Mycobacterium, and other groups of bacteria were more abundant in certain varieties. Specific microbiome development may be caused by physiological properties of the varieties, their pathogen/microbe resistance system, growth ability, accumulation of nutrients, organic matter production, and rhizodeposition [15]. Due to these factors, only a portion of microbial species/strains can colonize root tissue and potentially other parts of plants. Root exudates play a key role in microbial development in the root zone [71]. Exudates are the main negotiation mechanism between plants and microorganisms in the surrounding soil [72]. Plant genotypes with a certain composition of exudates have specific microbiomes in the rhizosphere [73]. Modern varieties with faster and stronger development of roots usually harbor more diverse microbial communities [74]. Reciprocally, microorganisms in the rhizosphere can promote plant growth by producing molecules that modulate the growth of plants such as fytohormones. Jasmonic acid, salicylic acid, ethylene, cytokinins, gibberellic acid, abscisic acid, auxins, and others are produced by various species of bacteria [75]. Microorganisms can also increase the availability of nutrients (e.g., phosphorus) and thus increase plant growth [76].
As the results show, a large part of the bacterial community was shared between the rhizosphere and root internal tissue despite significantly fewer species/OTUs within the roots. On the other hand, the fungal community contained distinct species/OTUs for the root tissue and the rhizosphere. The internalizing ability of fungi is generally lower than bacteria [77]. Yeast or yeast-like species were most common in root tissues with great variability between samples. Fungal communities in the rhizosphere were shaped similarly to bacterial ones, probably due to the root exudates. Microbial loads on the surface of seeds or bacteria internalized in the seed can directly affect the microbiome of developing plants, including the rhizosphere [78]. These microbes are naturally selected and already adapted to certain conditions of plant/variety. Despite surface sterilized seeds in this assay, it is still necessary to consider the possibility that distinct development of microbiomes across varieties was caused by their primary microbial load as the microorganisms are internalized within the seed [79].
Both biotic and abiotic stresses are usually associated with microbiome changes [80]. In this study, salinity stress caused significant changes in the community. However, the effect was dependent on variety. The effect of salinity on the root associated microbiome was significant only in the bacterial community. The fungal community did not respond to the salinity stress.
Many authors [33,81,82,83] found negative correlations between soil salinity and bacterial diversity. It is based on the elevated extracellular osmolarity that leads to damage of membranes as well as proteins and nucleic acids of the bacteria. The microbial diversity decreases as a consequence, because only some species are able to adapt to these conditions. The results of this study confirmed this hypothesis. Liu et al. [84] found Proteobacteria to be less sensitive to salinity as their frequency rose alongside a salinity gradient whereas Actinobacteria decreased.
Specific microbiome reactions were found mainly in varieties that were more resistant to salinity. In salinity conditions, sensitive varieties are stressed and would greatly change their growth. Comparing the habitus, plants of the sensitive varieties were smaller than resistant varieties in the assay and probably shortened their production of rhizodeposits. Resistant varieties with maintained growth still provided a high amount of metabolites resulting in the development of specific microbiomes. Fungi are generally less sensitive to osmotic stress, and they also grow significantly slower than bacteria [85]. The combination of such factors with relatively low levels of osmotic stress and short examined periods resulted in insignificant changes. Specific microbiomes develop on the roots of plants growing in saline environments [34]. From another viewpoint, specifically developed microbiomes can help plants survive in saline conditions [36]. Modification of microbiomes may be a viable way to enhance plant tolerance to salinity stress [86,87]. For example, inoculation of rice seeds with halotolerant microbiomes obtained from marine sediment or rice fields led to the improved growth of the rice plants under salinity stress [88]. According the results, plant variety must be considered when such an inoculation is applied in the field.
5. Conclusions
The results of this study showed that the rhizosphere and root internal microbiomes are significantly affected by lettuce variety. Moreover, the microbiome of roots reacts to osmotic stress differently in certain varieties, and it seems to be related with the variety resistance to osmotic stress. Different growth of resistant and sensitive varieties is likely to be the basis of microbiome changes in salinity conditions. Changes in plant microbiomes may have consequences for plant health, yield amount and quality, and consumer safety. However, further studies are needed to determine the reasons and outcomes of the specific changes in lettuce microbiomes under osmotic stress.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae8121174/s1, Table S1. Primers used for amplification of 16S rRNA gene and ITS2 region. Table S2. One-way ANOVA comparison of alpha diversity indices of bacterial microbiome in root and rhizosphere of six varieties of lettuce under normal conditions and salinity stress. Table S3. Pairwise comparison of root and rhizosphere microbiome between six varieties of lettuce. Table S4. Changes of genera abundance in bacterial community in roots of six varieties of lettuce under salinity stress. Only genera significantly changed in at least single variety are listed. Values are two-fold logs, ** p < 0.01, * p < 0.05. Table S5. Changes of genera abundance in bacterial community in rhizosphere of six varieties of lettuce under salinity stress. Only genera significantly changed in at least single variety are listed. Values are two-fold logs, ** p < 0.01, * p < 0.05. Table S6. One-way ANOVA comparison of alpha diversity indices of fungal microbiome in root and rhizosphere of six varieties of lettuce under normal conditions and salinity stress. Table S7. Changes of genera abundance in fungal community in rhizosphere of six varieties of lettuce under salinity stress. Only genera significantly changed in at least single variety are listed. Values are two-fold logs, ** p < 0.01, * p < 0.05.
Author Contributions
Conceptualization, J.Ž. and J.M.; methodology J.M.; software J.M.; validation, J.Ž., L.U. and R.O.; formal analysis, L.U., R.A. and J.M.; investigation, J.Ž., L.U., R.A., R.O. and J.M.; resources R.O. and J.M. data curation J.M.; writing—original draft preparation, J.Ž., L.U. and J.M. writing—review and editing, J.Ž. and D.M.; project administration J.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was financially supported by a project of the Ministry of Education, Science, Research and Sport of the Slovak Republic, grant no. VEGA 1/0661/19 “Plant microbiome and safe food”.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data are available from the correspondence author upon request. Sequence data were deposited in GenBank databases under BioProject accession No. PRJNA893639.
Acknowledgments
The authors would like to thank to Ivan Šimko, USDA Agricultural Research Service, U.S. Department of Agriculture, for providing the biological material and Katarína Ražná for the set-up of the assay-growing conditions.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Shatilov, M.V.; Razin, A.F.; Ivanova, M.I. Analysis of the world lettuce market. IOP Conf. Ser. Earth Environ. Sci. 2019, 395, 012053. [Google Scholar] [CrossRef]
- Ocean, N.; Howley, P.; Ensor, J. Lettuce be happy: A longitudinal UK study on the relationship between fruit and vegetable consumption and well-being. Soc. Sci. Med. 2019, 222, 335–345. [Google Scholar] [CrossRef]
- Shi, M.; Gu, J.; Wu, H.; Rauf, A.; Bin Emran, T.; Khan, Z.; Mitra, S.; Aljohani, A.S.M.; Alhumaydhi, F.A.; Al-Awthan, Y.S.; et al. Phytochemicals, Nutrition, Metabolism, Bioavailability, and Health Benefits in Lettuce—A Comprehensive Review. Antioxidants 2022, 11, 1158. [Google Scholar] [CrossRef]
- Still, D.W. Lettuce. In Vegetables; Kole, C., Ed.; Springer: Berlin/Heidelberg, Germany, 2007; pp. 127–140. [Google Scholar]
- FAOSTAT. Statistical Databases of the Food and Agriculture Organization of the United Nations. 2020. Available online: http://www.fao.org/faostat/en/#home (accessed on 2 October 2022).
- Brader, G.; Compant, S.; Vescio, K.; Mitter, B.; Trognitz, F.; Ma, L.-J.; Sessitsch, A. Ecology and Genomic Insights into Plant-Pathogenic and Plant-Nonpathogenic Endophytes. Annu. Rev. Phytopathol. 2017, 55, 61–83. [Google Scholar] [CrossRef]
- Lemanceau, P.; Blouin, M.; Muller, D.; Moënne-Loccoz, Y. Let the Core Microbiota Be Functional. Trends Plant Sci. 2017, 22, 583–595. [Google Scholar] [CrossRef]
- Babalola, O.O.; Fadiji, A.E.; Enagbonma, B.J.; Alori, E.T.; Ayilara, M.S.; Ayangbenro, A.S. The Nexus Between Plant and Plant Microbiome: Revelation of the Networking Strategies. Front. Microbiol. 2020, 11, 548037. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Samad, A.; Faist, H.; Sessitsch, A. A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 2019, 19, 29–37. [Google Scholar] [CrossRef]
- Mendes, R.; Garbeva, P.; Raaijmakers, J.M. The rhizosphere microbiome: Significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 2013, 37, 634–663. [Google Scholar] [CrossRef] [PubMed]
- Van Der Heijden, M.G.A.; Hartmann, M. Networking in the Plant Microbiome. PLoS Biol. 2016, 14, e1002378. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Grube, M.; Schloter, M.; Smalla, K. Unraveling the plant microbiome: Looking back and future perspectives. Front. Microbiol. 2014, 5, 148. [Google Scholar] [CrossRef]
- Qu, Q.; Zhang, Z.; Peijnenburg, W.J.G.M.; Liu, W.; Lu, T.; Hu, B.; Chen, J.-M.; Chen, J.; Lin, Z.; Qian, H. Rhizosphere Microbiome Assembly and Its Impact on Plant Growth. J. Agric. Food Chem. 2020, 68, 5024–5038. [Google Scholar] [CrossRef] [PubMed]
- Berg, G.; Erlacher, A.; Grube, M. The edible plant microbiome: Importance and health issues. In Principles of Plant-Microbe Interactions: Microbes for Sustainable Agriculture; Lugtenberg, B., Ed.; Springer International Publishing: Berlin/Heidelberg, Germany, 2015; pp. 419–426. [Google Scholar]
- Bulgarelli, D.; Schlaeppi, K.; Spaepen, S.; van Themaat, E.V.L.; Schulze-Lefert, P. Structure and Functions of the Bacterial Microbiota of Plants. Annu. Rev. Plant Biol. 2013, 64, 807–838. [Google Scholar] [CrossRef] [PubMed]
- Rheault, K.; Lachance, D.; Morency, M.-J.; Thiffault, É.; Guittonny, M.; Isabel, N.; Martineau, C.; Séguin, A. Plant Genotype Influences Physicochemical Properties of Substrate as Well as Bacterial and Fungal Assemblages in the Rhizosphere of Balsam Poplar. Front. Microbiol. 2020, 11, 575625. [Google Scholar] [CrossRef]
- Brown, S.P.; Grillo, M.A.; Podowski, J.C.; Heath, K.D. Soil origin and plant genotype structure distinct microbiome compartments in the model legume Medicago truncatula. Microbiome 2020, 8, 139. [Google Scholar] [CrossRef] [PubMed]
- Malacrinò, A.; Mosca, S.; Nicosia, M.G.L.D.; Agosteo, G.E.; Schena, L. Plant Genotype Shapes the Bacterial Microbiome of Fruits, Leaves, and Soil in Olive Plants. Plants 2022, 11, 613. [Google Scholar] [CrossRef]
- Li, Y.; Wu, X.; Chen, T.; Wang, W.; Liu, G.; Zhang, W.; Li, S.W.; Wang, M.; Zhao, C.; Zhou, H.; et al. Plant Phenotypic Traits Eventually Shape Its Microbiota: A Common Garden Test. Front. Microbiol. 2018, 9, 2479. [Google Scholar] [CrossRef]
- Leff, J.; Fierer, N. Bacterial Communities Associated with the Surfaces of Fresh Fruits and Vegetables. PLoS ONE 2013, 8, e59310. [Google Scholar] [CrossRef]
- Kyere, E.O.; Qiu, G.W.; Zain, S.N.; Palmer, J.; Wargent, J.J.; Fletcher, G.C.; Flint, S. A comparison of Listeria monocytogenes contamination in bagged and un-bagged lettuce in supermarkets. LWT 2020, 134, 110022. [Google Scholar] [CrossRef]
- Kirzinger, M.W.B.; Nadarasah, G.; Stavrinides, J. Insights into Cross-Kingdom Plant Pathogenic Bacteria. Genes 2011, 2, 980–997. [Google Scholar] [CrossRef]
- Yu, Y.-C.; Yum, S.-J.; Jeon, D.-Y.; Jeong, H.-G. Analysis of the Microbiota on Lettuce (Lactuca sativa L.) Cultivated in South Korea to Identify Foodborne Pathogens. J. Microbiol. Biotechnol. 2018, 28, 1318–1331. [Google Scholar] [CrossRef]
- Liao, C.; Wang, L. The Microbial Quality of Commercial Chopped Romaine Lettuce Before and After the “Use By” Date. Front. Microbiol. 2022, 13, 850720. [Google Scholar] [CrossRef] [PubMed]
- Banach, J.; Zwietering, M.; van der Fels-Klerx, H. Multi-criteria decision analysis to evaluate control strategies for preventing cross-contamination during fresh-cut lettuce washing. Food Control 2021, 128, 108136. [Google Scholar] [CrossRef]
- van Overbeek, L.; Duhamel, M.; Aanstoot, S.; van der Plas, C.L.; Nijhuis, E.; Poleij, L.; Russ, L.; van der Zouwen, P.; Andreo-Jimenez, B. Transmission of Escherichia coli from Manure to Root Zones of Field-Grown Lettuce and Leek Plants. Microorganisms 2021, 9, 2289. [Google Scholar] [CrossRef] [PubMed]
- Dao, J.; Stenchly, K.; Traoré, O.; Amoah, P.; Buerkert, A. Effects of Water Quality and Post-Harvest Handling on Microbiological Contamination of Lettuce at Urban and Peri-Urban Locations of Ouagadougou, Burkina Faso. Foods 2018, 7, 206. [Google Scholar] [CrossRef]
- Marshall, K.E.; Hexemer, A.; Seelman, S.L.; Fatica, M.K.; Blessington, T.; Hajmeer, M.; Kisselburgh, H.; Atkinson, R.; Hill, K.; Sharma, D.; et al. Lessons Learned from a Decade of Investigations of Shiga Toxin–Producing Escherichia coli Outbreaks Linked to Leafy Greens, United States and Canada. Emerg. Infect. Dis. 2020, 26, 2319–2328. [Google Scholar] [CrossRef]
- Raaijmakers, J.M.; Paulitz, T.; Steinberg, C.; Alabouvette, C.; Moënne-Loccoz, Y. The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 2008, 321, 341–361. [Google Scholar] [CrossRef]
- Shahid, S.A.; Zaman, M.; Heng, L. Soil salinity: Historical perspectives and a world overview of the problem. In Guideline for Salinity Assessment, Mitigation and Adaptation Using Nuclear and Related Techniques; Springer: Berlin/Heidelberg, Germany, 2018; pp. 43–53. [Google Scholar]
- Bernstein, L. Effects of Salinity and Sodicity on Plant Growth. Annu. Rev. Phytopathol. 1975, 13, 295–312. [Google Scholar] [CrossRef]
- Xu, C.; Mou, B. Evaluation of Lettuce Genotypes for Salinity Tolerance. HortScience 2015, 50, 1441–1446. [Google Scholar] [CrossRef]
- Guan, Y.; Jiang, N.; Wu, Y.; Yang, Z.; Bello, A.; Yang, W. Disentangling the role of salinity-sodicity in shaping soil microbiome along a natural saline-sodic gradient. Sci. Total Environ. 2020, 765, 142738. [Google Scholar] [CrossRef]
- Mukhtar, S.; Mirza, B.S.; Mehnaz, S.; Mirza, M.S.; Mclean, J.; Malik, K.A. Impact of soil salinity on the microbial structure of halophyte rhizosphere microbiome. World J. Microbiol. Biotechnol. 2018, 34, 136. [Google Scholar] [CrossRef]
- Yuan, Y.; Brunel, C.; van Kleunen, M.; Li, J.; Jin, Z. Salinity-induced changes in the rhizosphere microbiome improve salt tolerance of Hibiscus hamabo. Plant Soil 2019, 443, 525–537. [Google Scholar] [CrossRef]
- Yuan, Z.; Druzhinina, I.S.; Labbé, J.; Redman, R.; Qin, Y.; Rodriguez, R.; Zhang, C.; Tuskan, G.; Lin, F. Specialized Microbiome of a Halophyte and its Role in Helping Non-Host Plants to Withstand Salinity. Sci. Rep. 2016, 6, 32467. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, A.; Sarkar, S.; Cuadros-Orellana, S.; Bandopadhyay, R. Exopolysaccharides and biofilms in mitigating salinity stress: The biotechnological potential of halophilic and soil-inhabiting PGPR microorganisms. In Microorganisms in Saline Environments: Strategies and Functions; Springer: Cham, Switzerland, 2019; pp. 133–153. [Google Scholar] [CrossRef]
- Arif, I.; Batool, M.; Schenk, P.M. Plant Microbiome Engineering: Expected Benefits for Improved Crop Growth and Resilience. Trends Biotechnol. 2020, 38, 1385–1396. [Google Scholar] [CrossRef]
- Tarroum, M.; Ben Romdhane, W.; Ali, A.; Al-Qurainy, F.; Al-Doss, A.; Fki, L.; Hassairi, A. Harnessing the Rhizosphere of the Halophyte Grass Aeluropus littoralis for Halophilic Plant-Growth-Promoting Fungi and Evaluation of Their Biostimulant Activities. Plants 2021, 10, 784. [Google Scholar] [CrossRef]
- Arora, N.K.; Fatima, T.; Mishra, J.; Mishra, I.; Verma, S.; Verma, R.; Verma, M.; Bhattacharya, A.; Verma, P.; Mishra, P.; et al. Halo-tolerant plant growth promoting rhizobacteria for improving productivity and remediation of saline soils. J. Adv. Res. 2020, 26, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Ullah, S.; Bano, A. Isolation of plant-growth-promoting rhizobacteria from rhizospheric soil of halophytes and their impact on maize (Zea mays L.) under induced soil salinity. Can. J. Microbiol. 2015, 61, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Adhikari, N.D.; Simko, I.; Mou, B. Phenomic and Physiological Analysis of Salinity Effects on Lettuce. Sensors 2019, 19, 4814. [Google Scholar] [CrossRef] [PubMed]
- Wei, Z.; Julkowska, M.; Laloë, J.-O.; Hartman, Y.; de Boer, G.-J.; Michelmore, R.W.; van Tienderen, P.H.; Testerink, C.; Schranz, M.E. A mixed-model QTL analysis for salt tolerance in seedlings of crop-wild hybrids of lettuce. Mol. Breed. 2014, 34, 1389–1400. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Lauber, C.L.; Walters, W.A.; Berg-Lyons, D.; Lozupone, C.A.; Turnbaugh, P.J.; Fierer, N.; Knight, R. Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4516–4522. [Google Scholar] [CrossRef]
- Ihrmark, K.; Bödeker, I.T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Clemmensen, K.E.; et al. New primers to amplify the fungal ITS2 region—Evaluation by 454-sequencing of artificial and natural communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef]
- Větrovský, T.; Baldrian, P.; Morais, D. SEED 2: A user-friendly platform for amplicon high-throughput sequencing data analyses. Bioinformatics 2018, 34, 2292–2294. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 2016, e2584. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve Bayesian Classifier for Rapid Assignment of rRNA Sequences into the New Bacterial Taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in Performance and Usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef] [PubMed]
- Guindon, S.; Dufayard, J.-F.; Lefort, V.; Anisimova, M.; Hordijk, W.; Gascuel, O. New Algorithms and Methods to Estimate Maximum-Likelihood Phylogenies: Assessing the Performance of PhyML 3.0. Syst. Biol. 2010, 59, 307–321. [Google Scholar] [CrossRef] [PubMed]
- Hamady, M.; Lozupone, C.A.; Knight, R.A. Fast UniFrac: Facilitating high-throughput phylogenetic analyses of microbial communities including analysis of pyrosequencing and PhyloChip data. ISME J. 2009, 4, 17–27. [Google Scholar] [CrossRef] [PubMed]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Kindt, R.; Legendre, P.; Minchin, P.R.; O’hara, R.; Simpson, G.L.; Solymos, P.; Stevens, M.H.H.; Wagner, H. Package ‘Vegan’. Community Ecology Package, Version 2.5-6. 2019. Available online: https://CRAN.R-project.org/package=vegan (accessed on 2 October 2022).
- Zhao, S.; Guo, Y.; Sheng, Q.; Shyr, Y. Advanced Heat Map and Clustering Analysis Using Heatmap3. BioMed Res. Int. 2014, 2014, 986048. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Hassani, M.A.; Durán, P.; Hacquard, S. Microbial interactions within the plant holobiont. Microbiome 2018, 6, 58. [Google Scholar] [CrossRef]
- Vandenkoornhuyse, P.; Quaiser, A.; Duhamel, M.; Le Van, A.; Dufresne, A. The importance of the microbiome of the plant holobiont. New Phytol. 2015, 206, 1196–1206. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.; Naveed, M.; Mustafa, A.; Abbas, A. The good, the bad, and the ugly of rhizosphere microbiome. In Probiotics and Plant Health; Springer: Singapore, 2017; pp. 253–290. [Google Scholar] [CrossRef]
- Schreiter, S.; Ding, G.-C.; Heuer, H.; Neumann, G.; Sandmann, M.; Grosch, R.; Kropf, S.; Smalla, K. Effect of the soil type on the microbiome in the rhizosphere of field-grown lettuce. Front. Microbiol. 2014, 5, 144. [Google Scholar] [CrossRef] [PubMed]
- Iliev, I.; Apostolova, E.; Hadjieva, N.; Kostadinov, K.; Filipov, S.; Kostadinova, S.; Baev, V.; Gozmanova, M. Bacterial diversity and physiological activity of lettuce (Lactuca sativa) rhizosphere under bio-organic greenhouse management strategies. Int. J. Environ. Sci. Technol. 2021, 19, 9945–9956. [Google Scholar] [CrossRef]
- Cardinale, M.; Grube, M.; Erlacher, A.; Quehenberger, J.; Berg, G. Bacterial networks and co-occurrence relationships in the lettuce root microbiota. Environ. Microbiol. 2014, 17, 239–252. [Google Scholar] [CrossRef]
- Feng, Z.; Wu, P.; Xie, X.; Zhou, Y.; Zhu, H.; Yao, Q. Feather-Based Compost Drastically Regulates Soil Microbial Community and Lettuce Growth in a Subtropical Soil: The Possible Role of Amino Acids. J. Soil Sci. Plant Nutr. 2021, 21, 709–721. [Google Scholar] [CrossRef]
- Sun, Y.; Snow, D.; Walia, H.; Li, X. Transmission Routes of the Microbiome and Resistome from Manure to Soil and Lettuce. Environ. Sci. Technol. 2021, 55, 11102–11112. [Google Scholar] [CrossRef]
- Erlacher, A.; Cardinale, M.; Grosch, R.; Grube, M.; Berg, G. The impact of the pathogen Rhizoctonia solani and its beneficial counterpart Bacillus amyloliquefaciens on the indigenous lettuce microbiome. Front. Microbiol. 2014, 5, 175. [Google Scholar] [CrossRef]
- Erlacher, A.; Cardinale, M.; Grube, M.; Berg, G. Biotic Stress Shifted Structure and Abundance of Enterobacteriaceae in the Lettuce Microbiome. PLoS ONE 2015, 10, e0118068. [Google Scholar] [CrossRef]
- Luziatelli, F.; Ficca, A.G.; Colla, G.; Baldassarre Švecová, E.; Ruzzi, M. Foliar Application of Vegetal-Derived Bioactive Compounds Stimulates the Growth of Beneficial Bacteria and Enhances Microbiome Biodiversity in Lettuce. Front. Plant Sci. 2019, 10, 60. [Google Scholar] [CrossRef]
- Leonard, S.R.; Simko, I.; Mammel, M.K.; Richter, T.K.S.; Brandl, M.T. Seasonality, shelf life and storage atmosphere are main drivers of the microbiome and E. coli O157:H7 colonization of post-harvest lettuce cultivated in a major production area in California. Environ. Microbiome 2021, 16, 25. [Google Scholar] [CrossRef]
- Hunter, P.J.; Hand, P.; Pink, D.; Whipps, J.M.; Bending, G.D. Both Leaf Properties and Microbe-Microbe Interactions Influence Within-Species Variation in Bacterial Population Diversity and Structure in the Lettuce (Lactuca Species) Phyllosphere. Appl. Environ. Microbiol. 2010, 76, 8117–8125. [Google Scholar] [CrossRef]
- Rastogi, G.; Sbodio, A.; Tech, J.J.; Suslow, T.V.; Coaker, G.L.; Leveau, J.H.J. Leaf microbiota in an agroecosystem: Spatiotemporal variation in bacterial community composition on field-grown lettuce. ISME J. 2012, 6, 1812–1822. [Google Scholar] [CrossRef] [PubMed]
- Sasse, J.; Martinoia, E.; Northen, T. Feed Your Friends: Do Plant Exudates Shape the Root Microbiome? Trends Plant Sci. 2018, 23, 25–41. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Jiang, S.; Jiang, C.; Wu, C.; Gao, M.; Wang, Q. A review of root exudates and rhizosphere microbiome for crop production. Environ. Sci. Pollut. Res. 2021, 28, 54497–54510. [Google Scholar] [CrossRef] [PubMed]
- Micallef, S.A.; Shiaris, M.P.; Colón-Carmona, A. Influence of Arabidopsis thaliana accessions on rhizobacterial communities and natural variation in root exudates. J. Exp. Bot. 2009, 60, 1729–1742. [Google Scholar] [CrossRef] [PubMed]
- Shenton, M.; Iwamoto, C.; Kurata, N.; Ikeo, K. Effect of Wild and Cultivated Rice Genotypes on Rhizosphere Bacterial Community Composition. Rice 2016, 9, 42. [Google Scholar] [CrossRef]
- Eichmann, R.; Richards, L.; Schäfer, P. Hormones as go-betweens in plant microbiome assembly. Plant J. 2020, 105, 518–541. [Google Scholar] [CrossRef]
- De Souza, R.; Ambrosini, A.; Passaglia, L.M.P. Plant growth-promoting bacteria as inoculants in agricultural soils. Genet. Mol. Biol. 2015, 38, 401–419. [Google Scholar] [CrossRef]
- Reinhold-Hurek, B.; Bünger, W.; Burbano, C.S.; Sabale, M.; Hurek, T. Roots Shaping Their Microbiome: Global Hotspots for Microbial Activity. Annu. Rev. Phytopathol. 2015, 53, 403–424. [Google Scholar] [CrossRef]
- Berg, G.; Raaijmakers, J. Saving seed microbiomes. ISME J. 2018, 12, 1167–1170. [Google Scholar] [CrossRef]
- Kuźniar, A.; Włodarczyk, K.; Grządziel, J.; Woźniak, M.; Furtak, K.; Gałązka, A.; Dziadczyk, E.; Skórzyńska-Polit, E.; Wolińska, A. New Insight into the Composition of Wheat Seed Microbiota. Int. J. Mol. Sci. 2020, 21, 4634. [Google Scholar] [CrossRef] [PubMed]
- Flemer, B.; Gulati, S.; Bergna, A.; Raendler, M.; Cernava, T.; Witzel, K.; Berg, G.; Grosch, R. Biotic and abiotic stress factors induce microbiome shifts and enrichment of distinct beneficial bacteria in tomato roots. Phytobiomes J. 2022. [Google Scholar] [CrossRef]
- Okie, J.G.; Van Horn, D.J.; Storch, D.; Barrett, J.E.; Gooseff, M.; Kopsova, L.; Takacs-Vesbach, C.D. Niche and metabolic principles explain patterns of diversity and distribution: Theory and a case study with soil bacterial communities. Proc. R. Soc. B Boil. Sci. 2015, 282, 20142630. [Google Scholar] [CrossRef] [PubMed]
- Andronov, E.E.; Petrova, S.N.; Pinaev, A.G.; Pershina, E.V.; Rakhimgaliyeva, S.; Akhmedenov, K.M.; Gorobets, A.V.; Sergaliev, N.K. Analysis of the structure of microbial community in soils with different degrees of salinization using T-RFLP and real-time PCR techniques. Eurasian Soil Sci. 2012, 45, 147–156. [Google Scholar] [CrossRef]
- Ibekwe, A.M.; Ors, S.; Ferreira, J.F.; Liu, X.; Suarez, D.L.; Ma, J.; Ghasemimianaei, A.; Yang, C.-H. Functional relationships between aboveground and belowground spinach (Spinacia oleracea L., cv. Racoon) microbiomes impacted by salinity and drought. Sci. Total Environ. 2020, 717, 137207. [Google Scholar] [CrossRef]
- Liu, L.; Wu, Y.; Yin, M.; Ma, X.; Yu, X.; Guo, X.; Du, N.; Eller, F.; Guo, W. Soil salinity, not plant genotype or geographical distance, shapes soil microbial community of a reed wetland at a fine scale in the Yellow River Delta. Sci. Total Environ. 2023, 856, 159136. [Google Scholar] [CrossRef]
- Rath, K.M.; Maheshwari, A.; Bengtson, P.; Rousk, J. Comparative Toxicities of Salts on Microbial Processes in Soil. Appl. Environ. Microbiol. 2016, 82, 2012–2020. [Google Scholar] [CrossRef]
- Liu, H.; Brettell, L.E.; Qiu, Z.; Singh, B.K. Microbiome-Mediated Stress Resistance in Plants. Trends Plant Sci. 2020, 25, 733–743. [Google Scholar] [CrossRef]
- Verma, H.; Kumar, D.; Kumar, V.; Kumari, M.; Singh, S.K.; Sharma, V.K.; Droby, S.; Santoyo, G.; White, J.F.; Kumar, A. The Potential Application of Endophytes in Management of Stress from Drought and Salinity in Crop Plants. Microorganisms 2021, 9, 1729. [Google Scholar] [CrossRef]
- Santos, S.S.; Rask, K.A.; Vestergård, M.; Johansen, J.L.; Priemé, A.; Frøslev, T.G.; González, A.M.M.; He, H.; Ekelund, F. Specialized microbiomes facilitate natural rhizosphere microbiome interactions counteracting high salinity stress in plants. Environ. Exp. Bot. 2021, 186, 104430. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).