Differential Triggering of the Phenylpropanoid Biosynthetic Pathway Key Genes Transcription upon Cold Stress and Viral Infection in Tomato Leaves
Abstract
:Highlights
- Phenolic, flavonoid contents, and PAL enzyme activity were elevated after biotic and abiotic stresses.
- Phenolic moieties were generally more enhanced by cold stress as compared to viral infection.
- The transcription of most genes encoding for enzymes regulating both the core PBP and flavonoid synthesis was affected by biotic and abiotic stresses.
- The former was mainly triggered by cold stress, whereas the latter by viral infection.
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growing Conditions
2.2. Mechanical Inoculation of CMV and PVY
2.3. TP Content, TF Content, and PAL Enzyme Activity
2.4. RNA Extraction and Real-Time PCR
2.5. Phylogenetic Analysis of 4CL and CHS Proteins
2.6. Statistical Analysis
3. Results
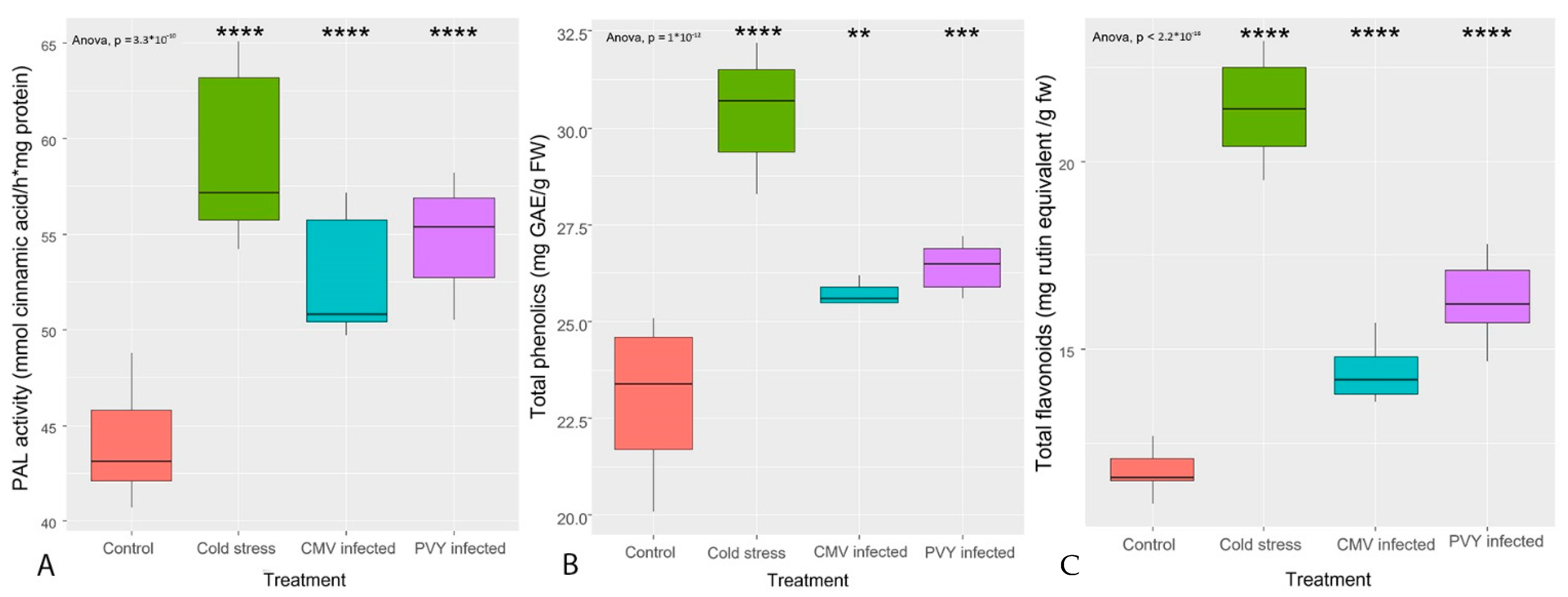
3.1. TP Content, TF Content, and PAL Enzyme Activity
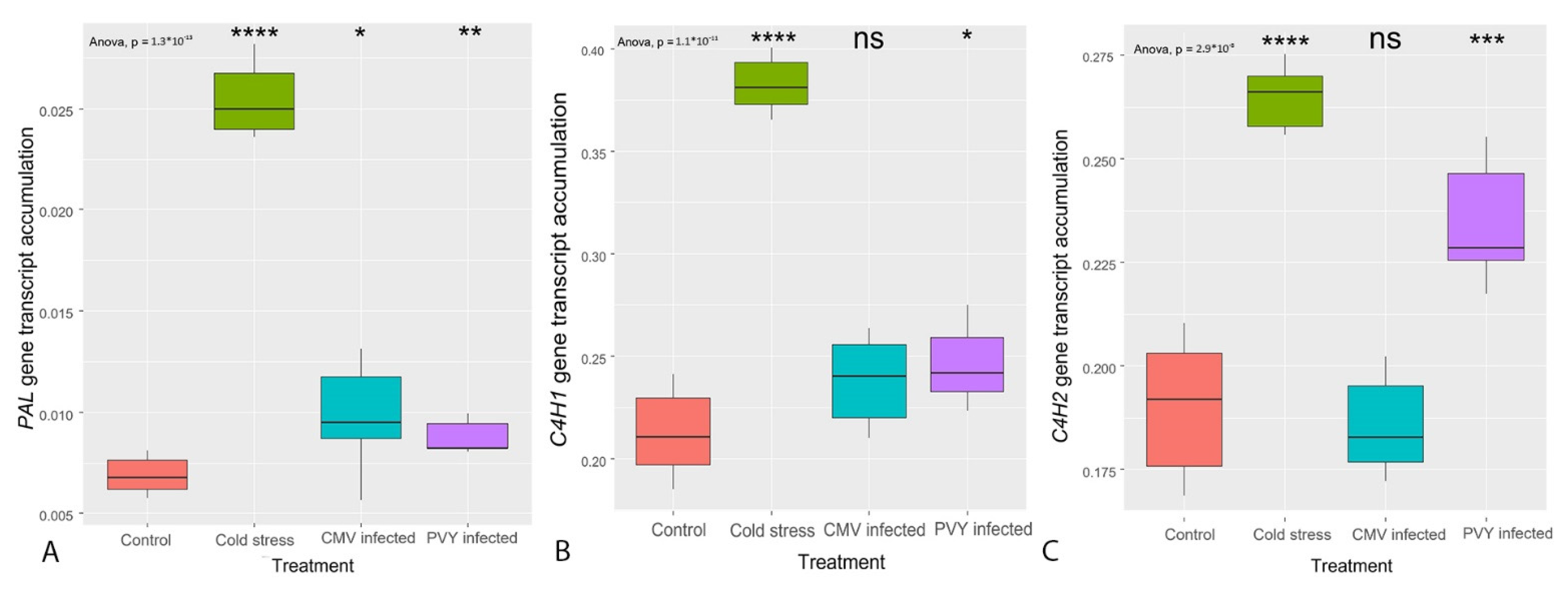
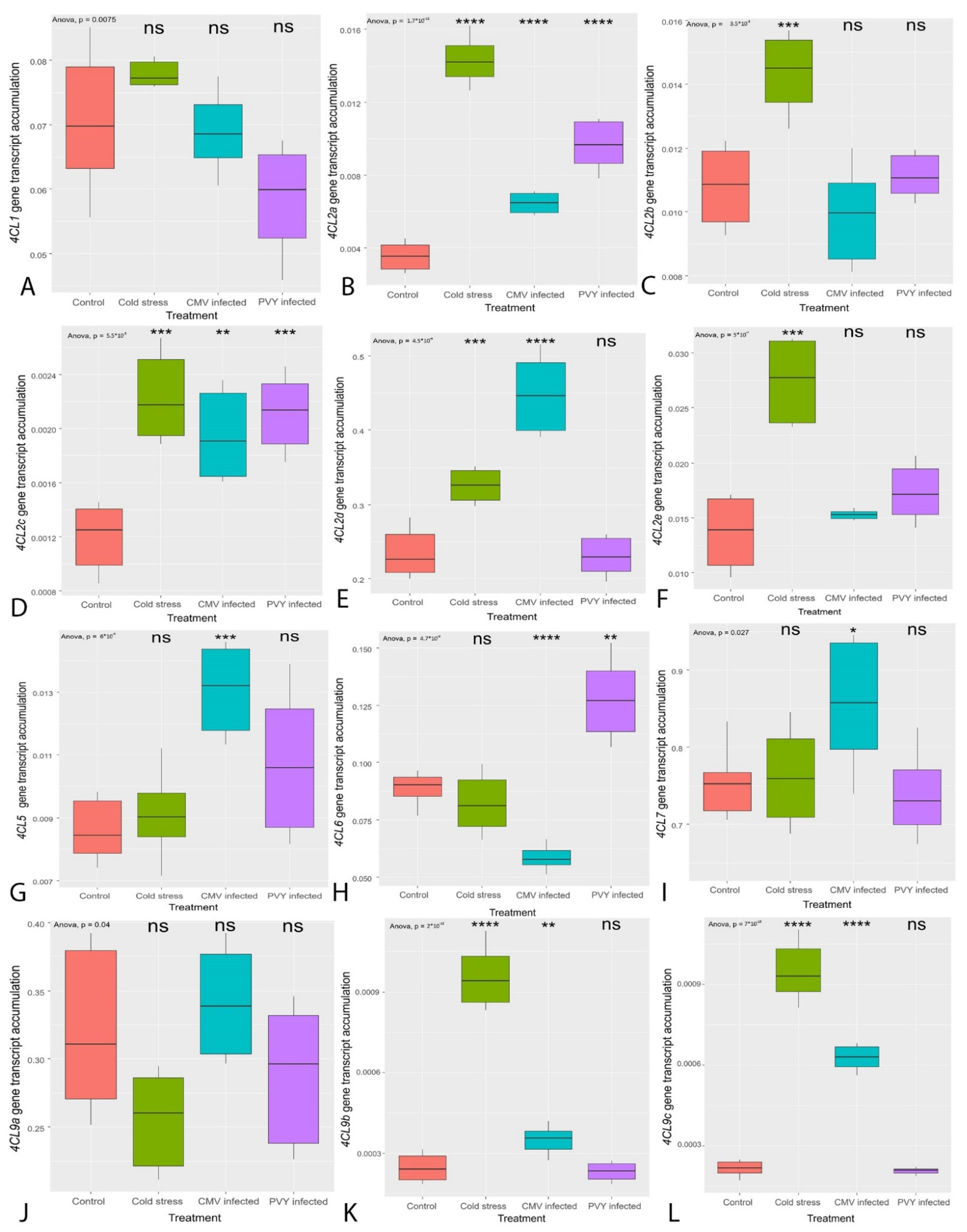
3.2. Transcription of the Genes Regulating the General PBP Pathway (PAL, C4H, 4CL)
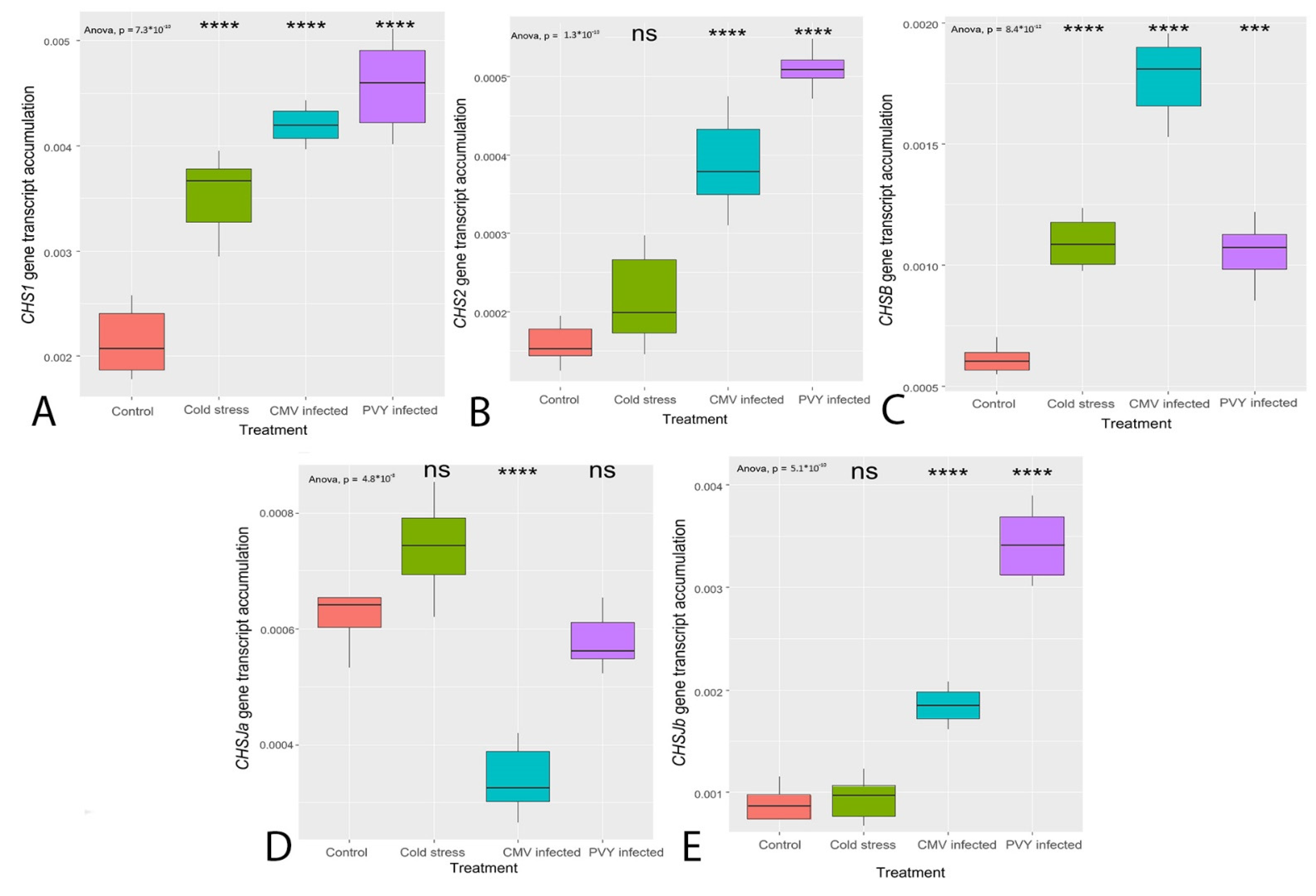
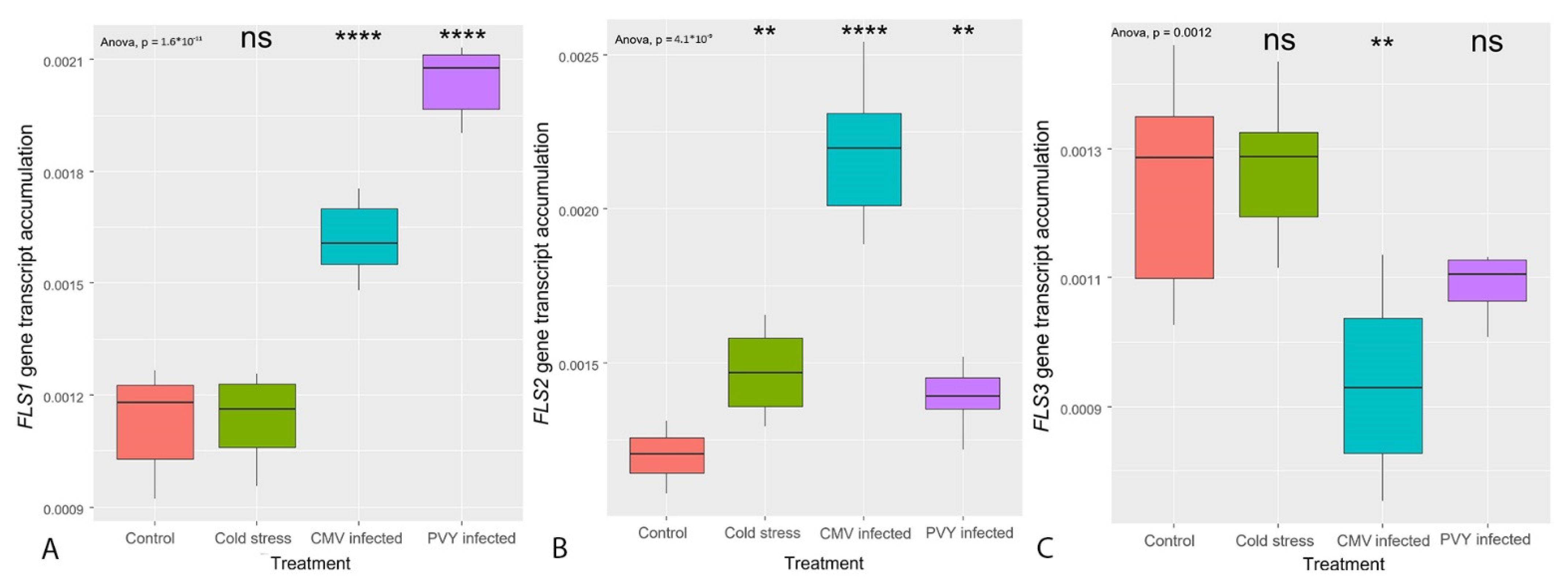
3.3. Transcription of the Genes Coding for the Enzymes Primarily Regulating Flavonoid Synthesis (CHS, FLS)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| CHS | chalcone synthase |
| CMV | cucumber mosaic virus |
| C4H | cinnamic acid 4-hydroxylase |
| FLS | flavonol synthase |
| GLRaV-3 | grapevine leafroll-associated virus 3 |
| GLRD | grapevine leafroll disease |
| PAL | phenylalanine ammonia-lyase |
| PBP | phenylpropanoid biosynthetic pathway |
| PVY | potato virus Y |
| ROS | reactive oxygen species |
| RT-PCR | reverse-transcription polymerase chain reaction |
| TF | total flavonoid |
| ToMV | tomato mosaic virus |
| TP | total phenolic |
| TSWV | tomato spotted wilt virus |
| TYLCV | tomato yellow leaf curl virus |
| 4CL | 4-coumarate-CoA ligase |
References
- Vogt, T. Phenylpropanoid biosynthesis. Mol. Plant 2010, 3, 2–20. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, W.J.; Kawaoka, A.; Tsai, C.J.; Lung, J.; Osakabe, K.; Ebinuma, H.; Chiang, V.L. Compartmentalized expression of two structurally and functionally distinct 4-coumarate:CoA ligase genes in aspen (Populus tremuloides). Proc. Nat. Acad. Sci. USA 1998, 95, 5407–5412. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betz, C.; McCollum, T.G.; Mayer, R.T. Differential expression of two cinnamate 4-hydroxylase genes in “Valencia” orange (Citrus sinensis Osbeck). Plant. Mol. Biol. 2001, 46, 741–748. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Yu, S.; Yu, J.; Zhan, Z.; Li, M.; Liu, G.; Wang, X.; Huang, L. Predicting the Function of 4-Coumarate:CoA Ligase (LJ4CL1) in Lonicera japonica. Int. J. Mol. Sci. 2014, 15, 2386–2399. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Kim, J.I.; Pysh, L.; Chapple, C. Four isoforms of Arabidopsis 4-coumarate:CoA ligase have overlapping yet distinct roles in phenylpropanoid metabolism. Plant Physiol. 2015, 169, 2409–2421. [Google Scholar] [PubMed] [Green Version]
- Lefevere, H.; Bauters, L.; Gheysen, G. Salicylic acid biosynthesis in plants. Front. Plant Sci. 2020, 11, 338. [Google Scholar] [CrossRef] [PubMed]
- Panche, A.N.; Diwan, A.D.; Chandra, S.R. Flavonoids: An overview. J. Nutr. Sci. 2016, 5, e47. [Google Scholar] [CrossRef] [Green Version]
- Kumar, S.; Pandey, A.K. Chemistry and biological activities of flavonoids: An overview. Sci. World J. 2013, 2013, 162750. [Google Scholar] [CrossRef] [Green Version]
- Tohge, T.; Watanabe, M.; Hoefgen, R.; Fernie, A.R. The evolution of phenylpropanoid metabolism in the green lineage. Crit. Rev. Biochem. Mol. Biol. 2013, 48, 123–152. [Google Scholar] [CrossRef]
- Zakaryan, H.; Arabyan, E.; Oo, A.; Zandi, K. Flavonoids: Promising natural compounds against viral infections. Arch. Virol. 2017, 162, 2539–2551. [Google Scholar] [CrossRef]
- Šamec, D.; Karalija, E.; Šola, I.; Vujcic Bok, V.; Salopek-Sondi, B. The role of polyphenols in abiotic stress response: The influence of molecular structure. Plants 2021, 10, 118. [Google Scholar] [CrossRef] [PubMed]
- Chatzistathis, T.; Fanourakis, D.; Aliniaeifard, S.; Kotsiras, A.; Delis, C.; Tsaniklidis, G. Leaf age-dependent effects of boron toxicity in two Cucumis melo varieties. Agronomy 2021, 11, 759. [Google Scholar] [CrossRef]
- Chen, Y.; Fanourakis, D.; Tsaniklidis, G.; Aliniaeifard, S.; Yang, Q.; Li, T. Low UVA intensity during cultivation improves the lettuce shelf-life, an effect that is not sustained at higher intensity. Postharvest Biol. Technol. 2021, 172, 111376. [Google Scholar] [CrossRef]
- Dixon, R.A.; Paiva, N.L. Stress-induced phenylpropanoid metabolism. Plant Cell 1995, 7, 1085–1097. [Google Scholar] [CrossRef]
- Król, A.; Amarowiczb, R.; Weidnera, S. The effects of cold stress on the phenolic compounds and antioxidant capacity of grapevine (Vitis vinifera L.) leaves. J. Plant Physiol. 2015, 189, 97–104. [Google Scholar] [CrossRef] [PubMed]
- Pellegrini, L.; Rohfritsch, O.; Fritig, B.; Legrand, M. Phenylalanine ammonia-lyase in tobacco: Molecular cloning and gene expression during the hypersensitive response to tobacco mosaic virus and the response to a fungal elicitor (fungi and wounding). Plant Physiol. 1994, 106, 877–886. [Google Scholar] [CrossRef] [Green Version]
- Rivero, R.M.; Ruiz, J.M.; Garcia, P.C.; López-Lefebre, L.R.; Sánchez, E.; Romero, L. Resistance to cold and heat stress: Accumulation of phenolic compounds in tomato and watermelon plants. Plant Sci. 2001, 160, 315–321. [Google Scholar] [CrossRef]
- Cushnie, T.P.T.; Lamb, A.J. Antimicrobial activity of flavonoids. Int. J. Antimicrob. Agents 2005, 26, 343–356. [Google Scholar] [CrossRef]
- Gutiérrez-Albanchez, E.; Gradillas, A.; García, A.; García-Villaraco, A.; Gutierrez-Mañero, F.J.; Ramos-Solano, B. Elicitation with Bacillus QV15 reveals a pivotal role of F3H on flavonoid metabolism improving adaptation to biotic stress in blackberry. PLoS ONE 2020, 15, e0232626. [Google Scholar] [CrossRef]
- Fini, A.; Brunetti, C.; Di Ferdinando, M.; Ferrini, F.; Tattini, M. Stress-induced flavonoid biosynthesis and the antioxidant machinery of plants. Plant Signal. Behav. 2011, 6, 709–711. [Google Scholar] [CrossRef]
- Tsaniklidis, G.; Pappi, P.; Tsafouros, A.; Charova, S.N.; Nikoloudakis, N.; Roussos, P.A.; Paschalidis, K.A.; Delis, C. Polyamine homeostasis in tomato biotic/abiotic stress cross-tolerance. Gene 2020, 727, 144230. [Google Scholar] [CrossRef] [PubMed]
- Mumford, A.; Barker, I.; Wood, K.R. The detection of tomato spotted wilt virus using the polymerase chain reaction. J. Virol. Met. 1994, 46, 303–311. [Google Scholar] [CrossRef]
- Faggioli, F.; Ferretti, L.; Albanese, G.; Sciarroni, R.; Pasquini, G.; Lumia, V.; Barba, M. Distribution of olive tree viruses in Italy as revealed by one-step RT-PCR. J. Plant Pathol. 2005, 87, 49–55. [Google Scholar]
- Dovas, C.I.; Efthimiou, K.; Katis, N.I. Generic detection and differentiation of tobamoviruses by a spot nested RTPCR-RFLP using dI-containing primers along with homologous dG-containing primers. J. Virol. Meth. 2004, 117, 137–144. [Google Scholar] [CrossRef] [PubMed]
- Nie, X. Reverse transcription loop-mediated isothermal amplification of DNA for detection of potato virus Y. Plant Disease 2005, 89, 605–610. [Google Scholar] [CrossRef] [Green Version]
- Singleton, V.L.; Orthofer, R.; La Muela-Raventos, R.M. Analysis of total phenols and other oxidation substrates and antioxidants by means of Folin-Cocalteu reagent. Methods Enzymol. 1999, 299, 152–178. [Google Scholar]
- Aryal, S.; Baniya, M.K.; Danekhu, K.; Kunwar, P.; Gurung, R.; Koirala, N. Total phenolic content, flavonoid content and antioxidantpotential of wild vegetables from Western Nepal. Plants 2019, 8, 96. [Google Scholar] [CrossRef] [Green Version]
- Assis, J.S.; Maldonado, R.; Munoz, T.; Escribano, M.I.; Merodio, C. Effect of high carbon dioxide concentration on PAL activity and phenolic contents in ripening cherimoya fruit. Postharvest Biol. Technol. 2001, 23, 33–39. [Google Scholar] [CrossRef] [Green Version]
- Maliogka, V.I.; Olmos, A.; Pappi, P.G.; Lotos, L.; Efthimiou, K.; Grammatikaki, G.; Thierry Candresse, T.; Katis, N.I.; Avgelis, A.D. A novel grapevine badnavirus is associated with the Roditis leaf discoloration disease. Virus Res. 2015, 203, 47–55. [Google Scholar] [CrossRef]
- Dereeper, A.; Guignon, V.; Blanc, G.; Audic, S.; Buffet, S.; Chevenet, F.; Dufayard, J.-F.; Guindon, S.; Lefort, V.; Lescot, M.; et al. Phylogeny.fr: Robust phylogenetic analysis for the non-specialist. Nucleic Acids Res. 2008, 36, W465–W469. [Google Scholar] [CrossRef]
- Atkinson, N.J.; Urwin, P.E. The interaction of plant biotic and abiotic stresses: From genes to the field. J. Exp. Bot. 2012, 63, 3523–3543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsaniklidis, G.; Chatzistathis, T.; Fanourakis, D.; Nikoloudakis, N.; Kotsiras, A.; Delis, C.; Tzortzakakis, E.A. Leaf antioxidant machinery stimulation by Meloidogyne javanica infestation: A case study on Cucumis melo seedlings. Plant Stress 2021, 1, 100002. [Google Scholar] [CrossRef]
- Sharma, A.; Shahzad, B.; Rehman, A.; Bhardwaj, R.; Landi, M.; Zheng, B. Response of phenylpropanoid pathway and the role of polyphenols in plants under abiotic stress. Molecules 2019, 24, 2452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agati, G.; Brunetti, C.; Fini, A.; Gori, A.; Guidi, L.; Landi, M.; Sebastiani, F.; Tattini, M. Are flavonoids effective antioxidants in plants? Twenty years of our investigation. Antioxidants 2020, 9, 1098. [Google Scholar] [CrossRef] [PubMed]
- Agati, G.; Azzarello, E.; Pollastri, S.; Tattini, M. Flavonoids as antioxidants in plants: Location and functional significance. Plant Sci. 2012, 196, 67–76. [Google Scholar] [CrossRef] [PubMed]
- Harborne, J.B.; Williams, C.A. Advances in flavonoid research since 1992. Phytochemistry 2000, 55, 481–504. [Google Scholar] [CrossRef]
- Selway, J.W.T. Antiviral activity of flavones and flavans. In Plant Flavonoids in Biology and Medicine: Biochemical, Pharmacological, and Structure–Activity Relationships, Proceedings of the a Symposium Held in Buffalo, New York, NY, USA, 22–26 July 1985; Cody, V., Middleton, E., Harborne, J.B., Eds.; Alan R. Liss Inc.: New York, NY, USA, 1986; pp. 521–536. [Google Scholar]
- Rezaie, R.; Abdollahi Mandoulakani, B.; Fattahi, M. Cold stress changes antioxidant defense system, phenylpropanoid contents and expression of genes involved in their biosynthesis in Ocimum basilicum L. Sci. Rep. 2020, 10, 5290. [Google Scholar] [CrossRef]
- Christie, P.J.; Alfenito, M.R.; Walbot, V. Impact of low-temperature stress on general phenylpropanoid and anthocyanin pathways: Enhancement of transcript abundance and anthocyanin pigmentation in maize seedlings. Planta 1994, 194, 541–549. [Google Scholar] [CrossRef]
- Schneider, K.; Hövel, K.; Witzel, K.; Hamberger, B.; Schomburg, D.; Kombrink, E.; Stuible, H.P. The substrate specificity-determining amino acid code of 4-coumarate:CoA ligase. Proc. Natl. Acad. Sci. USA 2003, 100, 8601–8606. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Han, Y.; Li, D.; Lin, Y.; Cai, Y. Systematic analysis of the 4-Coumarate:Coenzyme A Ligase (4CL) related genes and expression profiling during fruit development in the Chinese Pear. Genes 2016, 7, 89. [Google Scholar] [CrossRef] [Green Version]
- Lavhale, S.G.; Kalunke, R.M.; Giri, A.P. Structural, functional and evolutionary diversity of 4-coumarate-CoA ligase in plants. Planta 2018, 248, 1063–1078. [Google Scholar] [CrossRef] [PubMed]
- Duan, X.; Chen, X.; Wang, S.; Zhang, X. Transcriptome sequencing and analysis of chilling tolerance mutant tomato under low temperature. Russ. J. Plant Physiol. 2019, 66, 110–118. [Google Scholar] [CrossRef]
- Tao, X.; Wu, Q.; Aalim, H.; Li, L.; Mao, L.; Luo, Z.; Ying, T. Effects of exogenous abscisic acid on bioactive components and antioxidant capacity of postharvest tomato during ripening. Molecules 2020, 25, 1346. [Google Scholar] [CrossRef] [Green Version]
- Schulz, E.; Tohge, T.; Zuther, E.; Fernie, A.R.; Hincha, D.K. Flavonoids are determinants of freezing tolerance and cold acclimation in Arabidopsis Thaliana. Sci. Rep. 2016, 6, 34027. [Google Scholar] [CrossRef]
- Kogovšek, P.; Pompe-Novak, M.; Petek, M.; Fragner, L.; Weckwerth, W.; Gruden, K. Primary metabolism, phenylpropanoids and antioxidant pathways are regulated in potato as a response to Potato virus Y infection. PLoS ONE 2016, 11, e0146135. [Google Scholar] [CrossRef]
- Lou, L.N.; Su, X.J.; Liu, X.H.; Liu, Z. Transcriptome analysis of Luffa cylindrica (L.) Roem response to infection with Cucumber mosaic virus (CMV). Gene 2020, 737, 144451. [Google Scholar] [CrossRef] [PubMed]
- Aseel, D.G.; Rashad, Y.; Hammad, S.M. Arbuscular mycorrhizal fungi trigger transcriptional expression of flavonoid and chlorogenic acid biosynthetic pathways genes in tomato against Tomato Mosaic Virus. Sci. Rep. 2019, 9, 9692. [Google Scholar] [CrossRef] [Green Version]
- Gutha, L.R.; Casassa, L.F.; Harbertson, J.F.; Naidu, R.A. Modulation of flavonoid biosynthetic pathway genes and anthocyanins due to virus infection in grapevine (Vitis vinifera L.) leaves. BMC Plant Biol. 2010, 10, 187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sade, D.; Shriki, O.; Cuadros-Inostroza, A.; Tohge, T.; Semel, Y.; Haviv, Y.; Willmitzer, L.; Fernie, A.R.; Czosnek, H.; Brotman, Y. Comparative metabolomics and transcriptomics of plant response to Tomato yellow leaf curl virus infection in resistant and susceptible tomato cultivars. Metabolomics 2015, 11, 81–97. [Google Scholar] [CrossRef]
- Fernández-Calvino, L.; Osorio, S.; Hernández, M.L.; Hamada, I.B.; Del Toro, F.J.; Donaire, L.; Yu, A.; Bustos, R.; Fernie, A.R.; Martínez-Rivas, J.M. Virus-induced alterations in primary metabolism modulate susceptibility to Tobacco rattle virus in Arabidopsis. Plant Physiol. 2014, 166, 1821–1838. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niinemets, U. Mild versus severe stress and BVOCs: Thresholds, priming and consequences. Trends Plant Sci. 2010, 15, 145–153. [Google Scholar] [CrossRef] [PubMed]

Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pappi, P.; Nikoloudakis, N.; Fanourakis, D.; Zambounis, A.; Delis, C.; Tsaniklidis, G. Differential Triggering of the Phenylpropanoid Biosynthetic Pathway Key Genes Transcription upon Cold Stress and Viral Infection in Tomato Leaves. Horticulturae 2021, 7, 448. https://doi.org/10.3390/horticulturae7110448
Pappi P, Nikoloudakis N, Fanourakis D, Zambounis A, Delis C, Tsaniklidis G. Differential Triggering of the Phenylpropanoid Biosynthetic Pathway Key Genes Transcription upon Cold Stress and Viral Infection in Tomato Leaves. Horticulturae. 2021; 7(11):448. https://doi.org/10.3390/horticulturae7110448
Chicago/Turabian StylePappi, Polyxeni, Nikolaos Nikoloudakis, Dimitrios Fanourakis, Antonios Zambounis, Costas Delis, and Georgios Tsaniklidis. 2021. "Differential Triggering of the Phenylpropanoid Biosynthetic Pathway Key Genes Transcription upon Cold Stress and Viral Infection in Tomato Leaves" Horticulturae 7, no. 11: 448. https://doi.org/10.3390/horticulturae7110448
APA StylePappi, P., Nikoloudakis, N., Fanourakis, D., Zambounis, A., Delis, C., & Tsaniklidis, G. (2021). Differential Triggering of the Phenylpropanoid Biosynthetic Pathway Key Genes Transcription upon Cold Stress and Viral Infection in Tomato Leaves. Horticulturae, 7(11), 448. https://doi.org/10.3390/horticulturae7110448









