Potential Metabolic Pathways and Related Processes Involved in Pericarp Browning for Postharvest Pomegranate Fruits
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Evaluation of Browning Index
2.3. Malondialdehyde Concentration Measurement
2.4. Electrolyte Leakage Determination
2.5. Determination of Superoxide Dismutase, Catalase, Ascorbate Peroxidase and Polyphenol Oxidase
2.6. Ascorbic Acid and Glutathione and Total Phenolics Contents
2.7. RNA Extraction, Library Preparation and RNA-Seq
2.8. Data Analysis, Annotation and Differential Expression Analysis
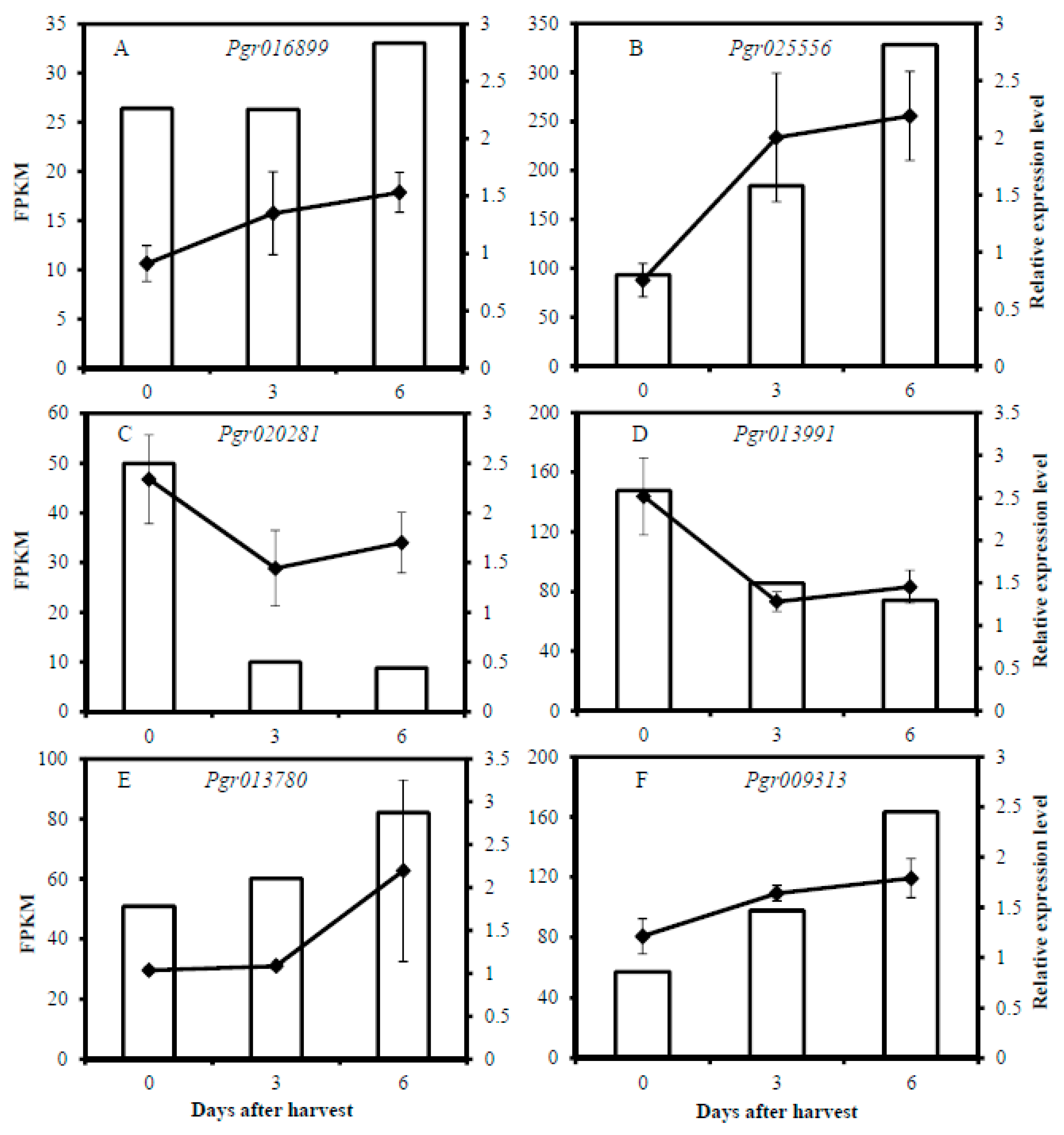
2.9. Quantitative Real-Time PCR (qRT-PCR)
2.10. Statistical Analyses
3. Results
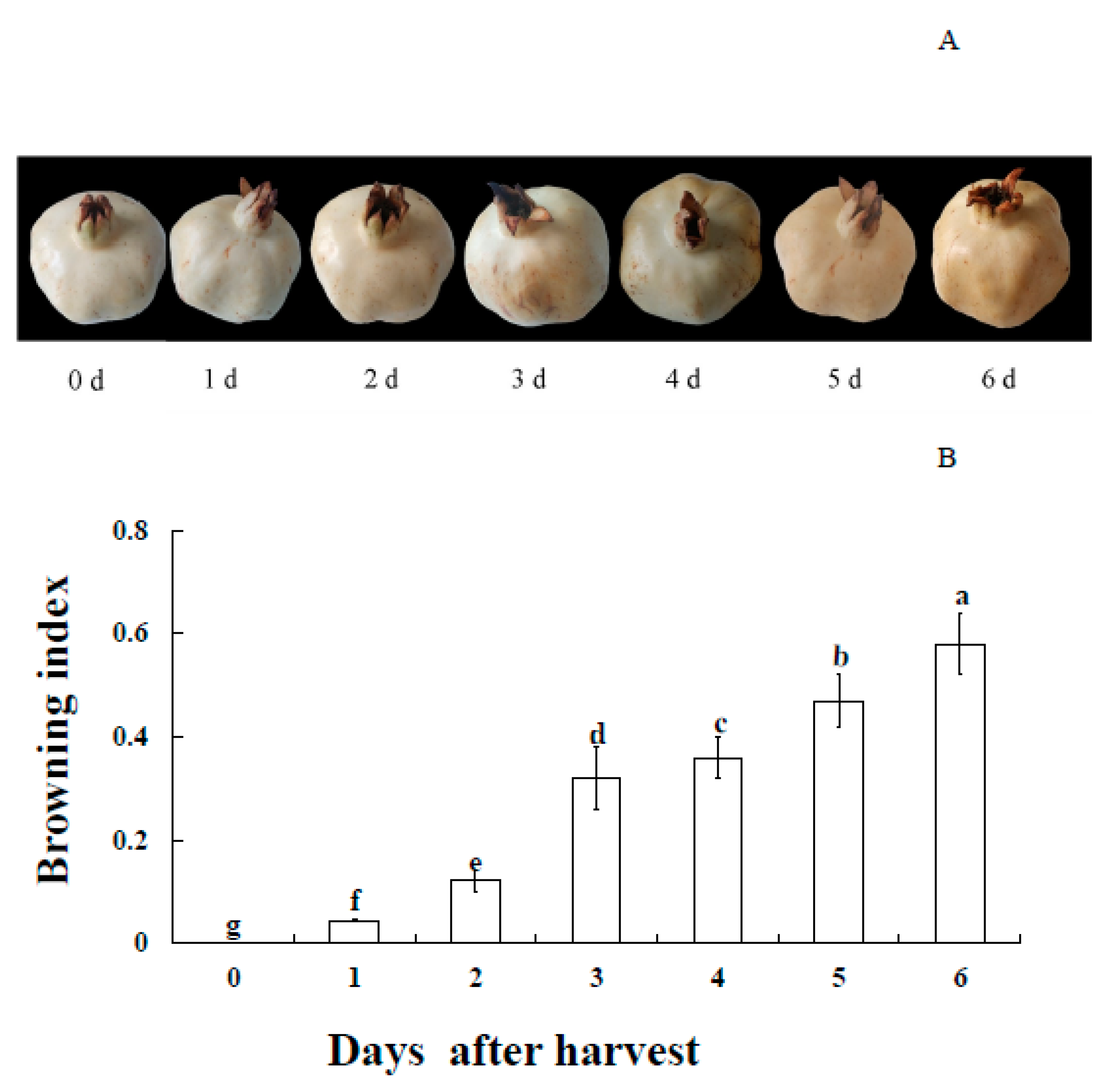
3.1. Appearance and BI Changes of Pericarp of Postharvest Fruit
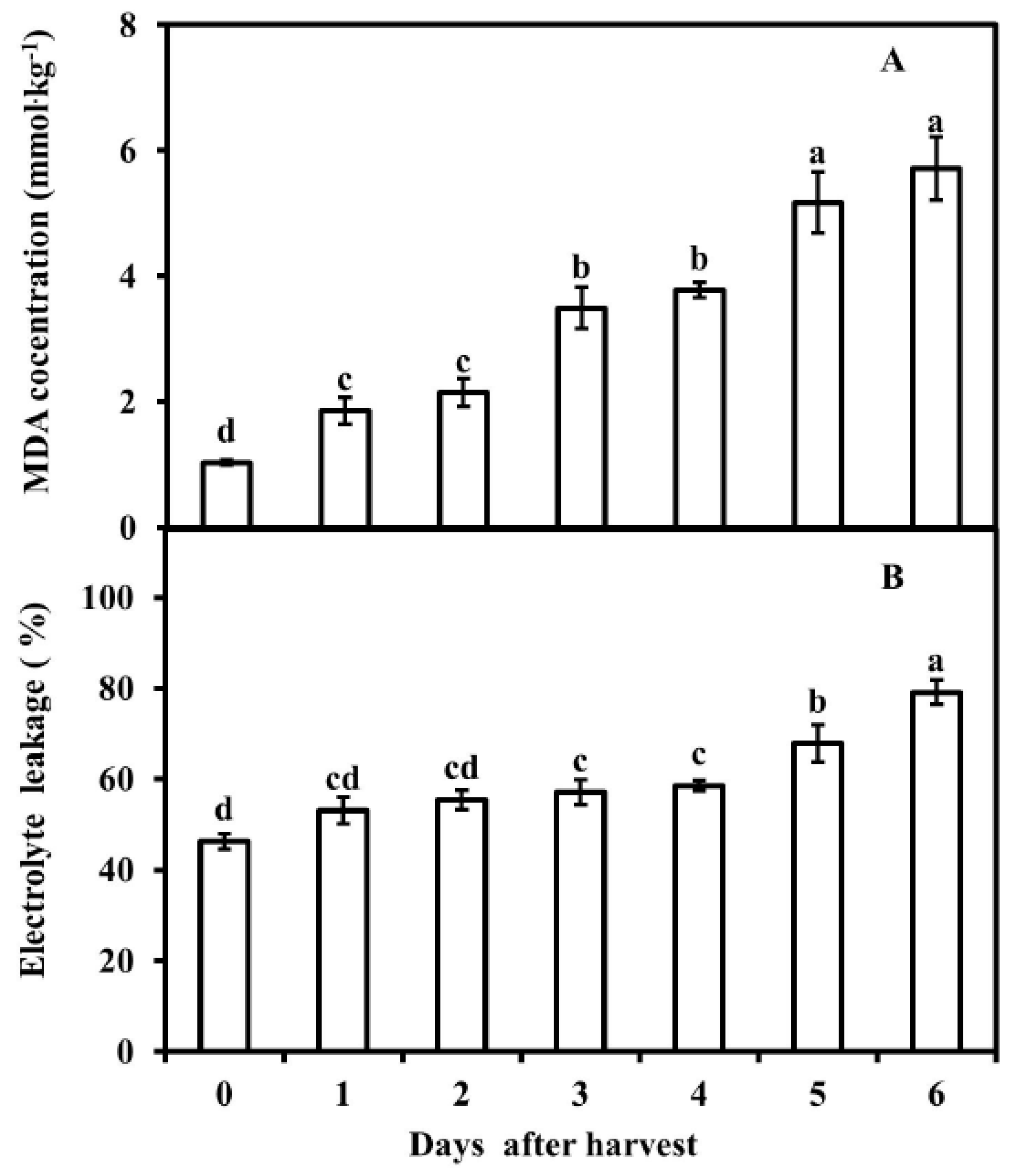
3.2. MDA Concentration and Electrolyte Leakage of Pericarp of Postharvest Fruit
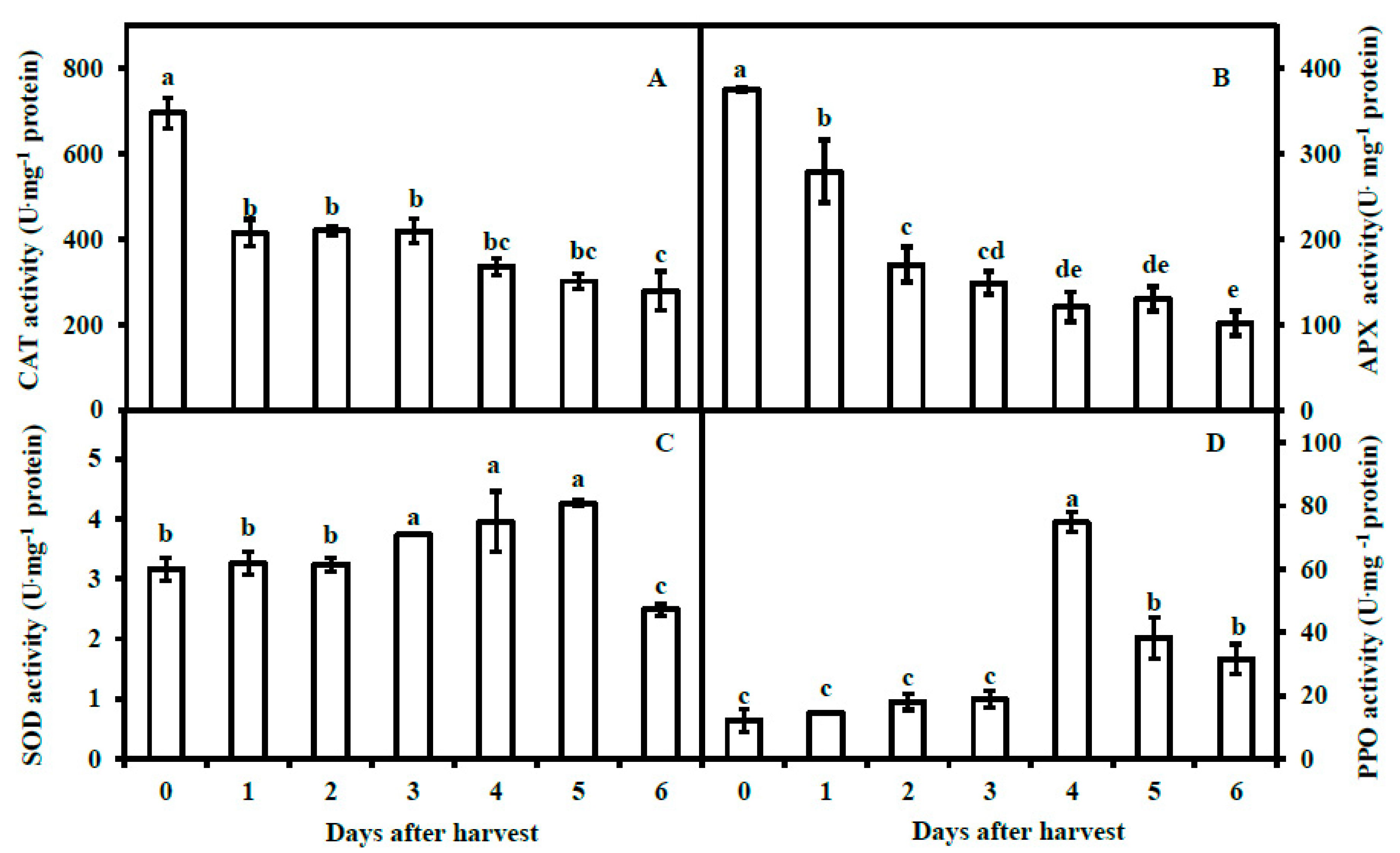
3.3. Activities of Antioxidant Enzymes and Polyphenol Oxidase of Pericarp of Postharvest Fruit
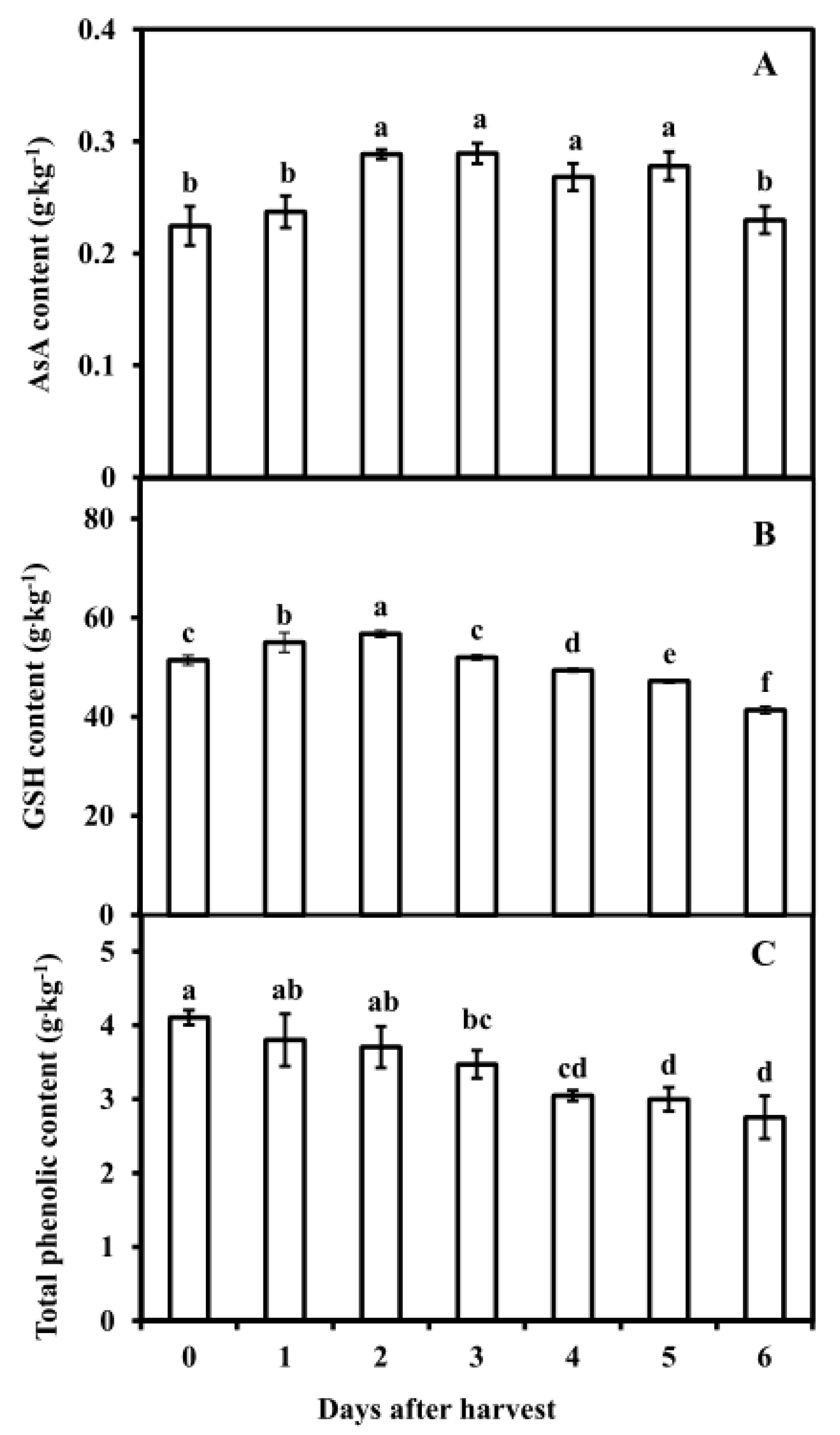
3.4. Antioxidant Substance Contents in Pericarp of Postharvest Fruit
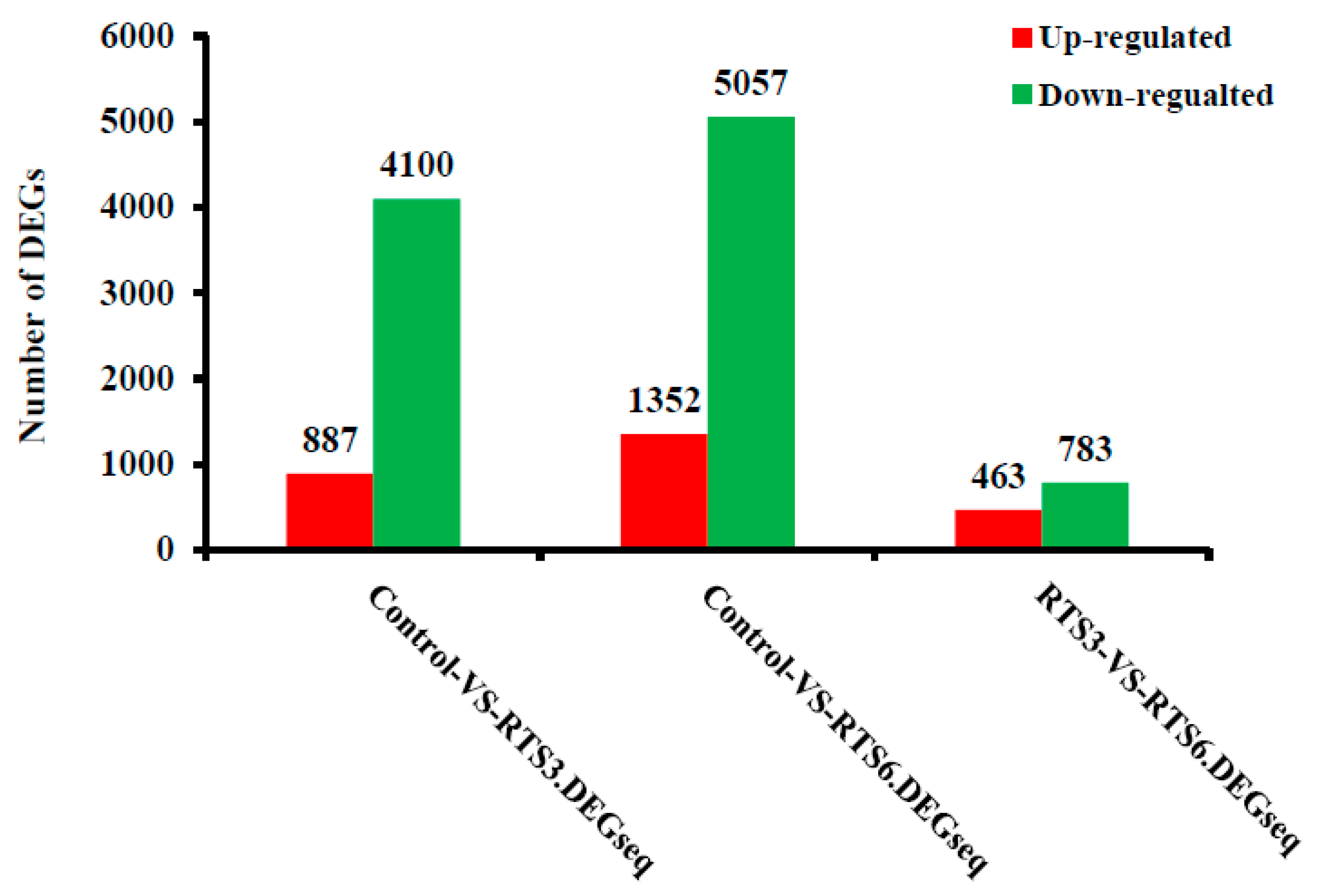
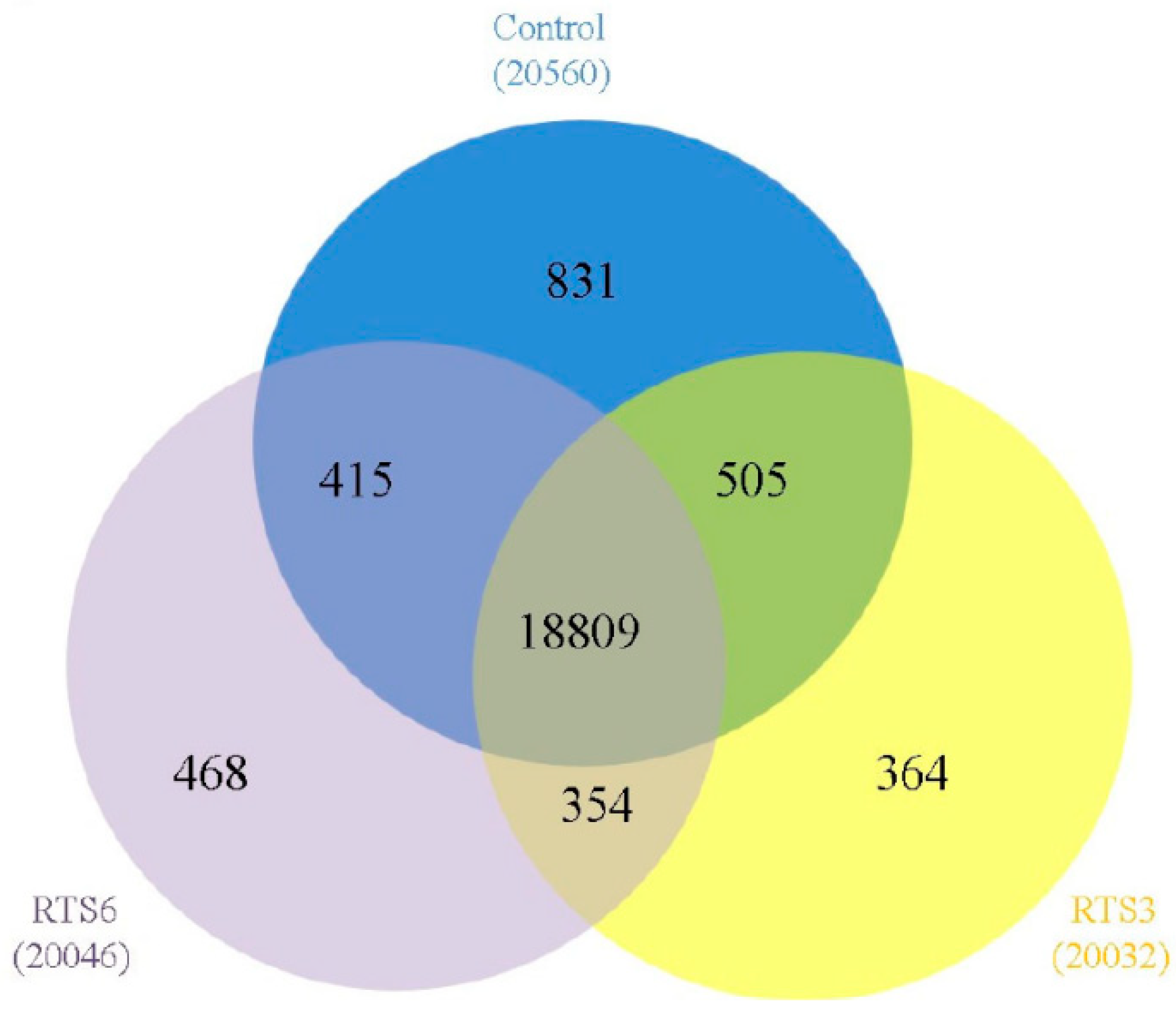
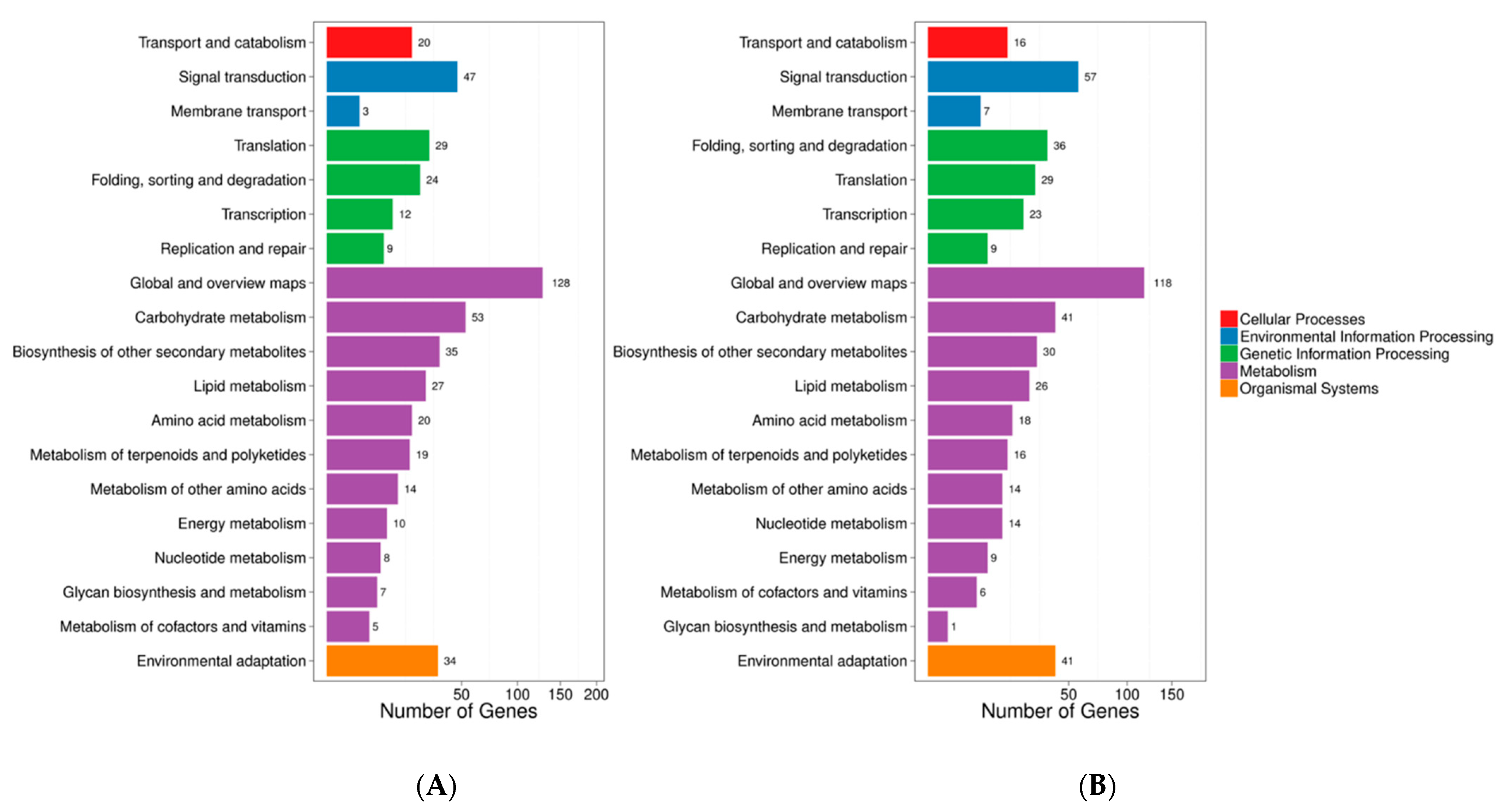
3.5. Transcriptome Profiling of Browning Pericarp
3.6. Selection of Candidate Genes Involved in Pericarp Browning
3.6.1. Genes Involved in Energy-Related Pathways
3.6.2. Genes Involved in Lipid Metabolism
3.6.3. Genes Involved in Sugar and Starch Metabolism
3.6.4. Genes Involved in Ethylene Biosynthesis and Signal Transduction
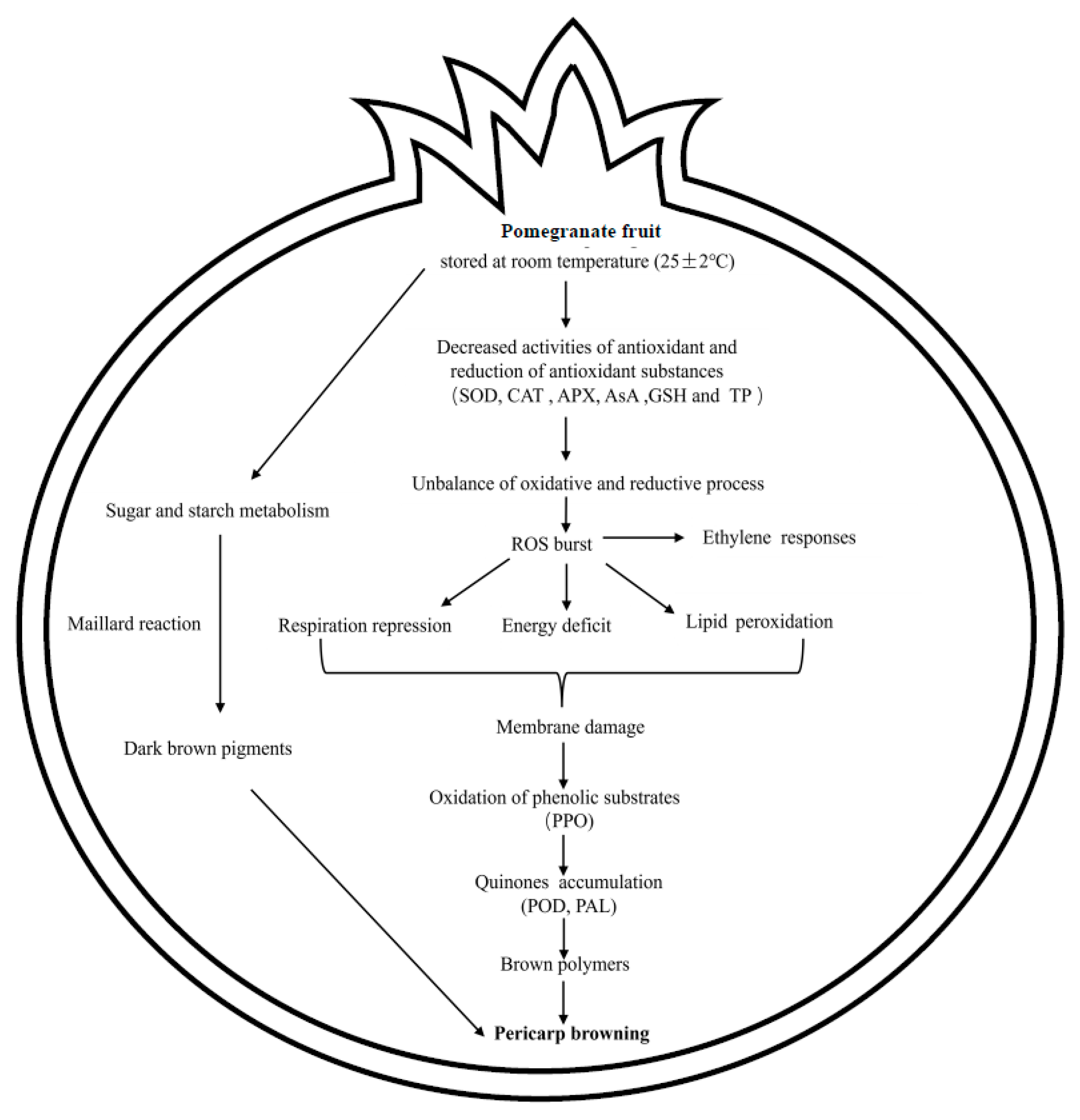
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Seeram, N.P.; Schulman, R.N.; Heber, D. Pomegranates: Ancient Roots to Modern Medicine; CRC Press: Boca Raton, FL, USA, 2006; pp. 63–183. [Google Scholar]
- Qin, G.H.; Xu, C.Y.; Ming, R.; Tang, H.B.; Guyot, R.; Kramer, E.M.; Hu, Y.D.; Yi, X.K.; Qi, Y.J.; Xu, X.Y.; et al. The pomegranate (Punica granatum L.) genome and the genomics of punicalagin biosynthesis. Plant J. 2017, 91, 1108–1128. [Google Scholar] [CrossRef]
- Ioannou, I.; Ghoul, M. Prevention of enzymatic browning in fruit and vegetables. Eur. J. Sci. 2013, 9, 310–341. [Google Scholar] [CrossRef]
- Wei, W.L.; Xu, C.; Wu, L.; Wang, J.S.; Ren, J.S.; Qu, X.G. Non-enzymatic-browning-reaction: A versatile route for production of nitrogen-doped carbon dots with tunable multicolor luminescent display. Sci. Rep. 2014, 4, 3564. [Google Scholar] [CrossRef] [PubMed]
- Hong, P.K.; Betti, M. Non-enzymatic browning reaction of glucosamine at mild conditions: Relationship between colour formation, radical scavenging activity and α-dicarbonyl compounds production. Food Chem. 2016, 212, 234–243. [Google Scholar] [CrossRef] [PubMed]
- Sun, J.Z.; Lin, H.T.; Zhang, S.; Lin, Y.F.; Wang, H.; Lin, M.S.; Hung, Y.C.; Chen, Y.H. The roles of ROS production-scavenging system in Lasiodiplodia theobromae (Pat.) Griff. & Maubl.-induced pericarp browning and disease development of harvested longan fruit. Food Chem. 2018, 247, 16–22. [Google Scholar] [CrossRef]
- Ali, S.; Khan, A.S.; Nawaz, A.; Anjum, M.A.; Naz, S.; Ejaz, S.; Hussain, S. Aloe vera gel coating delays postharvest browning and maintains quality of harvested litchi fruit. Postharvest Biol. Technol. 2019, 157, 110960. [Google Scholar] [CrossRef]
- Lin, Y.F.; Lin, H.T.; Zhang, S.; Chen, Y.H.; Chen, M.Y.; Lin, Y.X. The role of active oxygen metabolism in hydrogen peroxide-induced pericarp browning of harvested longan fruit. Postharvest Biol. Technol. 2014, 96, 42–48. [Google Scholar] [CrossRef]
- Lamikanra, O.; Watson, M.A. Effects of ascorbic acid on peroxidase and polyphenoloxidase activities in fresh-cut cantaloupe melon. J. Food Sci. 2001, 66, 1283–1286. [Google Scholar] [CrossRef]
- Degl’Innocenti, E.; Pardossi, A.; Tognoni, F.; Guidi, L. Physiological basis of sensitivity to enzymatic browning in ‘lettuce’, ‘escarole’ and ‘rocket salad’ when stored as fresh-cut products. Food Chem. 2007, 104, 209–215. [Google Scholar] [CrossRef]
- Holderbaum, D.F.; Kon, T.; Kudo, T.; Guerra, M.P. Enzymatic browning, polyphenol oxidase activity, and polyphenols in four apple cultivars: Dynamics during fruit development. HortScience 2010, 45, 1150–1154. [Google Scholar] [CrossRef]
- Banerjee, A.; Suprasanna, P.; Variyar, S.; Sharma, A. Gamma irradiation inhibits wound induced browning in shredded cabbage. Food Chem. 2015, 173, 38–44. [Google Scholar] [CrossRef]
- Zhu, H.S.; Liu, J.T.; Wen, Q.F.; Chen, M.D.; Wang, B.; Zhang, Q.R.; Xue, Z.Z. De novo sequencing and analysis of the transcriptome during the browning of fresh cut Luffa cylindrica ’Fusi-3′ fruit. PLoS ONE 2017, 12, e0187117. [Google Scholar] [CrossRef]
- Ali, S.; Nawaz, A.; Ejaz, S.; Haider, S.T.A.; Alam, M.W.; Javed, H.U. Effects of hydrogen sulfide on postharvest physiology of fruits and vegetables: An overview. Sci. Hortic. 2019, 243, 290–299. [Google Scholar] [CrossRef]
- Zhang, Z.; Huber, D.J.; Qu, H.; Yun, Z.; Wang, H.; Huang, Z.; Huang, H.; Jiang, Y. Enzymatic browning and antioxidant activities in harvested litchi fruit as influenced by apple polyphenols. Food Chem. 2015, 171, 191–199. [Google Scholar] [CrossRef] [PubMed]
- Chi, M.; Bhagwat, B.; Lane, W.D.; Tang, G.L.; Su, Y.G.; Sun, R.; Oomah, B.D.; Wiersma, P.A.; Xiang, Y. Reduced polyphenol oxidase gene expression and enzymatic browning in potato (Solanum tuberosum L.) with artificial microRNAs. BMC Plant Biol. 2014, 14, 62. [Google Scholar] [CrossRef]
- Liu, P.; Xue, C.; Wu, T.T.; Heng, W.; Jia, B.; Ye, Z.F.; Liu, L.; Zhu, L.W. Molecular analysis of the processes of surface brown spot (SBS) formation in pear fruit (Pyrus bretschneideri Rehd. Cv. Dangshansuli) by de novo transcriptome assembly. PLoS ONE 2013, 8, e74217. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, X.; Tan, T.M.; Xu, C.C.; Huang, S.P.; Tan, J.; Zhang, M.; Wang, C.L.; Xie, C.H. Genome-wide transcriptome profiling reveals novel insights into Luffa cylindrica browning. Biochem. Biophys. Res. Commun. 2015, 463, 1243–1249. [Google Scholar] [CrossRef]
- Zhang, Y.L.; Zhang, R.G. Study on the mechanism of browning of pomegranate (Punica granatum L. cv. Ganesh) peel in different storage conditions. Agric. Sci. China 2008, 7, 65–73. [Google Scholar] [CrossRef]
- Feng, L.J.; Yin, Y.L.; Yang, X.M.; Wang, C.Z.; Jiao, Q.Q. Changes of browning and its related enzyme activities in pomegranate peel during fruit development period. J. Nucl. Agric. Sci. 2017, 31, 821–827. (In Chinese) [Google Scholar] [CrossRef]
- Qi, X.X.; Qin, G.H. Study on the factors influencing postharvest browning of the pomegranate peel. J. Fruit Sci. 2017, 34, 735–743. (In Chinese) [Google Scholar] [CrossRef]
- Sun, J.; You, X.R.; Li, L.; Peng, H.X.; Su, W.Q.; Li, C.B.; He, Q.G.; Liao, B. Effects of a phospholipase D inhibitor on postharvest enzymatic browning and oxidative stress of litchi fruit. Postharvest Biol. Technol. 2011, 62, 288–294. [Google Scholar] [CrossRef]
- Zhu, L.Q.; Zhou, J.; Zhu, S.H.; Guo, L.H. Inhibition of browning on the surface of peach slices by short-term exposure to nitric oxide and ascorbic acid. Food Chem. 2009, 114, 174–179. [Google Scholar] [CrossRef]
- Cao, J.K.; Jiang, W.B.; Zhao, Y.M. Experiment Guidance of Postharvest Physiology and Biochemistry of Fruits and vegetables; China Light Industry Press: Beijing, China, 2007; pp. 101–103. [Google Scholar]
- Zhang, R.G.; Guo, X.C.; Zhang, Y.L.; Tian, C.R. Influence of modified atmosphere treatment on post-harvest reactive oxygen metabolism of pomegranate peels. Nat. Prod. Res. 2018, 34, 740–744. [Google Scholar] [CrossRef] [PubMed]
- Wojdyło, A.; Oszmiański, J.; Bielicki, P. Polyphenolic composition, antioxidant activity, and polyphenol oxidase (PPO) activity of quince (Cydonia oblonga Miller) varieties. J. Agric. Food Chem. 2013, 61, 2762–2772. [Google Scholar] [CrossRef] [PubMed]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Cai, Y.T.; Cao, S.F.; Yang, Z.F.; Zheng, Y.H. MeJA regulates enzymes involved in ascorbic acid and glutathione metabolism and improves chilling tolerance in loquat fruit. Postharvest Biol. Technol. 2011, 59, 324–326. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Y.Y.; Fan, X.L.; Wang, J.D.; Liang, L.Y.; Yan, S.J.; Xiao, L.X. Relationship between activated oxygen metabolism and browning of “Yali” pears during storage. J. Food Process. Preserv. 2020, 44, e14392. [Google Scholar] [CrossRef]
- Ainsworth, E.A.; Gillespie, K.M. Estimation of total phenolic contents and other oxidation substrates in plant tissue using Folin-Ciocalteu reagent. Nat. Protoc. 2007, 2, 875–877. [Google Scholar] [CrossRef]
- Kumar, R.; Ichihashi, Y.; Kimura, S.; Chitwood, D.H.; Headland, L.R.; Peng, J.; Maloof, J.N.; Sinha, N.R. A high-throughput method for Illumina RNA-Seq library preparation. Front. Plant Sci. 2012, 3, 202. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Langmead, B.; Salzberg, S. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 357–359. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Dewey, C.N. RSEM: Accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 2011, 12, 323. [Google Scholar] [CrossRef]
- Wang, L.K.; Feng, Z.X.; Wang, X.; Wang, X.W.; Zhang, X.G. DEGseq: An R package for identifying differentially expressed genes from RNA-seq data. Bioinformatics 2010, 26, 136–138. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Luo, Y.G. Enzymatic browning and its control in fresh-cut produce. Stewart Postharvest Rev. 2007, 3, 1–7. [Google Scholar] [CrossRef]
- Chumyam, A.; Faiyue, B.; Saengnil, K. Reduction of enzymatic browning of fresh-cut guava fruit by exogenous hydrogen peroxide-activated peroxiredoxin/thioredoxin system. Sci. Hortic. 2019, 255, 260–268. [Google Scholar] [CrossRef]
- Tang, T.T.; Xie, X.F.; Ren, X.; Wang, W.J.; Tang, X.M.; Zhang, J.; Wang, Z.D. A difference of enzymatic browning unrelated to PPO from physiology, targeted metabolomics and gene expression analysis in Fuji apples. Postharvest Biol. Technol. 2020, 170, 111323. [Google Scholar] [CrossRef]
- Duan, X.W.; Liu, T.; Zhang, D.; Su, X.G.; Lin, H.T.; Jiang, Y.M. Effect of pure oxygen atmosphere on antioxidant enzyme and antioxidant activity of harvested litchi fruit during storage. Food Res. Int. 2011, 44, 1905–1911. [Google Scholar] [CrossRef]
- Silva, V.L.D.; Cerqueira, M.R.F.; Lowinsohn, D.; Matos, M.A.C.; Matos, R.C. Amperometric detection of ascorbic acid in honey using ascorbate oxidase immobilised on amberlite IRA-743. Food Chem. 2012, 133, 1050–1054. [Google Scholar] [CrossRef]
- Wang, Y.S.; Luo, Z.S.; Khan, Z.U.; Mao, L.C.; Ying, T.J. Effect of nitric oxide on energy metabolism in postharvest banana fruit in response to chilling stress. Postharvest Biol. Technol. 2015, 108, 21–27. [Google Scholar] [CrossRef]
- Wang, J.W.; Jiang, Y.G.; Li, G.D.; Lv, M.; Zhou, X.; Zhou, Q.; Fu, W.W.; Zhang, L.; Chen, Y.F.; Ji, S.J. Effect of low temperature storage on energy and lipid metabolisms accompanying peel browning of ‘Nanguo’ pears during shelf life. Postharvest Biol. Technol. 2018, 139, 75–81. [Google Scholar] [CrossRef]
- Saha, B.; Borovskii, G.; Panda, S.K. Alternative oxidase and plant stress tolerance. Plant Signal. Behav. 2016, 11, e1256530. [Google Scholar] [CrossRef] [PubMed]
- Ding, C.H.; Chachin, K.; Ueda, Y.; Wang, C.Y. Inhibition of loquat enzymatic browning by sulfhydryl compounds. Food Chem. 2002, 76, 213–218. [Google Scholar] [CrossRef]
- Espley, R.V.; Leif, D.; Plunkett, B.; Mcghie, T.; Henry-Kirk, R.; Hall, M.; Johnston, J.W.; Punter, M.P.; Boldingh, H.; Nardozza, S.; et al. Red to brown: An elevated anthocyanic response in apple drives ethylene to advance maturity and fruit flesh browning. Front. Plant Sci. 2019, 10, 1248. [Google Scholar] [CrossRef] [PubMed]
- Luna, M.C.; Tudela, J.A.; Martínez-Sánchez, A.; Allende, A.; Marín, A.; Gil, M.L. Long-term deficit and excess of irrigation influences quality and browning related enzymes and phenolic metabolism of fresh-cut iceberg lettuce (Lactuca sativa L.). Postharvest Biol. Technol. 2012, 73, 37–45. [Google Scholar] [CrossRef]
- Lin, Y.F.; Lin, Y.X.; Lin, H.T.; Zhang, S.; Chen, Y.H.; Shi, J. Inhibitory effects of propyl gallate on browning and its relationship to active oxygen metabolism in pericarp of harvested longan fruit. LWT—Food Sci. Technol. 2015, 60, 1122–1128. [Google Scholar] [CrossRef]
- Elfalleh, W.; Hannachi, H.; Tlili, N.; Yahia, Y.; Nasri, N.; Ferchichi, A. Total phenolic contents and antioxidant activities of pomegranate peel, seed, leaf and flower. J. Med. Plants Res. 2012, 6, 4724–4730. [Google Scholar] [CrossRef]
- Pereyra, L.; Roura, S.I.; del Valle, C.E. Phenylalanine ammonia lyase activity in minimally processed Romaine lettuce. LWT—Food Sci. Technol. 2005, 38, 67–72. [Google Scholar] [CrossRef]
- Dong, T.; Shi, J.; Jiang, C.Z.; Feng, Y.; Cao, Y.; Wang, Q. A short-term carbon dioxide treatment inhibits the browning of fresh-cut burdock. Postharvest Biol. Technol. 2015, 110, 96–102. [Google Scholar] [CrossRef]
- Ramakrishna, A.; Ravishankar, G.A. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal. Behav. 2011, 6, 1720–1731. [Google Scholar] [CrossRef]
- Chen, M.Y.; Lin, H.T.; Zhang, S.; Lin, Y.F.; Chen, Y.H.; Lin, Y.X. Effects of adenosine triphosphate (ATP) treatment on postharvest physiology, quality and storage behavior of longan fruit. Food Bioprocess Technol. 2015, 8, 971–982. [Google Scholar] [CrossRef]
- Yang, S.; Su, X.; Prasad, K.N.; Yang, B.; Cheng, G.; Chen, Y.; Yang, E.; Jiang, Y. Oxidation and peroxidation of postharvest banana fruit during softening. Pak. J. Bot. 2008, 40, 2023–2029. [Google Scholar]
- Jiang, Y.M.; Jiang, Y.L.; Qu, H.X.; Duan, X.W.; Luo, Y.B.; Jiang, W.B. Energy aspects in ripening and senescence of harvested horticultural crops. Stewart Postharvest Rev. 2007, 3, 1–5. [Google Scholar] [CrossRef]
- McWeeny, D.J.; Knowles, M.E.; Hearne, J.F. The chemistry of non-enzymic browning in foods and its control by sulphites. J. Sci. Food Agric. 1974, 25, 735–746. [Google Scholar] [CrossRef]
- Lievonen, S.M.; Laaksonen, T.J.; Roos, Y.H. Nonenzymatic browning in food models in the vicinity of the glass transition: Effects of fructose, glucose, and xylose as reducing sugar. J. Agric. Food Chem. 2002, 50, 7034–7041. [Google Scholar] [CrossRef]
- Korbel, E.; Attal, E.H.; Grabulos, J.; Lluberas, E.; Durand, N.; Morel, G.; Goli, T.; Brat, P. Impact of temperature and water activity on enzymatic and nonenzymatic reactions in reconstituted dried mango model system. Eur. Food Res. Technol. 2013, 237, 39–46. [Google Scholar] [CrossRef]
- Fukuoka, N.; Miyata, M.; Hamada, T.; Takeshita, E. Histochemical observations and gene expression changes related to internal browning in tuberous roots of sweet potato (Ipomea batatas). Plant Sci. 2018, 274, 476–484. [Google Scholar] [CrossRef]
- Mellidou, I.; Buts, K.; Hatoum, D.; Ho, Q.T.; Johnston, J.W.; Watkins, C.B.; Schaffer, R.J.; Gapper, N.E.; Giovannoni, J.J.; Rudell, D.R.; et al. Transcriptomic events associated with internal browning of apple during postharvest storage. BMC Plant Biol. 2014, 14, 328. [Google Scholar] [CrossRef]
| Sample | Total Raw Reads (Mb) | Total Clean Reads (Mb) | Total Bases (Gb) | Clean Reads Q30(%) | Total Mapping Ratio (%) |
|---|---|---|---|---|---|
| Control_1 | 55.52 | 44.12 | 6.62 | 94.16 | 85.07 |
| Control_2 | 53.88 | 44.11 | 6.62 | 94.29 | 86.59 |
| Control_3 | 53.88 | 44.33 | 6.65 | 94.56 | 86.99 |
| RTS3_1 | 55.52 | 44.90 | 6.74 | 94.19 | 87.57 |
| RTS3_2 | 53.89 | 44.58 | 6.69 | 94.27 | 88.59 |
| RTS3_3 | 55.52 | 45.02 | 6.75 | 94.30 | 86.41 |
| RTS6_1 | 57.15 | 45.08 | 6.76 | 94.00 | 83.90 |
| RTS6_2 | 55.52 | 44.60 | 6.69 | 94.48 | 85.49 |
| RTS6_3 | 53.88 | 44.39 | 6.69 | 94.20 | 87.42 |
| Gene ID | Length (bp) | FPKM | Gene Description | ||
|---|---|---|---|---|---|
| Control | RTS3 | RTS6 | |||
| Genes encode enzymes involved in phenolic compound metabolism | |||||
| Pgr016899 | 2849 | 26.42 | 26.31 | 33.04 | polyphenol oxidase |
| Pgr024170 | 1086 | 0.02 | 0.18 | 0.38 | peroxidase |
| Pgr025556 | 2687 | 93.05 | 184.02 | 328.36 | phenylalanine ammonia-lyase |
| Genes involved ascorbic acid-glutathione metabolism and encoding antioxidant enzymes | |||||
| Pgr019084 | 2443 | 27.85 | 12.23 | 12.63 | L-galactono-1,4-lactone dehydrogenase |
| Pgr026685 | 1317 | 0.34 | 0 | 0.02 | L-ascorbate oxidase |
| Pgr020281 | 1704 | 49.92 | 10.01 | 8.87 | copper/zinc superoxide dismutase |
| Pgr020981 | 1471 | 8.92 | 3.04 | 3.36 | superoxide dismutase [Fe] 3 |
| Pgr014250 | 1917 | 3467.51 | 1691.19 | 1540.47 | catalase isozyme 3 |
| Pgr025048 | 1365 | 310.49 | 122.97 | 95.11 | L-ascorbate peroxidase 2, cytosolic |
| BGI_novel_G000299 | 1128 | 1.95 | 0.66 | 0.63 | PREDICTED: putative L-ascorbate peroxidase 6 isoform X8 |
| Genes involved in energy-related pathway | |||||
| Pgr016265 | 729 | 25.67 | 14.94 | 9.47 | NADH dehydrogenase (ubiquinone) activity |
| Pgr016267 | 873 | 18.94 | 7.75 | 4.06 | NADH dehydrogenase (ubiquinone) activity |
| Pgr013991 | 1234 | 147.68 | 85.29 | 74.04 | ATP synthase, mitochondrial |
| Pgr012837 | 1041 | 26.78 | 48.72 | 38.53 | alternative oxidase activity |
| Pgr009832 | 1413 | 444.53 | 189.83 | 212.89 | glyceraldehyde 3-phosphate dehydrogenase |
| Pgr022301 | 1616 | 218.53 | 68.49 | 45.48 | glyceraldehyde 3-phosphate dehydrogenase |
| Pgr007501 | 1035 | 193.95 | 64.26 | 42.07 | malate dehydrogenase, mitochondrial |
| Pgr002381 | 1050 | 102.24 | 43.67 | 43.29 | malate dehydrogenase, mitochondrial |
| Pgr017364 | 1627 | 2.47 | 0.46 | 0.19 | alcohol dehydrogenase 1 |
| BGI_novel_G000840 | 1471 | 1.19 | 0.41 | 0.65 | alcohol dehydrogenase 1 isoform X2 |
| Pgr009655 | 2284 | 58.94 | 23.29 | 15.28 | pyruvate dehydrogenase E1 component alpha subunit |
| Pgr002364 | 3888 | 43.74 | 14.12 | 8.95 | phosphoenolpyruvate carboxylase, housekeeping isozyme |
| Pgr024145 | 4230 | 27.76 | 12.64 | 13.8 | phosphoenolpyruvate carboxylase 4 |
| Genes involved in lipid metabolism | |||||
| Pgr027677 | 2583 | 0.13 | 0.3 | 0.5 | phospholipase D |
| Pgr013780 | 2754 | 50.9 | 60.16 | 82.13 | lipoxygenase |
| Genes involved in sugar and starch metabolism | |||||
| Pgr009313 | 3111 | 57.36 | 97.78 | 163.44 | sucrose synthase |
| Pgr008953 | 1905 | 28.3 | 13.11 | 10.76 | ADP-glucose pyrophosphorylase family protein |
| Pgr018115 | 2529 | 111.59 | 45.33 | 50.01 | granule-bound starch synthase |
| Genes involved ethylene biosynthesis and signal transduction | |||||
| Pgr012839 | 1349 | 14.7 | 0.22 | 0.04 | 1-aminocyclopropane-1-carboxylate oxidase |
| Pgr011853 | 1173 | 4.11 | 0.47 | 0.65 | 1-aminocyclopropane-1-carboxylate oxidase |
| Pgr012022 | 1763 | 10.41 | 65.17 | 58.36 | ethylene-responsive transcription factor |
| Pgr013421 | 1199 | 0.44 | 0.95 | 1.64 | AP2-like ethylene-responsive transcription factor |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Qi, X.; Zhao, J.; Jia, Z.; Cao, Z.; Liu, C.; Li, J.; Su, Y.; Pan, Y.; He, C.; Xu, Y.; et al. Potential Metabolic Pathways and Related Processes Involved in Pericarp Browning for Postharvest Pomegranate Fruits. Horticulturae 2022, 8, 924. https://doi.org/10.3390/horticulturae8100924
Qi X, Zhao J, Jia Z, Cao Z, Liu C, Li J, Su Y, Pan Y, He C, Xu Y, et al. Potential Metabolic Pathways and Related Processes Involved in Pericarp Browning for Postharvest Pomegranate Fruits. Horticulturae. 2022; 8(10):924. https://doi.org/10.3390/horticulturae8100924
Chicago/Turabian StyleQi, Xiaoxiao, Jianrong Zhao, Zhenyu Jia, Zhen Cao, Chunyan Liu, Jiyu Li, Ying Su, Yongbao Pan, Cong He, Yiliu Xu, and et al. 2022. "Potential Metabolic Pathways and Related Processes Involved in Pericarp Browning for Postharvest Pomegranate Fruits" Horticulturae 8, no. 10: 924. https://doi.org/10.3390/horticulturae8100924
APA StyleQi, X., Zhao, J., Jia, Z., Cao, Z., Liu, C., Li, J., Su, Y., Pan, Y., He, C., Xu, Y., & Qin, G. (2022). Potential Metabolic Pathways and Related Processes Involved in Pericarp Browning for Postharvest Pomegranate Fruits. Horticulturae, 8(10), 924. https://doi.org/10.3390/horticulturae8100924








