Occurrence of Grapevine Leafroll-Associated Virus-3 (GLRaV-3), Complete Nucleotide Sequence and Cultivar Susceptibility to a GLRaV-3 Isolate from Shaanxi Province of China
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Virus Detection by RT-PCR
2.3. Sequencing of Full Genome of GLRaV-3-Sau
2.4. Micrografting and Virus Localization
2.5. Analyses of Relative Levels of GLRaV-3 mRNA
2.6. Experiment Design and Data Analysis
3. Results
3.1. Occurrence of GLRaV-3
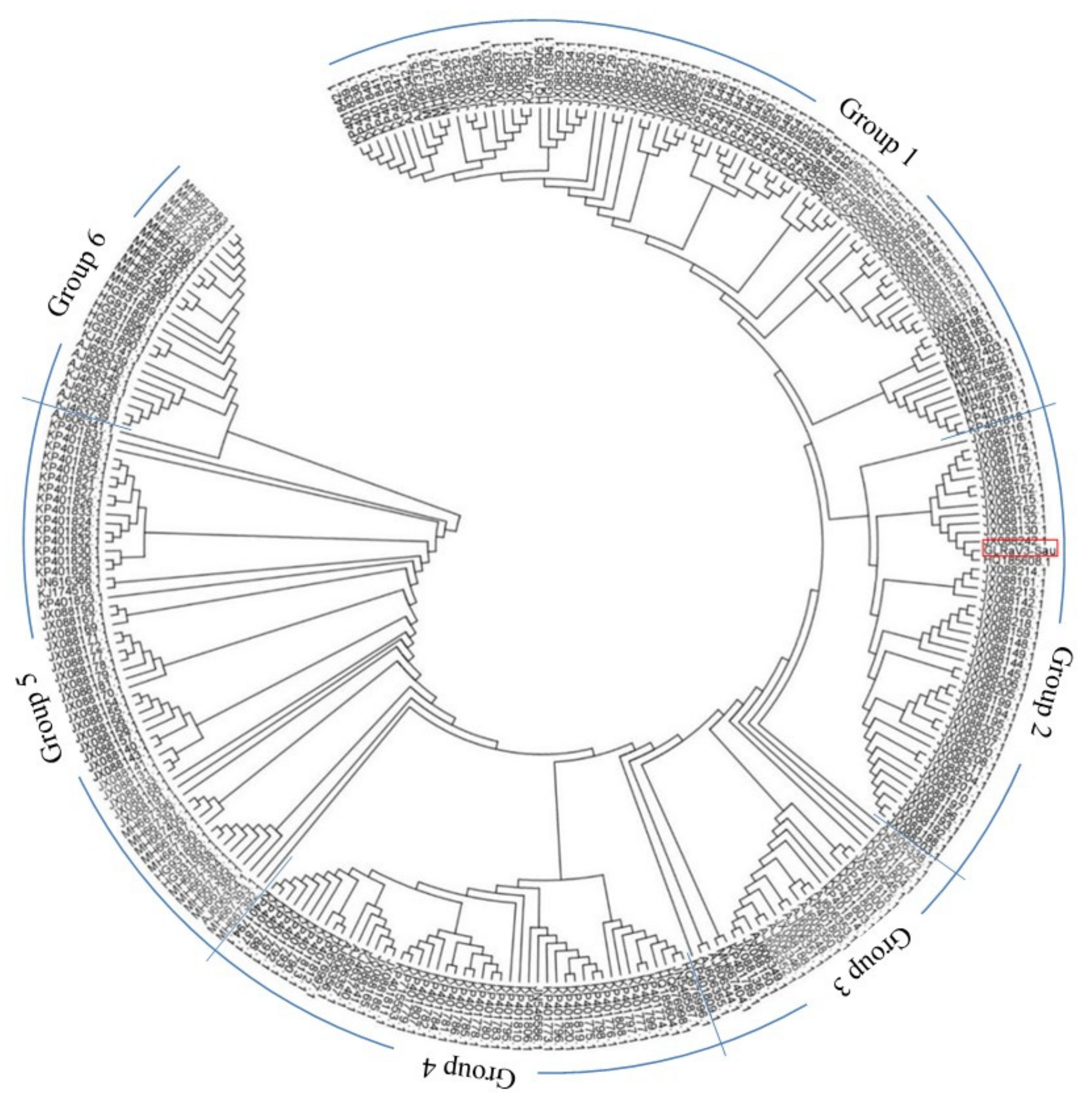
3.2. Full Genome of GLRaV-3
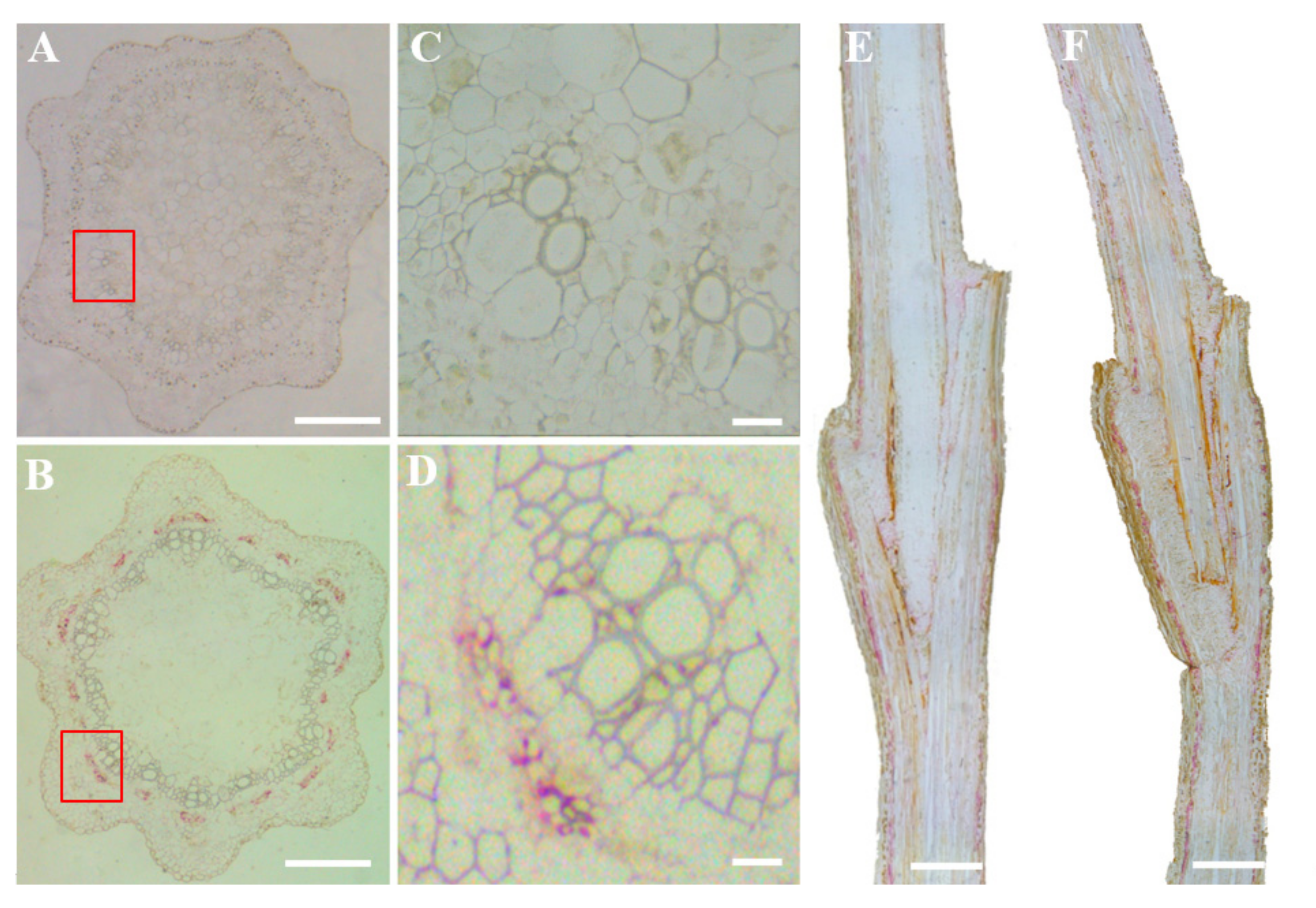
3.3. Distribution Pattern of GLRaV-3 in Micrografts
3.4. Analysis of Susceptibility to GLRaV-3
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Constable, F.E.; Connellan, J.; Nicholas, P.; Rodoni, B.C. The reliability of woody indexing for detection of grapevine virus associated diseases in three different climatic conditions in Australia. Aust. J. Grape Wine Res. 2013, 19, 74–80. [Google Scholar] [CrossRef]
- Maree, H.J.; Almeida, R.P.P.; Bester, R.; Chooi, K.M.; Cohen, D.; Dolja, V.V.; Fuchs, M.F.; Golino, D.A.; Jooste, A.E.C.; Martelli, G.P.; et al. Grapevine leafroll-associated virus 3. Front. Microbiol. 2013, 4, 82. [Google Scholar] [CrossRef]
- Naidu, R.A.; Maree, H.J.; Burger, J.T. Grapevine leafroll disease and associated viruses: A unique pathosystem. Annu. Rev. Phytopathol. 2015, 53, 613–634. [Google Scholar] [CrossRef] [PubMed]
- Cabaleiro, C.; Segura, A. Effect of grapevine leafroll-associated virus-3 in a commercial cv. “Albarino” vineyard. Investig. Agrar. Prod. Prot. Veg. 1996, 11, 451–463. [Google Scholar] [CrossRef]
- Cabaleiro, C.; Piniro, A.; Segura, A. Photosynthesis in grapevines infected with grapevine leafroll virus (GLRaV-3). In Proceedings of the 12th Meeting of the International Council for the Study of Viruses and Virus-like Diseases of the Grapevine (ICVG), Lisbon, Portugal, 28 September–2 October 1997; de Sequera, O.A., de Sequera, J.S., Santos, M.T., Eds.; pp. 153–154. [Google Scholar]
- Mannini, F.; Mollo, A.; Credi, R. Field performance and wine quality modification in a clone of Vitis vinifera cv. “Dolcetto” after GLRaV-3 elimination. Am. J. Enol. Vitic. 2012, 63, 144–147. [Google Scholar] [CrossRef]
- Goheen, A.C.; Pearson, R.C.; Goheen, A.C. Compendium of Grape Diseases; APS Press: St Paul, MN, USA, 1988; p. 52. [Google Scholar]
- Brar, H.S.; Singh, Z.; Swinny, E.; Cameron, I. Girdling and grapevine leafroll-associated viruses affect berry weight, colour development and accumulation of anthocyanins in ‘Crimson Seedless’ grapes during maturation and ripening. Plant Sci. 2008, 175, 885–897. [Google Scholar] [CrossRef]
- Akbas, B.; Kunter, B.; Ilhan, D. Influence of leafroll on local grapevine cultivars in agroecological conditions of Central Anatolia region. HortScience 2009, 36, 97–104. [Google Scholar] [CrossRef]
- Martelli, G.P.; Walter, B.; Hadid, A.; Khetarpal, R.K.; Koganezawa, H. Plant Virus Disease Control; APS Press: St Paul, MN, USA, 1998; pp. 150–166. [Google Scholar]
- Bi, W.L.; Hao, X.Y.; Cui, Z.H.; Pathirana, R.; Volk, G.M.; Wang, Q.C. Shoot tip cryotherapy for efficient eradication of grapevine leafroll-associated virus-3 from diseased grapevine in vitro plants. Ann. Appl. Biol. 2018, 3, 261–270. [Google Scholar] [CrossRef]
- Hao, X.Y.; Bi, W.L.; Cui, Z.H.; Pan, C.; Xu, Y.; Wang, Q.C. Development, histological observations and Grapevine leafroll associated virus-3 localisation in in vitro grapevine micrografts. Ann. Appl. Biol. 2017, 170, 379–390. [Google Scholar] [CrossRef]
- Zee, F.; Gonsalves, D.; Goheen, A.; Kim, K.S.; Pool, R.; Lee, R.F. Cytopathology of leafroll-diseased grapevines and the purification and serology of associated closterovirus-like particles. Phytopathology 1987, 77, 1427–1434. [Google Scholar] [CrossRef]
- Bertin, S.; Cavalieri, V.; Gribaudo, I.; Sacco, D.; Marzachi, C.; Bosco, D. Transmission of Grapevine virus A and Grapevine leafroll-associated virus 1 and 3 by Heliococcus bohemicus (Hemiptera: Pseudococcidae) Nymphs From Plants With Mixed Infections. J. Econ. Entomol. 2016, 4, 1504–1511. [Google Scholar] [CrossRef] [PubMed]
- Ling, K.S.; Ling, H.Y.; Zhu, D. Gonsalves Complete nucleotide sequence and genome organization of Grapevine leafroll-associated virus 3, type member of the genus Ampelovirus. J. Gen. Virol. 2004, 85, 2099–2102. [Google Scholar] [CrossRef] [PubMed]
- Engel, E.A.; Girardi, C.; Escobar, P.F.; Arredondo, V.; Dominguez, C.; Perez-Acle, T.; Valenzuela, P.D.T. Genome analysis and detection of a Chilean isolate of Grapevine leafroll associated virus-3. Virus Genes 2008, 37, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Maree, H.J.; Freeborough, M.J.; Burger, J.T. Complete nucleotide sequence of a South African isolate of grapevine leafroll-associated virus 3 reveals a 5′ UTR of 737 nucleotides. Arch. Virol. 2008, 4, 755–757. [Google Scholar] [CrossRef]
- Jarugula, S.; Gowda, S.; William, O.; Naidu, R.A. 3′-coterminal subgenomic RNAs and putative cisacting elements of Grapevine leafroll-associated virus 3 reveals ‘unique’ features of gene expression strategy in the genus Ampelovirus. J. Virol. 2010, 7, 180. [Google Scholar] [CrossRef]
- Jooste, A.E.C.; Maree, H.J.; Bellstedt, D.U.; Goszczynski, D.E.; Pietersen, G.; Burger, J.T. Three genetic grapevine leafroll-associated virus 3 variants identified from South African vineyards show high variability in their 5′ UTR. Arch. Virol. 2010, 155, 1997–2006. [Google Scholar] [CrossRef]
- Jooste, A.E.C.; Pietersen, G.; Burger, J.T. Distribution of grapevine leafroll associated virus-3 variants in South African vineyards. Eur. J. Plant Pathol. 2011, 3, 371–381. [Google Scholar] [CrossRef]
- Bester, R.; Maree, H.J.; Burger, J.T. Complete nucleotide sequence of a new strain of grapevine leafroll-associated virus 3 in South Africa. Arch. Virol. 2012, 9, 1815–1819. [Google Scholar] [CrossRef]
- Chooi, K.M.; Cohen, D.; Pearson, M.N. Generic and sequence-variant specific molecular assays for the detection of the highly variable Grapevine leafroll-associated virus 3. J. Virol. Methods 2013, 1, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Farooq, A.B.U.; Ma, Y.X.; Wang, Z.Q.; Zhuo, N.; Xu, W.X.; Wang, G.P.; Hong, N. Genetic diversity analyses reveal novel recombination events in Grapevine leafroll-associated virus 3 in China. Virus Res. 2013, 1, 15–21. [Google Scholar] [CrossRef]
- Fei, F.; Lyu, M.D.; Li, J.; Fan, Z.F.; Cheng, Y.Q. Complete nucleotide sequence of a Chinese isolate of Grapevine leafroll-associated virus 3 reveals a 5′ UTR of 802 nucleotides. Virus Genes 2013, 1, 182–185. [Google Scholar] [CrossRef]
- OIV. 2019 Report on the World Vitivinicultural Situation [R/OL]. Available online: http://www.oiv.int/en/oiv-life/oiv-2019-report-on-the-world-vitivinicultural-situation (accessed on 31 October 2019).
- Liu, M.H.; Li, M.J.; Qi, H.H.; Guo, R.; Liu, X.M.; Wang, Q.; Cheng, Y.Q. Occurrence of grapevine leafroll-associated viruses in China. Plant Dis. 2013, 97, 1339–1345. [Google Scholar] [CrossRef][Green Version]
- Cui, Z.H.; Bi, W.L.; Liu, J.; Pan, C.; Wang, Q.C. Abiotic stress improves in vitro biological indexing of Grapevine leafroll-associated virus-3 in red grapevine cultivars. Aust. J. Grape Wine Res. 2015, 21, 490–495. [Google Scholar] [CrossRef]
- Turturo, C.; Saldarelli, P.; Dong, Y.; Digiaro, M.; Minafra, A.; Savino, V.; Martelli, G.P. Genetic variability and population structure of Grapevine leafroll-associated virus 3 isolates. J. Gen. Virol. 2005, 86, 217–224. [Google Scholar] [CrossRef]
- Zhao, L.; Hao, X.G.; Liu, P.; Wu, Y.F. Complete sequence of an Apple stem grooving virus (ASGV) isolate from China. Virus Genes 2012, 45, 596–599. [Google Scholar] [CrossRef]
- Cui, Z.H.; Bi, W.L.; Hao, X.Y.; Li, P.M.; Duan, Y.; Walker, M.A.; Xu, Y.; Wang, Q.C. Drought stress enhances up-regulation of anthocyanin biosynthesis in Grapevine leafroll-associated virus 3 infected in vitro grapevine (Vitis vinifera) leaves. Plant Dis. 2017, 101, 1606–1615. [Google Scholar] [CrossRef]
- Bester, R.; Pepler, P.T.; Burger, J.T.; Maree, H.J. Relative quantitation goes viral: An RT-qPCR assay for a grapevine virus. J. Virol. Methods. 2014, 210, 67–75. [Google Scholar] [CrossRef] [PubMed]
- Osman, F.; Rowhani, A. Application of a spotting sample preparation technique for the detection of pathogens in woody plants by RT-PCR and real-time PCR (TaqMan). J. Virol. Methods 2006, 133, 130–136. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2 (-Delta Delta C (T)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Koonin, E.V.; Dolja, V.V.; Morris, T.J. Evolution and taxonomy of positive-strand RNA viruses: Implications of comparative analysis of aminoacid sequences. Crit. Rev. Biochem. Mol. Biol. 1993, 28, 375–430. [Google Scholar] [CrossRef] [PubMed]
- Dolja, V.V.; Kreuze, J.F.; Valkonen, J.P.T. Comparative and functional genomics of closteroviruses. Virus Res. 2006, 117, 38–51. [Google Scholar] [CrossRef]
- Alzhanova, D.V.; Prokhnevsky, A.; Peremyslov, V.V.; Dolja, V.V. Virion tails of Beet yellows virus: Coordinated assembly by three structural proteins. Virology 2007, 359, 220–226. [Google Scholar] [CrossRef]
- Peng, C.W.; Dolja, V.V. Leader proteinase of the beet yellows closterovirus: Mutation analysis of the function in genome amplification. J. Virol. 2000, 74, 9766–9770. [Google Scholar] [CrossRef][Green Version]
- Peng, C.W.; Napuli, A.J.; Dolja, V.V. Leader proteinase of the beet yellows virus functions in long-distance transport. J. Virol. 2003, 77, 2843–2849. [Google Scholar] [CrossRef]
- Vanden, B.E.; Omelchenko, M.V.; Bekkelund, A.; Leihne, V.; Koonin, E.V.; Dolja, V.V. Viral AlkB proteins repair RNA damage by oxidative demethylation. Nucleic Acids Res. 2008, 36, 5451–5461. [Google Scholar] [CrossRef]
- Liu, Y.P.; Peremyslov, V.V.; Medina, V.; Dolja, V.V. Tandem leader proteases of Grapevine leafroll-associated virus-2: Host-specific functions in the infection cycle. Virology 2009, 383, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Alzhanova, D.V.; Hagiwara, Y.; Peremyslov, V.V.; Dolja, V.V. Genetic analysis of the cell-to-cell movement of beet yellows closterovirus. Virology 2000, 268, 192–200. [Google Scholar] [CrossRef][Green Version]
- Saenz, P.; Cervera, M.T.; Dallot, S.; Quiot, L.; Quiot, J.B.; Riechmann, J.L.; Garcıa, J.A. Identification of a pathogenicity determinant of Plum pox virus in the sequence encoding the C-terminal region of protein P3+6K1. J. Gen. Virol. 2000, 81, 557–566. [Google Scholar] [CrossRef] [PubMed]
- Hjulsager, C.K.; Olsen, B.S.; Jensen, D.M.K.; Cordea, M.I.; Krath, B.N.; Johansen, I.E.; Lund, O.S. Multiple determinants in the coding region of Pea seed-borne mosaic virus P3 are involved in virulence against sbm-2 resistance. Virology 2006, 355, 52–61. [Google Scholar] [CrossRef]
- Rajamäki, M.L.; Valkonen, J.P.T. Localization of a potyvirus and the viral genome-linked protein in upper non-inoculated leaves at an early stage of systemic infection. Mol. Plant Microbe Interact. 2003, 16, 25–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.C.; Valkonen, J.P.T. Elimination of two viruses which interact synergistically from sweetpotato by shoot tip culture and cryotherapy. J. Virol. Methods. 2008, 154, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.R.; Li, B.Q.; Feng, C.H.; Wang, Q.C. Culture of shoot tips from adventitious shoots can eradicate Apple stem pitting virus but fails in Apple stem grooving virus. Plant Cell Tissue Organ Cult. 2016, 125, 283–291. [Google Scholar] [CrossRef]
- Martelli, G.P. Major graft-transmissible diseases of grapevines: Nature diagnosis, and sanitation. In Proceedings of the ASEV 5th Anniversary Annual Meeting, Seattle, WA, USA, 19–23 June 2000. [Google Scholar]
- Walker, M.A.; Meredith, C.P. The genetics of resistance to grapevine fanleaf virus in Vitis vinifera. Vitis 1990, 29, 228–238. [Google Scholar]
- Cui, Z.H.; Aguero, C.B.; Wang, Q.C.; Walker, M.A. Validation of micrografting to identify incompatible interactions of rootstocks with virus-infected scions of Cabernet Franc. Aust. J. Grape Wine Res. 2019, 25, 268–275. [Google Scholar] [CrossRef]
- Rast, H.E.; James, D.; Habili, N.; Masri, S.A. Genome organization and characterization of a novel variant of Grapevine leafroll-associated virus 3. In Proceedings of the 17th Congress of the International Council for the Study of Virus and Virus-like Diseases of the Grapevine (ICVG), Davis, CA, USA, 8–11 October 2012; pp. 61–63. [Google Scholar]
- Diaz-Lara, A.; Klaassen, V.; Stevens, K.; Sudarshana, M.R.; Rowhani, A.; Maree, H.J.; Chooi, K.M.; Blouin, A.G.; Habili, N.; Song, Y.; et al. Characterization of grapevine leafroll-associated virus 3 genetic variants and application towards RT-qPCR assay design. PLoS ONE. 2018, 13, e0208862. [Google Scholar] [CrossRef] [PubMed]
- Voncina, D.; Almeida, R.P.P. Screening of some Croatian autochthonous grapevine varieties reveals a multitude of viruses, including novel ones. Arch Virol. 2018, 163, 2239–2243. [Google Scholar] [CrossRef] [PubMed]
- Seah, Y.M.; Sharma, A.M.; Zhang, S.; Almeida, R.P.; Duffy, S. A divergent variant of Grapevine leafroll-associated virus 3 is present in California. Virol. J. 2012, 9, 235. [Google Scholar] [CrossRef] [PubMed]
- Prator, C.A.; Kashiwagi, C.M.; Voncina, D.; Almeida, R.P.P. Infection and Colonization of Nicotiana benthamiana by Grapevine leafroll-associated virus 3. Virology 2017, 510, 60–66. [Google Scholar] [CrossRef] [PubMed]




| Type and Cultivars of Grapevine | Grapevine-Growing Region | Mean | ||
|---|---|---|---|---|
| Wine Grape | Xianyang | Xi’an | Weinan | |
| ‘Cabernet Sauvignon’ | 90.0 | 86.7 | 100.0 | 92.2 |
| ‘Chardonay’ | 86.7 | 83.3 | 93.3 | 87.8 |
| ‘Riesling’ | 63.3 | 56.7 | 90.0 | 70.0 |
| Table grape | ||||
| ‘Hutai’ | 80.0 | 73.3 | 66.7 | 73.3 |
| ‘Red Globe’ | 76.7 | 60.0 | 80.0 | 72.2 |
| ‘Shine Muscat’ | 83.3 | 83.3 | 93.3 | 86.6 |
| ‘Summer Black’ | 70.0 | 66.7 | 86.7 | 74.5 |
| Mean | 78.6 | 72.9 | 87.1 | |
| Graft Combinations | Weeks after Micrografting | |||
|---|---|---|---|---|
| 2 | 3 | 4 | 5 | |
| C(+V)/Ch(−V) | 70 | 100 | 100 | 100 |
| C(+V)/TS(−V) | 60 | 100 | 100 | 100 |
| C(+V)/HN(−V) | 20 | 40 | 60 | 100 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hao, X.; Jiao, B.; Wang, Y.; Shang, B.; Xu, Y. Occurrence of Grapevine Leafroll-Associated Virus-3 (GLRaV-3), Complete Nucleotide Sequence and Cultivar Susceptibility to a GLRaV-3 Isolate from Shaanxi Province of China. Horticulturae 2022, 8, 73. https://doi.org/10.3390/horticulturae8010073
Hao X, Jiao B, Wang Y, Shang B, Xu Y. Occurrence of Grapevine Leafroll-Associated Virus-3 (GLRaV-3), Complete Nucleotide Sequence and Cultivar Susceptibility to a GLRaV-3 Isolate from Shaanxi Province of China. Horticulturae. 2022; 8(1):73. https://doi.org/10.3390/horticulturae8010073
Chicago/Turabian StyleHao, Xinyi, Bolei Jiao, Yunlei Wang, Boxing Shang, and Yan Xu. 2022. "Occurrence of Grapevine Leafroll-Associated Virus-3 (GLRaV-3), Complete Nucleotide Sequence and Cultivar Susceptibility to a GLRaV-3 Isolate from Shaanxi Province of China" Horticulturae 8, no. 1: 73. https://doi.org/10.3390/horticulturae8010073
APA StyleHao, X., Jiao, B., Wang, Y., Shang, B., & Xu, Y. (2022). Occurrence of Grapevine Leafroll-Associated Virus-3 (GLRaV-3), Complete Nucleotide Sequence and Cultivar Susceptibility to a GLRaV-3 Isolate from Shaanxi Province of China. Horticulturae, 8(1), 73. https://doi.org/10.3390/horticulturae8010073






