Abstract
Wuyi Mountain in Southeast China is the origin of black tea and oolong tea. It is also considered the ‘treasure trove of tea cultivars’ because of its rich tea germplasm resources. In the present study, the population structure and genetic diversity of 137 tea germplasms from Wuyi Mountain and its adjacent areas were analyzed by SNPs. The information index (I), observed heterozygosity (Ho), expected heterozygosity (He) and fixation index (F) polymorphisms of the selected SNPs were high, stable and reliable. Ho had an average of 0.389, while He had an average of 0.324, indicating that Wuyi Mountain tea germplasms had rich genetic diversity. The AMOVA results showed that genetic variation came mainly from intrapopulation variation, accounting for 66% of the total variation. The differences in the Fst and Nei values of tea germplasm between Wuyi Mountain and its adjacent areas are similar to the geographical differences. Multiple analyses based on high-quality SNPs found that the landraces of tea plants on Wuyi Mountain had different genetic backgrounds from the wild-type landraces and the landraces of Wuyi Mountain tea plants underwent population differentiation. This study provides a basis for the effective protection and utilization of tea germplasms on Wuyi Mountain and lays a foundation for identifying potential parents to optimize tea cultivation.
1. Introduction
Tea is the most popular beverage globally, aside from water [1]. There are six tea categories based on the degree of fermentation and processing techniques, i.e., black tea, dark tea, oolong tea, yellow tea, white tea and green tea. Among them, black tea is the most consumed tea in the world, while oolong tea has the most variable fragrant aromas and tastes. Both of these teas originated on Wuyi Mountain, Southeast China [2,3]. It is also recorded in “All about Tea” that the earliest Chinese tea transported to Europe was produced on Wuyi Mountain [4]. Tea is manufactured from the fresh leaves of tea plants (Camellia sinensis (L.) Kuntze). The tea plant is an economically important crop worldwide and it originated in China [5,6]. Rich tea germplasm resources are the basis of producing various kinds of tea and forming products that are both diverse and high quality. As an essential tea-producing province, Fujian Province has wealthy tea germplasm resources. In particular, Wuyi Mountain, located in the northwest of Fujian Province, has a long history of tea production and is considered the origin of the world’s natural and cultured teas. It preserves the world’s most complete, typical and largest central subtropical forest ecosystem within the same latitude and breeds rich tea germplasm resources, known as the ‘treasure trove of tea cultivars’. The core area is the Wuyi Mountain Nature Reserve, which straddles Wuyishan City, Jianyang Area and Guangze County. At present, the relationship between the richness of germplasm resources and the germplasm distribution of the tea plant on Wuyi Mountain requires further investigation.
Tea germplasm resources are not only the material basis for the development of the tea industry, but also the basis for tea science research [7]. The exploration and protection of excellent tea germplasm resources have become increasingly prominent, such that, as a result, understanding genetic diversity among and within populations is vital for developing effective and efficient protection measures [8,9,10]. Molecular markers are based on differences in DNA sequences at the genomic level. They are less affected by the outside world, have higher stability and accuracy and have become one of the most widely used characteristics in identifying plant germplasm resources. The molecular markers used in tea plants mainly include RAPD, AFPL, ISSR and SSR [11,12,13,14], which can effectively evaluate the genetic diversity of tea germplasm resources. SNP analysis is a third-generation molecular marker technology that has also been successfully applied to analyze tea germplasm diversity and provides accurate results, independent of target species or populations [15]. Compared with other methods, SNP analysis has the advantages of automation, high throughput and high genetic stability [16]. In addition, due to the diallel hybridization of SNPs, the experimental error rate is reduced and the resulting accuracy of the experiment is further improved [17,18]. Previously, SNP analysis was used to accurately and efficiently determine the genetic relationship among oolong tea germplasms in China [19]. Furthermore, we constructed a molecular identity card of tea cultivars based on SNP information and basic information [20]. In addition, the diversity of Yunxiao tea germplasm resources in Fujian is more accurately understood by SNP technology [21]. Analyses of single nucleotide polymorphisms reveal the genetic structure of tea germplasm and Japanese landraces [22].
In this study, the population structure, genetic diversity and genetic distance of 137 tea germplasm resources from Wuyi Mountain and its adjacent areas were analyzed by SNP. These findings provide a valuable basis for further understanding the genetic relationship and composition of Wuyi Mountain tea germplasm resources and provide a scientific reference for protecting and utilizing Wuyi Mountain tea germplasm resources.
2. Materials and Methods
2.1. Sample Collection
The samples were collected from Wuyi Mountain and its adjacent areas, resulting in a total of 137 tea plant germplasms. Detailed information about these samples is shown in Table 1, consisting of 87 tea samples collected from Wuyi Mountain (WY) and 50 tea samples from adjacent areas: 15 from Eastern Fujian (EF), 15 from Southern Fujian (SF) and 20 from wild-type Fujian (FWL). Voucher specimens were deposited in the Herbarium, Fujian Agriculture and Forestry University (FAFU). Figure 1 shows the original sample distribution of the tested tea plant germplasms. Samples were obtained from tea plant buds or tender leaves, kept in −20 °C refrigerator for backup.

Table 1.
List of 137 tea samples used in SNP genotyping.
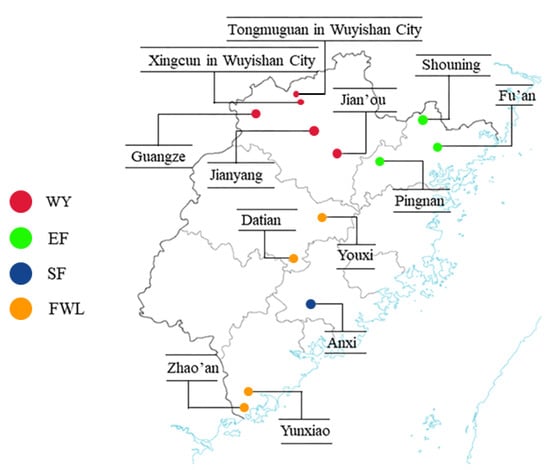
Figure 1.
Sample distribution of Fujian tea plant accessions used in this study [23].
2.2. DNA Extraction
Genomic DNA was extracted from samples using a new plant genome extraction kit (DP320, TIANGEN, Beijing, China) according to the kit’s instructions [19]. Fresh tea buds or leaves (ca. 0.1 g) were cut and placed in a grinding pan. Add liquid nitrogen and grind quickly. Next, 400 µL of lysis solution (buffer LP1) and 6 µL of RNase (10 mg/mL) were added to the tubes. After the cracked sample is added to the test tube, the tube was placed into an shaker instrument (WS350B, WIGGENS, Beijing, China) to be completely lysed. The next extracted steps were performed in accordance with the supplier’s instructions. Genomic DNA in the tea tissue was extracted using centrifugal adsorption columns that specifically bind DNA. Then, the DNA quality was analyzed by the a NanoDrop nucleic acid sequencing instrument (NanoDrop 2000/2000C, Thermo, Waltham, WA, USA), which requires a DNA concentration of >50 µL/mL. Successful extraction material was stored in a −20 °C refrigerator.
2.3. Single Nucleotide Polymorphism Markers and Genotyping
In the early stages of this study, the express sequence tag (EST) of tea was downloaded from the database of the National Center of Biological Information (NCBI) (http://www.ncbi.nlm.nih.gov/) (accessed on 15 July 2021). For the mining and selection of single nucleotide polymorphisms of tea, 96 SNP loci for genotyping of tea germplasm resources were ultimately screened out [16]. Genotyping was performed with Fluidigm 96.96 Dynamic ArrayTM IFC (Integrated Fluidic Circuit) chip (Fluidigm® Corp., South San Francisco, CA, USA), with Fluidigm 96.96 SNP genotyping workflow reference (Fluidigm, PN100-3912) for experimental response. The primers were synthesized by the Fluidigm Company (USA) and included allele-specific primers (ASPs), specific target amplification (STA) primers and locus-specific primers (LSPs). Preamplification of DNA called STA is required on PCR instruments (ABI VERITI, South San Francisco, CA, USA). The sample and primer solution were added to the IFC chip according to the SNP genotyping method. Then, samples were automatically mixed and detected by the Juno instrument (Fluidigm, South San Francisco, CA, USA). Finally, 96.96 IFC fluorescence images were obtained by EP1TM (Fluidigm® Corp., USA) [21].
2.4. Data Analysis
Data collection was performed by the EP1 instrument. The data were exported and analyzed using Fluidigm SNP Genotyping Analysis software (https://www.fluidigm.com/software) (accessed on 5 January 2022), followed by GenAlEx6.503 software to analyze population differentiation, allele frequency, information index (I), observed heterozygosity (Ho), expected heterozygosity (He), fixation index (F) and minor allele frequency (MAF). GenAlEx6.503 software was used for genetic distance calculation, Fst and AMOVA analysis. PCoA plot analysis was obtained from genetic distance measurements in GenAIEx. The population structure was analyzed by Structure 2.3.4 software and the optimal K value. Finally, the genetic structure analysis map was obtained by Clumpp version 1.1.2 and Distruct version 1.1. For the hierarchical clustering tree, MEGA5.05 software was applied to calculate the sequence data and then the hierarchical clustering method was used to construct graphs [19,20].
3. Results
3.1. Screening and Analysis of the Polymorphic Loci
As a result of screening 96 SNP markers, 47 specific loci with strong polymorphisms that were suitable for genotyping tea germplasms from Wuyi Mountain and its adjacent areas were obtained, accounting for 49.0% of all loci. The selected 47 SNPs and associated allele information are shown in Table 2. Statistically, the I for these SNP markers ranged from 0.103 to 0.691, with an average of 0.483. The Ho was 0.029~0.888, with an average of 0.389. The He was 0.062~0.497, with an average of 0.324. The F ranged from approximately −0.829 to 0.873, with an average of −0.111. The MAF ranged from 0.058 to 0.485, with an average of 0.312. The averages of Ho and He were close, indicating that Wuyi Mountain tea germplasm resources had relatively high genetic diversity. In addition, when the loci were screened, the MAF value of the secondary allele frequency was ensured to be ≥ 0.05. The suballele frequencies of 47 polymorphic SNPs are shown in Figure 2. The genotypic DNA fingerprinting of tea germplasms based on SNP array is listed in Table 3, showing the truncated spectra of 20 loci of 22 samples. The full loci information is in Table S1.

Table 2.
Allele information of 47 polymorphic SNP markers.
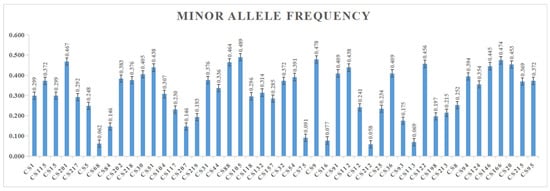
Figure 2.
Minor allele frequency of 47 polymorphic SNP markers.

Table 3.
Genotypic DNA fingerprinting of tea germplasms based on SNP arrays (showing truncated spectra of 20 loci of 22 samples).
3.2. Genetic Relationship Analysis of Test Samples
According to the principal coordinate analysis, the first principal component, the second principal component and the third principal component accounted for 30.24%, 11.42% and 5.65% of the total variation, respectively. The PCoA diagram (Figure 3) shows the genetic relationship of the tested samples. This result suggests that WY tea germplasms interacted with other tested germplasms. As a result, the 137 tea samples were clustered into three main groups (I, II and Ⅲ). Among these, 20 landraces from the FWL clustered together to form a relatively close group Ⅲ. Group Ⅱ contained 57 landraces from WY, whereas group I contained 30 landraces from WY, 15 from EF and 15 from SF. Interestingly, the landrace tea samples of WY were clearly divided into two groups: group I and group II. Among them, group I contained 30 samples of WY area, 28 of which were from Xingcun of Wuyishan City and 2 of which were from the city of Jian’ou; group II contained 57 samples, 9 of which were from Tongmuguan in Wuyishan City and 33 of which were from Guangze, while the remainder were from Jianyang. Group I was composed of part of WY germplasms and those of EF and SF, while group II was relatively independent. Overall, the germplasms of tea plants in WY interacted more with the genetic material of landraces in EF and SF but did not interact with FWL.
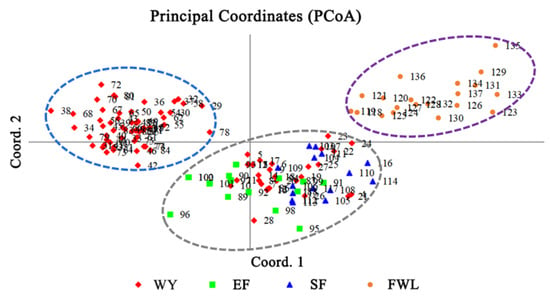
Figure 3.
PCoA plot of 137 samples based on the top three principal components with different colors representing the groups.
3.3. Population Structure Analysis of Samples
To verify the results of principal component analysis, Structure-Clumpp-Distruct software was used for population structure analysis. The best K value was determined through the simulation model using Structure Harvester (http://taylor0.biology.ucla.edu/struct_harvest/) (accessed on 3 May 2022), K = 3. The results of population structure classification are based on the model (Figure 4). The experimental results revealed that the 137 tested tea germplasms were divided into three populations. The FWL landraces were aggregated into a single pool. While the landraces of tea plants in EF, SF and some of WY clustered into one population, other WY landraced germplasms formed a population. The map also suggests that there were significant differences among landraces of WY, which could be further divided into two groups, consistent with those of principal component analysis (Figure 3). When compared with the clustering results of the PCoA map, the characteristics of landraces in some parts of WY were closer to those of EF and SF landraces. Among them, the germplasms of landrace tea plants in SF and some landraces of tea plants in WY belong to oolong tea varieties. Therefore, it is speculated that the genotypes of these varieties are relatively similar. Objectively, the germplasm resources of the tea plant WY landraces have high genetic richness.
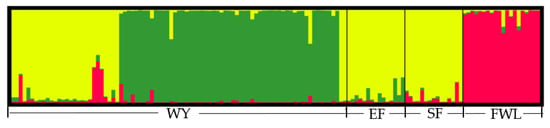
Figure 4.
Model-based structure of the 137 tea germplasms.
3.4. Hierarchical Clustering Diagram Describing Kinship
The hierarchical clustering tree generated by MEGA software in this study showed that 137 tea germplasms could also be divided into three groups (I, II and Ⅲ) (Figure 5), consistent with the results of the PCoA (Figure 3) and groups I and II are on a branch. Among these, group Ⅲ mainly includes all tea plant germplasms in the FWL area along with a few other landraces. Groups I and II consist of tea plant landraces of the WY, EF and SF areas: group I includes a majority of landraces in EF and SF and part of landraces in WY and group II contains most of the landraces in WY. The tea plant landraces of WY are clustered in two different groups and the members of the groups are basically consistent with the PCoA. Genotypic hybridization (gray region of group I) exists in tea germplasms of the EF, SF and WY areas. However, the tree suggests that some of the landraces in the WY area are clustered independently (cyan area of group II), which is consistent with the results of PCoA and population structure.
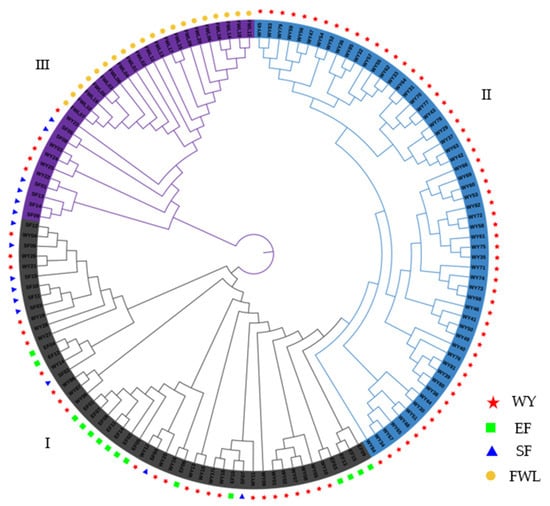
Figure 5.
Hierarchical clustering tree of the 137 tea germplasms.
3.5. Population Differentiation Analysis
Fst analysis and AMOVA were used for genetic differentiation analysis of four areas and Nei’s analysis was used to calculate the genetic distance between areas. The results reveal that the Fst value was between 0.053 and 0.144 and the populational degree of differentiation of landrace FWL and EF was the highest, followed by FWL and WY (Table 4). The Fst value between the FWL area and the other areas was higher than that between the WY, EF and SF areas. According to Fst analysis, the Fst values between the WY area and EF and SF areas were small, only 0.053 and 0.074, respectively, indicating that the degree of differentiation between the WY landraces and EF and SF landraces was small. The Nei’s genetic distance values ranged from 0.067 to 0.188, while the distribution of genetic distance values among the four areas was the same as that of geographical differences (Table 4). The AMOVA results showed that the genetic differentiation among the four areas accounted for only 34% of the total variation, while the genetic variation within the areas accounted for 66% of the total variation (Table 5), indicating that the individual germplasms in the test areas had varying degrees of genetic differentiation, yet rich genetic diversity.

Table 4.
Matrix of pairwise Nei’s genetic distance and Fst among the four areas.

Table 5.
AMOVAs of four areas and 137 tea germplasms were examined in this study.
4. Discussion
4.1. SNP Screening and Genetic Richness Analysis
The genetic diversity analysis of germplasms was determined using the included test samples and marker methods [24]. As perennial evergreen woody plants, tea plants contain a large number of bioactive substances and have a relatively large genome. In the past decade, high-quality genomes of tea plants have been reported, including Yunkang10 [25], Shuchazao [26], wild tea plants [27], Longjing43 [28], Tieguanyin [29], Biyun [30] and Huangdan [31], which dramatically improves the efficiency of functional and comparative genomics. SNPs are single nucleotide polymorphism sites that exist widely in biological genomes, which are abundant and provide a relative advantage in plant identification [17,32]. SNPs with MAF ≥ 0.05 were screened for subsequent analysis to ensure the accuracy and validity of the results. In this study, 47 pairs of SNP loci were used to analyze the genetic diversity and genetic relationship of 137 tea plants from Wuyi Mountain and its adjacent areas. Among all failed primer pairs, some marker pairs identified only one SNP in all samples and some SNP marker pair failures may be caused by EST sequence errors or may be due to flanking sequence polymorphisms [19]. The I, Ho, He and F polymorphisms of these 47 pairs of SNP loci were high, stable and reliable, which was conducive to the study of tea population germplasms. According to the concept of heterozygosity, the closer Ho is to He, the higher the genetic diversity of the population [33,34]. In this study, the average Ho of the tested tea populations was 0.368, while the average He was 0.301. Thus, the average values of the two were relatively similar, indicating that Wuyi Mountain tea germplasms had relatively high genetic richness.
4.2. Genetic Relationship Analysis of Tea Plant Population between Wuyi Mountain and Its Adjacent Areas
Previous studies found that the landrace population of tea plants showed rich genetic diversity and a relatively close relationship [35,36]. Based on high-quality SNPs, this study conducted multiple analyses of PCoA, population structure and hierarchical clustering of 137 tea plant samples and found that the tea plant landraces of Fujian wild type (FWL) were clearly clustered together and distinguishable from the landraces of Wuyi Mountain (WY). This may be because tea plant landraces of the WY are cultivated, while the FWL landraces are wild type. Previous studies have found similar results, such as that wild tea plants and cultivated tea plants belong to different groups [37]. The results revealed that the landraces in WY might have different genetic backgrounds from the landraces in FWL. In addition, the landraces of tea plants in WY, mainly Xingcun in Wuyishan City and Jian’ou City, were closely related to Eastern Fujian (EF) and Southern Fujian (SF) landraces (group I in Figure 3 and Figure 5). This is related to the frequent germplasm exchange among the three tea-producing regions of Wuyi Mountain, eastern Fujian and southern Fujian throughout history. The geographical proximity between Wuyi Mountain and eastern Fujian is conducive to the germplasm exchange of tea plants and is consistent with the geographical distribution [38]. The tea plant landrace in southern Fujian is oolong tea, which is the same as the tea germplasms in some areas of Wuyi Mountain (Xingcun in Wuyishan City). This result is also similar to the results of close interaction between northern Fujian and southern Fujian in a study of the genetic diversity in oolong tea [19]. The results suggest that breeders may have introduced seeds or crossbred their plants with tea landraces from other adjacent areas, which leads to frequent genetic material exchange.
The AMOVA results showed that the genetic variation within the four test areas accounted for only 66% of the total variation. However, the genetic variation between areas accounted for only 34%. The main genetic variation came from intra-area differentiation, which was similar to the results observed by Zhu et al. [39] and Yao et al. [40]. Therefore, the results of AMOVA suggest higher genetic diversity among WY tea plant germplasms. Differences in geographic distance are WY to EF < WY to SF < WY to FWL. According to the results of Fst and Nei, Fst and Nei values were 0.053, 0.067 between WY and EF, 0.074, 0.096 between WY and SF and 0.143, 0.183 between WY and FWL, respectively. On the Fst and Nei values, WY and EF < WY and SF < WY and FWL. The same difference is true between other areas. The genetic distance and genetic differentiation among the four areas increased with increasing distance, which was consistent with the geographical difference [41]. These results explain the hierarchical clustering tree analysis; even samples from the same area, such as WY tea plant landraces, are not completely together. Thus, the frequent introduction of excellent tea varieties for cultivation and breeding from different regions may promote the exchange of genetic materials. Geographical differences also have a larger impact on genetic diversity, which has also been found in crops, such as potato [42] and sweet potato [43].
4.3. Genetic Diversity Analysis of Tea Plant Population in Wuyi Mountain
Xia et al. (2020) reported that phylogenetic analysis of 81 tea plant germplasm sequences from different sources revealed that they could be divided into three well-differentiated tea plant populations [26]. In this study, the results of principal component analysis, population structure and hierarchical clustering tree analysis showed that the tea plant landraces of Wuyi Mountain were divided into two different groups. Among them, the tea plant germplasms of Xingcun in Wuyishan City and Jian’ou City were clustered into group I, which is consistent with the fact that the special germplasm oolong tea from Jian’ou City was introduced to Wuyishan City very early. In contrast, the tea plant germplasms from Tongmuguan in Wuyishan City, Guangze County and Jianyang Area were clustered into group II. This indicates that the tea plant landraces of Wuyi Mountain have undergone population differentiation. This is consistent with the previous AMOVA results that the genetic variation is mainly from within the areas. The results indicate that tea plants have undergone genetic differentiation to different degrees in the process of long-term production and domestication. Previous studies also demonstrated that China-type tea, Chinese Assam-type tea and Indian Assam-type tea might result from independent domestication events [44]. During domestication, disease resistance and flavor of tea germplasms were selectively promoted [28]. Similar results have been found in other crops: wild oil-tea camellia [45], wild potato [46] and Arabidopsis [47]. From the hierarchical clustering tree, group II had more far genetic relationships with other germplasms yet less interaction, which may be the result of being the original tea germplasms in the core area of Wuyi Mountain; group I contains more gene exchange, which is also confirmed by population structure analysis.
5. Conclusions
The results show that the landraces of tea plants on Wuyi Mountain had different genetic backgrounds from the wild-type landrace. The SNP-based molecular characterization of the Wuyi Mountain tea germplasms in this study revealed large variations within samples and rich genetic diversity. The patterns of population structure and genetic diversity varied across model-based groups. The tea plant landraces of Wuyi Mountain can be divided into two groups. This study provides a basis for the effective protection and utilization of tea germplasms on Wuyi Mountain and lays a foundation for identifying potential parents to optimize tea cultivation.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae8100932/s1, Table S1: Genotypic DNA fingerprinting of tea germplasms based on SNP arrays.
Author Contributions
N.Y., W.Y. and C.L. conceived and designed the research. S.H., H.W., C.C. and Y.Z. cultivated and provided the tea plant germplasm, C.L., Z.W. and P.W. (Pan Wang) performed the experiments. C.L. and P.W. (Pengjie Wang) analyzed the data. C.L., wrote the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Fujian Province “2011 Collaborative Innovation Center” Chinese Oolong Tea Industry Innovation Center special project (J2015-75), the Scientific Research Project of General Administration of Customs, China (2020HK187), the Agricultural Guidance Project of Fujian Provincial Science and Technology Department (2021N0024) and the Fund for Excellent Master’s Dissertations of Fujian Agriculture and Forestry University (1122YS01010).
Data Availability Statement
Not applicable.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mondael, T.K.; Bhattacharya, A.; Laxmikumaran, M.; Ahuja, P.S. Recent advances of tea (Camellia sinensis) biotechnology. Plant Cell Tissue Organ Cult. 2004, 76, 195–254. [Google Scholar] [CrossRef]
- Yao, S.S.; Guo, W.F.; Lu, Y.; Jiang, Y.X. Flavor characteristics of lapsang souchong and smoked lapsang souchong, a special chinese black tea with pine smoking process. J. Agric. Food Chem. 2005, 53, 8688–8693. [Google Scholar] [CrossRef]
- Zeng, L.T.; Zhou, X.C.; Su, X.G.; Yang, Z.Y. Chinese oolong tea: An aromatic beverage produced under multiple stresses. Trends Food Sci. Technol. 2020, 106, 242–253. [Google Scholar] [CrossRef]
- Ye, N.X. Introduction to Tea Science, 2nd ed.; China Agriculture Press: Beijing, China, 2021; pp. 19–20. [Google Scholar]
- Wang, P.J.; Jin, S.; Chen, X.J.; Wu, L.Y.; Zheng, Y.C.; Yue, C.; Guo, Y.C.; Zhang, X.T.; Yang, J.F.; Ye, N.X. Chromatin accessibility and translational landscapes of tea plants under chilling stress. Hortic. Res. 2021, 8, 96–110. [Google Scholar] [CrossRef]
- Lu, L.T.; Chen, H.F.; Wang, X.J.; Zhao, Y.C.; Yao, X.Z.; Xiong, B.; Deng, Y.L.; Zhao, D.G. Genome-level diversification of eight ancient tea populations in the Guizhou and Yunnan regions identifies candidate genes for core agronomic traits. Hortic. Res. 2021, 8, 190–199. [Google Scholar] [CrossRef]
- Wang, P.J.; Gu, M.Y.; Shao, S.X.; Chen, X.M.; Hou, B.H.; Ye, N.X.; Zhang, X.T. Changes in non-volatile and volatile metabolites associated with heterosis in tea plants (Camellia sinensis). J. Agric. Food Chem. 2022, 70, 3067–3078. [Google Scholar] [CrossRef] [PubMed]
- Guney, M.; Kafkas, S.; Keles, H.; Zarifikhosroshahi, M.; Gundesli, M.A.; Ercisli, S.; Necas, T.; Bujdoso, G. Genetic diversity among some walnut (Juglans regia L.) genotypes by SSR markers. Sustainability 2021, 13, 6830. [Google Scholar] [CrossRef]
- Barut, M.; Nadeem, M.A.; Karaköy, T.; Baloch, F.S. DNA fingerprinting and genetic diversity analysis of world quinoa germplasm using iPBS-retrotransposon marker system. Turk. J. Agric. For. 2020, 44, 479–491. [Google Scholar] [CrossRef]
- Savaş, T.G.; Yücel, G.; Kaygisiz, A.T.; Ateş, D.; Eşİyok, D.; Tanyolaç, M.B.; Tuna, M. Molecular cytogenetic characterization of common bean (Phaseolus vulgaris L.) accessions. Turk. J. Agric. For. 2020, 44, 612–630. [Google Scholar] [CrossRef]
- Chen, L.; Yamaguchi, S. RAPD markers for discriminating tea germplasms at the inter-specific level in China. Plant Breed. 2010, 124, 404–409. [Google Scholar] [CrossRef]
- Chen, L.; Gao, Q.K.; Chen, D.M.; Xu, C.J. The use of RAPD markers for detecting genetic diversity, relationship and molecular identification of chinese elite tea genetic resources [Camellia sinensis (L.) O. Kuntze] preserved in a tea germplasm repository. Biodivers. Conserv. 2005, 14, 1433–1444. [Google Scholar] [CrossRef]
- Liu, B.Y.; Li, Y.Y.; Tang, Y.C.; Wang, L.Y.; Cheng, H.; Wang, P.S. Assessment of genetic diversity and relationship of tea germplasm in Yunnan as revealed by ISSR markers. Acta Agron. 2010, 36, 391–400. [Google Scholar] [CrossRef]
- Tan, L.Q.; Peng, M.; Xu, L.; Wang, L.Y.; Chen, S.X.; Zou, Y.; Qi, G.N.; Cheng, H. Fingerprinting 128 chinese clonal tea cultivars using SSR markers provides new insights into their pedigree relationships. Tree Genet. Genom. 2015, 11, 90. [Google Scholar] [CrossRef]
- Wang, P.; Fang, J.; Lin, H.; Yang, W.; Yu, J.; Hong, Y.; Jiang, M.; Gu, M.; Chen, Q.; Zheng, Y.; et al. Genomes of single and double-petal jasmines (Jasminum sambac) provide insights into their divergence time and structural variations. Plant Biotechnol. J. 2022, 20, 1232–1234. [Google Scholar] [CrossRef] [PubMed]
- Fang, W.P.; Meinhardt, L.W.; Tan, H.W.; Zhou, L.; Mischke, S.; Zhang, D.P. Varietal identification of tea (Camellia sinensis) using nanofluidic array of single nucleotide polymorphism (SNP) markers. Hortic. Res. 2014, 1, 14035. [Google Scholar] [CrossRef] [PubMed]
- Buckler, E.S.; Thornsberry, J. Plant molecular diversity and applications to genomics. Curr. Opin. Plant Biol. 2002, 5, 107–111. [Google Scholar] [CrossRef]
- Rafalski, A. Applications of single nucleotide polymorphisms in crop genetics. Curr. Opin. Plant Biol. 2002, 5, 94–100. [Google Scholar] [CrossRef]
- Lin, Y.; Yu, W.T.; Zhou, L.; Fan, X.J.; Wang, F.Q.; Wang, P.J.; Fang, W.P.; Cai, C.P.; Ye, N.X. Genetic diversity of oolong tea (Camellia sinensis) germplasms based on the nanofluidic array of single-nucleotide polymorphism (SNP) markers. Tree Genet. Genom. 2020, 16, 3. [Google Scholar] [CrossRef]
- Fan, X.J.; Yu, W.T.; Cai, C.P.; Li, Y.; Wang, Z.H.; Fang, W.P.; Zhang, J.M.; Ye, N.X. Construction of molecular id for tea cultivars by using of single nucleotide polymorphism (SNP) Markers. Sci. Agric. Sin. 2021, 54, 1751–1772. [Google Scholar] [CrossRef]
- Wang, Z.H.; Yu, W.T.; Fang, D.Y.; Cai, J.Y.; Wang, J.H.; Fan, X.J.; Liu, C.G.; Xu, B.; Ye, N.X. EST-SNP marker-based genetic analysis on tea germplasms of Yunxiao in Fujian. Fujian J. Agric. Sci. 2021, 36, 1431–1438. [Google Scholar] [CrossRef]
- Yamashita, H.; Katai, H.; Kawaguchi, L.; Nagano, A.J.; Nakamura, Y.; Morita, A.; Ikka, T. Analyses of single nucleotide polymorphisms identified by ddRAD-seq reveal genetic structure of tea germplasm and japanese landraces for tea breeding. PLoS ONE 2019, 14, e0220981. [Google Scholar] [CrossRef] [PubMed]
- Fujian Province Standard Map Service. Available online: http://bzdt.fjmap.net/ (accessed on 27 August 2022).
- Mohammadi, S.A.; Prasanna, B.M. Analysis of genetic diversity in crop plants—Salient statistical tools and considerations. Crop Sci. 2003, 43, 1235–1248. [Google Scholar] [CrossRef]
- Xia, E.H.; Zhang, H.B.; Sheng, J.; Li, K.; Zhang, Q.J.; Kim, C.H.; Zhang, Y.; Liu, Y.; Zhu, T.; Li, W.; et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant 2017, 10, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.H.; Tong, W.; Hou, Y.; An, Y.L.; Chen, L.B.; Wu, Q.; Liu, Y.L.; Yu, J.; Li, F.D.; Li, R.P.; et al. The reference genome of tea plant and resequencing of 81 diverse accessions provide insights into its genome evolution and adaptation. Mol. Plant 2020, 13, 1013–1026. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.Y.; Zhang, Y.J.; Qiu, H.J.; Guo, Y.F.; Wan, H.L.; Zhang, X.L.; Scossa, F.; Alseekh, S.; Zhang, Q.H.; Wang, P.; et al. Genome assembly of wild tea tree DASZ reveals pedigree and selection history of tea varieties. Nat. Commun. 2020, 11, 3719. [Google Scholar] [CrossRef]
- Wang, X.C.; Feng, H.; Chang, Y.X.; Ma, C.; Wang, L.Y.; Hao, X.Y.; Li, A.L.; Cheng, H.; Wang, L.; Cui, P.; et al. Population sequencing enhances understanding of tea plant evolution. Nat. Commun. 2020, 11, 4447. [Google Scholar] [CrossRef]
- Zhang, X.T.; Chen, S.; Shi, L.Q.; Gong, D.P.; Zhang, S.C.; Zhao, Q.; Zhan, D.L.; Vasseur, L.; Wang, Y.B.; Yu, J.X.; et al. Haplotype-resolved genome assembly provides insights into evolutionary history of the tea plant Camellia sinensis. Nat. Genet. 2021, 53, 1250–1259. [Google Scholar] [CrossRef]
- Zhang, Q.J.; Li, W.; Li, K.; Nan, H.; Shi, C.; Zhang, Y.; Dai, Z.Y.; Lin, Y.L.; Yang, X.; Tong, Y.; et al. The chromosome-level reference genome of tea tree unveils recent bursts of non-autonomous LTR retrotransposons in driving genome size evolution. Mol. Plant 2020, 13, 935–938. [Google Scholar] [CrossRef]
- Wang, P.J.; Yu, J.X.; Jin, S.; Chen, S.; Yue, C.; Wang, W.L.; Gao, S.L.; Cao, H.L.; Zheng, Y.C.; Gu, M.Y.; et al. Genetic basis of high aroma and stress tolerance in the oolong tea cultivar genome. Hortic. Res. 2021, 8, 107–121. [Google Scholar] [CrossRef]
- Ganal, M.W.; Altmann, T.; Roder, M.S. SNP identification in crop plants. Curr. Opin. Plant Biol. 2009, 12, 211–217. [Google Scholar] [CrossRef]
- Hansson, B.; Westerberg, L. On the correlation between heterozygosity and fitness in natural populations. Mol. Ecol. 2002, 11, 2467–2474. [Google Scholar] [CrossRef] [PubMed]
- Slate, J.; Davi, P.; Dodds, K.G.; Veenvliet, B.A.; Glass, B.C.; Broad, T.E.; Mcewan, J.C. Understanding the relationship between the inbreeding coefficient and multilocus heterozygosity: Theoretical expectations and empirical data. Heredity 2004, 93, 255–265. [Google Scholar] [CrossRef] [PubMed]
- Tan, L.Q.; Wang, X.Y.; Li, H.; Liu, G.Q.; Zou, Y.; Chen, S.X.; Li, P.W.; Tang, Q. Systematic evaluation of landrace tea populations in Northern Sichuan, China, Based on Morphology, DNA Markers, and Biochemistry Analyses. Hortscience 2018, 53, 1095–1101. [Google Scholar] [CrossRef]
- Tamaki, I.; Kuze, T.; Hirota, K.; Mizuno, M. Genetic variation and population demography of the landrace population of Camellia sinensis in Kasuga, Gifu Prefecture, Japan. Genet. Resour. Crop Evol. 2016, 63, 823–831. [Google Scholar] [CrossRef]
- Niu, S.Z.; Song, Q.F.; Koiwa, H.; Qiao, D.H.; Zhao, D.G.; Chen, Z.W.; Liu, X.; Wen, X.P. Genetic diversity, linkage disequilibrium, and population structure analysis of the tea plant (Camellia sinensis) from an origin center, Guizhou plateau, using genome-wide SNPs developed by genotyping-by-sequencing. BMC Plant Biol. 2019, 19, 328. [Google Scholar] [CrossRef]
- Zhao, Z.F.; Song, Q.F.; Bai, D.C.; Niu, S.Z.; He, Y.Q.; Qiao, D.H.; Chen, Z.W.; Li, C.Y.; Luo, J.; Li, F. Population structure analysis to explore genetic diversity and geographical distribution characteristics of cultivated-type tea plant in Guizhou Plateau. BMC Plant Biol. 2022, 22, 55. [Google Scholar] [CrossRef]
- Zhu, C.; Zhang, H.T.; Chang, X.J.; Wang, Z.; Xu, C.T.; Zhang, X.H.; Lin, Y.L.; Lai, Z.X.; Guo, Y.Q. Analysis of genetic diversity of fifty-three Camellia sinensis in Fujian Province by ISSR. Chin. J. Trop. Crops 2017, 38, 1303–1310. [Google Scholar] [CrossRef]
- Yao, M.Z.; Ma, C.L.; Qiao, T.T.; Jin, J.Q.; Chen, L. Diversity distribution and population structure of tea germplasms in China revealed by EST-SSR markers. Tree Genet. Genom. 2012, 8, 205–220. [Google Scholar] [CrossRef]
- Huang, F.Y.; Duan, J.H.; Lei, Y.; Liu, Z.; Kang, Y.K.; Luo, Y.; Chen, Y.G.; Li, Y.Y.; Liu, S.Q.; Li, S.J.; et al. Genetic diversity, population structure and core collection analysis of Hunan tea plant germplasm through genotyping-by-sequencing. Beverage Plant Res. 2022, 2, 5. [Google Scholar] [CrossRef]
- Su, W.J.; Wang, L.J.; Lei, J.; Chai, S.S.; Liu, Y.; Yang, Y.Y.; Yang, X.S.; Jiao, C.H. Genome-wide assessment of population structure and genetic diversity and development of a core germplasm set for sweet potato based on specific length amplified fragment (SLAF) sequencing. PLoS ONE 2017, 12, e0172066. [Google Scholar] [CrossRef]
- Wadl, P.A.; Olukolu, B.A.; Branham, S.E.; Jarret, R.L.; Yencho, G.C.; Jackson, D.M. Genetic diversity and population structure of the USDA sweetpotato (Ipomoea batatas) germplasm collections using GBSpoly. Front. Plant Sci. 2018, 9, 1166–1178. [Google Scholar] [CrossRef]
- Meegahakumbura, M.K.; Wambulwa, M.C.; Thapa, K.K.; Li, M.M.; Möller, M.; Xu, J.C.; Yang, J.B.; Liu, B.Y.; Ranjitkar, S.; Liu, J.; et al. Indications for three independent domestication events for the tea plant (Camellia sinensis (L.) O. Kuntze) and new insights into the origin of tea germplasm in China and India revealed by nuclear microsatellites. PLoS ONE 2016, 11, e0155369. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.I.; Yang, X.Q.; Huang, X.M.; Duan, S.; Long, C.; Chen, J.K.; Rong, J. Leaf transcriptome analysis of a subtropical evergreen broadleaf plant, wild oil-tea camellia (Camellia oleifera), revealing candidate genes for cold acclimation. BMC Genom. 2017, 18, 211–224. [Google Scholar] [CrossRef] [PubMed]
- Hardigan, M.A.; Bamberg, J.; Buell, C.R.; Douches, D.S. Taxonomy and genetic differentiation among wild and cultivated germplasm of solanum sect Petota. Plant Genome 2015, 8, plantgenome2014.06.0025. [Google Scholar] [CrossRef] [PubMed]
- Leinonen, P.H.; Sandring, S.; Quilot, B.; Clauss, M.J.; Mitchell-Olds, T.; Agren, J.; Savolainen, O. Local adaptation in European populations of Arabidopsis lyrata (Brassicaceae). Am. J. Bot. 2019, 96, 1129–1137. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).