Assessment of Genetic Diversity by Morphological, Biochemical, and Molecular Markers in Gloriosa superba Ecotypes Collected from Different Agro-Climatic Zones in India
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material
2.2. Morphological Data
2.3. Estimation of Colchicine Content
2.4. DNA Isolation
2.5. SSR Analysis
2.6. Statistical Analysis of Phenotypic and Genotypic Data
3. Results
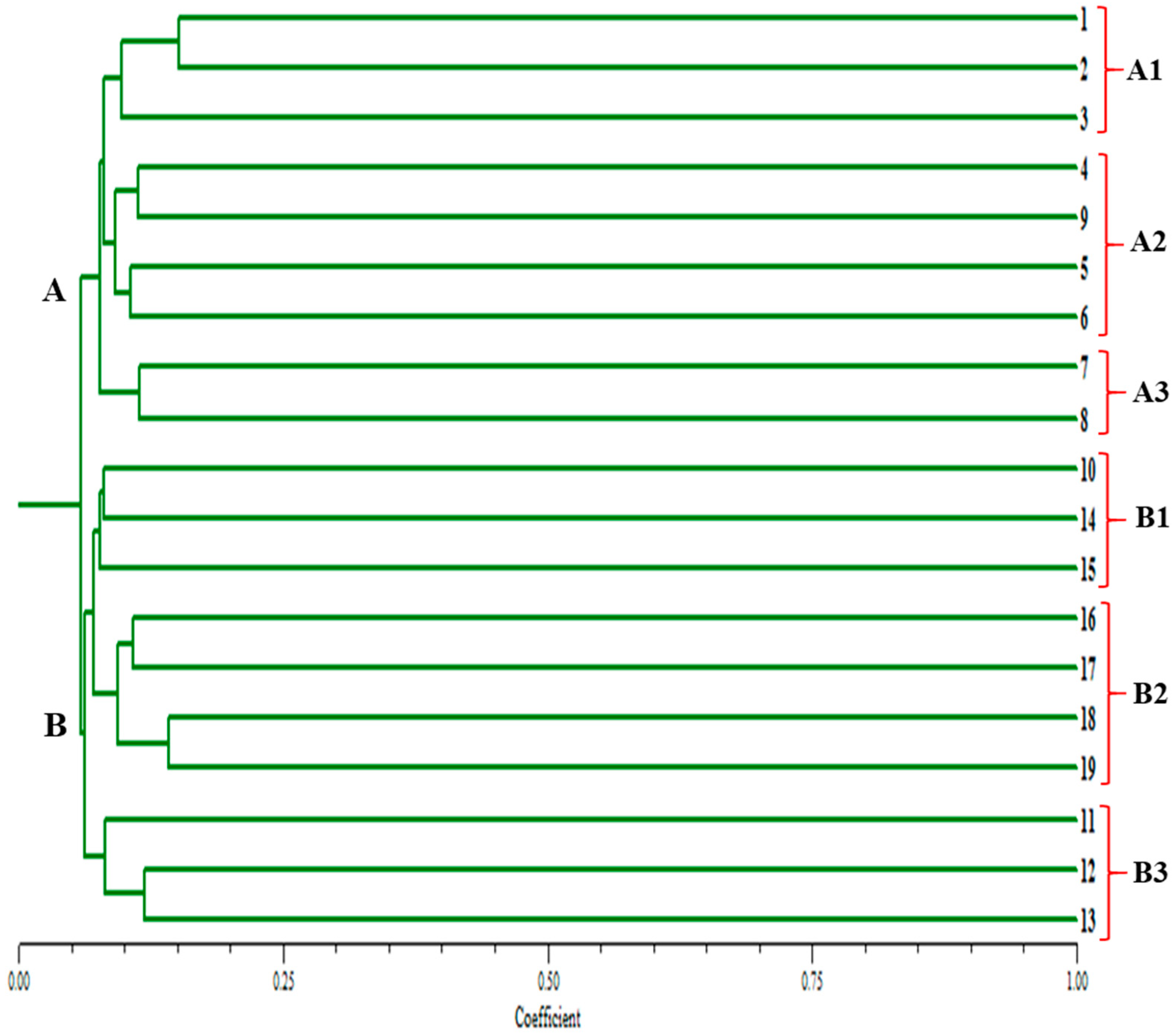
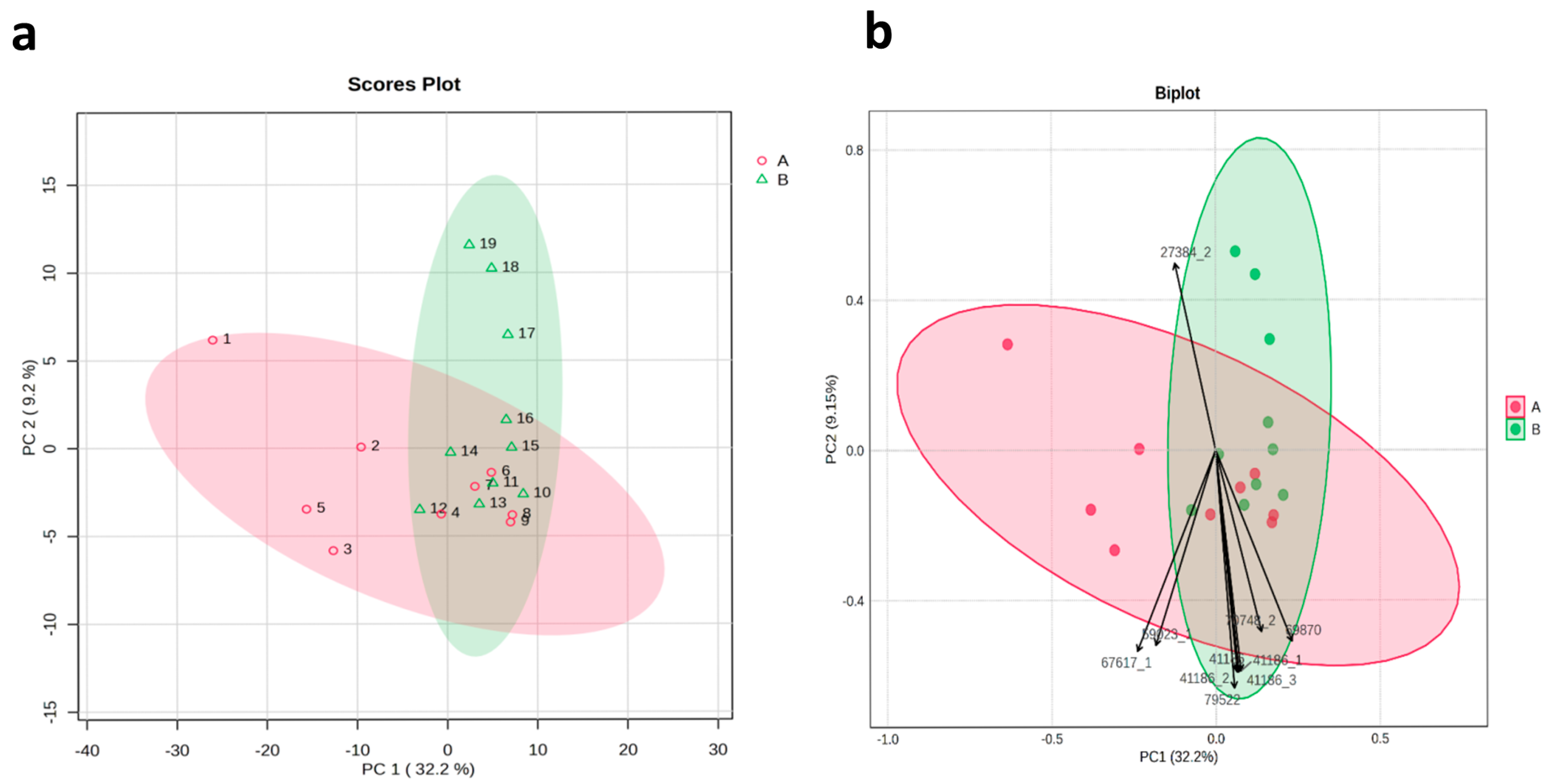
3.1. Morphological and Biochemical Study
3.1.1. Hierarchical Clustering Analysis
3.1.2. Principal Component Analysis (PCA) and Phenotypic Correlations
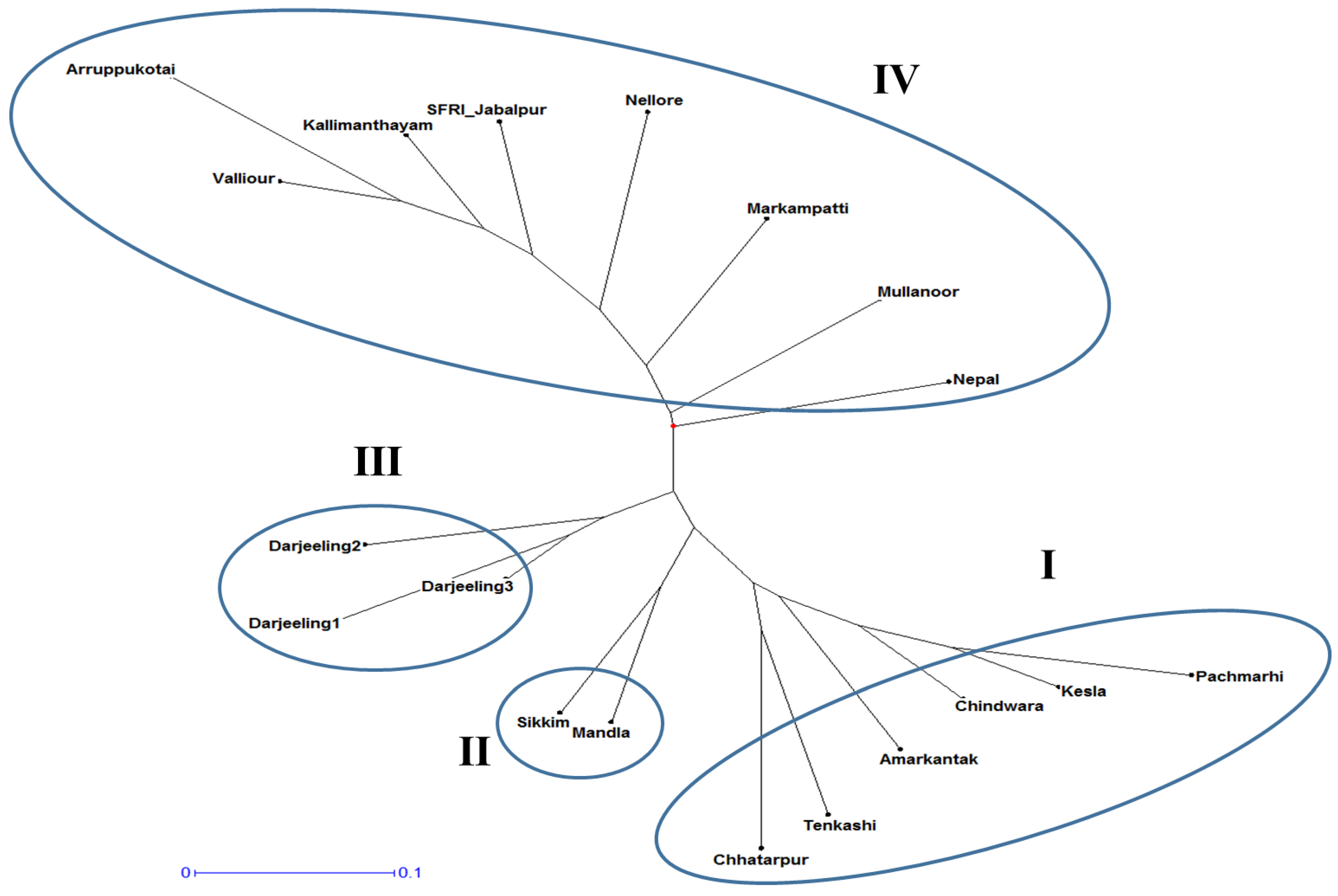
3.2. SSR Analysis and Polymorphism Study
3.2.1. Similarity Matrix
3.2.2. Cluster Analysis
3.2.3. Principal Component Analysis and SSR Markers
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Padmapriya, S.; Rajamani, K.; Sathiyamurthy, V. Glory Lily (Gloriosa superba L.)—A Review. Int. J. Curr. Pharm. Rev. Res. 2015, 7, 43–49. [Google Scholar]
- Ghosh, S.; Ghosh, B.; Jha, S. Polymorphism in Gloriosa superba. Plant Genet. Resour. 2009, 7, 9–15. [Google Scholar] [CrossRef]
- Rajadurai, K. Enhancing the Bio Productivity of Gloriosa superba L. through Mutatic Genetic Manipulation. Ph.D. Thesis, Tamil Nadu Agricultural University, Coimbatore, India, 2001. [Google Scholar]
- Selvarasu, A.; Rajamani, K. Analysis of Variability, Correlation and Path Coefficients in Induced Mutants of Glory Lily (Gloriosa superba L.). Int. J. Plant Breed. 2013, 7, 69–75. [Google Scholar]
- Das, M.; Prakash Sahu, S.; Tiwari, A. De novo Transcriptome Assembly and Mining of EST-SSR Markers in Gloriosa superba. J. Genet. 2020, 99, 77. [Google Scholar] [CrossRef]
- Liang, Y.; Gao, Q.; Li, F.; Du, Y.; Wu, J.; Pan, W.; Wang, S.; Zhang, X.; Zhang, M.; Song, X.; et al. The Giant Genome of Lily Provides Insights into the Hybridization of Cultivated Lilies. Nat. Commun. 2025, 16, 45. [Google Scholar] [CrossRef] [PubMed]
- Chitra, R.; Rajamani, K. Evaluation of Different Glory Lily (Gloriosa superba L.) Genotypes for Vegetative, Floral and Yield Characters. Agric. Sci. Dig. 2009, 29, 190–193. [Google Scholar]
- Arun Kumar, P.; Elangaimannan, R. Evaluation of Gloriosa superba for Yield Attributing Characters and Quantification of Colchicine Originated from Different Agro Climatic Zones of Tamil Nadu and Andhra Pradesh. Int. J. Pharm. Clin. Res. 2017, 9, 183–192. [Google Scholar]
- Chitra, R.; Rajamani, K. Assessment of Genetic Diversity of Gloriosa superba L. Accessions Detected by Random Amplified Polymorphic DNA Analysis. J. Med. Plants Res. 2013, 7, 2122–2127. [Google Scholar] [CrossRef]
- Babu, V.; Pai, D.; Shaik, T.; Muthusamy, A.; Satyamoorthy, K. Low Intraspecific Diversity Suggesting Genetic Drift in Gloriosa superba L. (Liliaceae) in Konkan Coast of Karnataka, India. Ann. Plant Sci. 2013, 2, 524–531. [Google Scholar]
- Mahajan, Y.A.; Shinde, B.A.; Mulani, F.A.; Gade, A.B.; Kasodekar, A.K.; Thulasiram, H.V.; Kadoo, N.Y.; Nikam, T.D. Diversity Assessment of Gloriosa superba Accessions from Western Ghats of India Based on Morphological Traits, ISSR Markers and Metabolite Content. J. Appl. Res. Med. Aromat. Plants 2022, 30, 100388. [Google Scholar] [CrossRef]
- Fang, H.; Guo, Q.; Shen, H.; Li, Y. Genetic Diversity Evaluation of Chrysanthemum Indicum L. by Medicinal Compounds and Molecular Biology Tools. Biochem. Syst. Ecol. 2012, 41, 26–34. [Google Scholar] [CrossRef]
- Kremer, D.; Bolarić, S.; Ballian, D.; Bogunić, F.; Stešević, D.; Karlović, K.; Kosalec, I.; Vokurka, A.; Vuković Rodríguez, J.; Randić, M.; et al. Morphological, Genetic and Phytochemical Variation of the Endemic Teucrium arduini L. (Lamiaceae). Phytochemistry 2015, 116, 111–119. [Google Scholar] [CrossRef]
- Fang, M.-F.; Li, J.; Zhou, T.-H.; Yang, J.; Zhao, G.-F. Genetic Diversity in Natural Populations of the Medicinal Herb Polygala Tenuifolia Willd. and Its Implications for Conservation. Biochem. Syst. Ecol. 2012, 44, 400–406. [Google Scholar] [CrossRef]
- Falconer, D. Introduction to Quantitative Genetics; Pearson Education: Noida, India, 1996. [Google Scholar]
- Liu, C.-H.; Liu, T.-S.; Luo, C.-Q.; Zhang, J.; Zeng, X.-Y.; Cui, L.; Xie, L.-J. Determination of Forsythiaside B and Poliumoside in Different Origin and Parts from Callicarpa Kwangtungensis. Zhongguo Zhong Yao Za Zhi 2013, 38, 3324–3326. [Google Scholar] [PubMed]
- Liu, G.-D.; Chen, G.-L.; Li, W.; Li, C.-X. Genetic and Phytochemical Diversities of Cynomorium Songaricum Rupr. in Northwest China Indicated by ISSR Markers and HPLC-Fingerprinting. Biochem. Syst. Ecol. 2013, 48, 34–41. [Google Scholar] [CrossRef]
- Fadaei Heidari, E.; Rahimmalek, M.; Mohammadi, S.; Hossein Ehtemam, M. Genetic Structure and Diversity of Ajowan (Trachyspermum ammi) Populations Based on Molecular, Morphological Markers, and Volatile Oil Content. Ind. Crops Prod. 2016, 92, 186–196. [Google Scholar] [CrossRef]
- Tiwari, V.; Meena, B.; Nair, K.N.; Upreti, D.K.; Tamta, S.; Rana, T.S. Assessment of Genetic Diversity and Population Structure of Bergenia stracheyi (Saxifragaceae) in the Western Himalaya (India). Biochem. Syst. Ecol. 2017, 70, 205–210. [Google Scholar] [CrossRef]
- Zheng, P.; Zhang, K.; Wang, Z. Genetic Diversity and Gentiopicroside Content of Four Gentiana Species in China Revealed by ISSR and HPLC Methods. Biochem. Syst. Ecol. 2011, 39, 704–710. [Google Scholar] [CrossRef]
- Tomar, R.S.; Parakhia, M.V.; Rathod, V.M.; Thakkar, J.R.; Padhiyar, S.M.; Thummar, V.D.; Dalal, H.; Kothari, V.V.; Kheni, J.; Dhingani, R.M.; et al. Molecular Mapping and Identification of QTLs Responsible for Charcoal Rot Resistance in Castor (Ricinus communis L.). Ind. Crops Prod. 2017, 95, 184–190. [Google Scholar] [CrossRef]
- Hayashi, T.; Yoshida, K.; Sano, K. Formation of Alkaloids in Suspension-Cultured Colchicum Autumnale. Phytochemistry 1988, 27, 1371–1374. [Google Scholar] [CrossRef]
- Porebski, S.; Bailey, L.G.; Baum, B.R. Modification of a CTAB DNA Extraction Protocol for Plants Containing High Polysaccharide and Polyphenol Components. Plant Mol. Biol. Report. 1997, 15, 8–15. [Google Scholar] [CrossRef]
- Ogunkanmi, L.A.; Ogundipe, O.T.; Ng, N.Q.; Fatokun, C.A. Genetic Diversity in Wild Relatives of Cowpea (Vigna unguiculata) as Revealed by Simple Sequence Repeats (SSR) Markers. J. Food Agric. Environ. 2008, 6, 132–137. [Google Scholar]
- Devi, K.D.; Punyarani, K.; Singh, N.S.; Devi, H.S. An Efficient Protocol for Total DNA Extraction from the Members of Order Zingiberales- Suitable for Diverse PCR Based Downstream Applications. SpringerPlus 2013, 2, 669. [Google Scholar] [CrossRef] [PubMed]
- Pacheco, A.; Rodriguez, F.; Alvarado, G.; Crossa, J.; Burgueno, J. ADEL-R, Version 2.0; Analysis and Design of Experiments with R for Windows; International Maize and Wheat Improvement Center (CIMMYT): Mexico City, Mexico, 2017. [Google Scholar]
- Tsilanizara, A.; Diop, C.M.; Nimal, B.; Detoc, M.; Lunéville, L.; Chiron, M.; Huynh, T.D.; Brésard, I.; Eid, M.; Klein, J.C.; et al. DARWIN: An Evolution Code System for a Large Range of Applications. J. Nucl. Sci. Technol. 2000, 37, 845–849. [Google Scholar] [CrossRef]
- Pang, Z.; Xu, L.; Viau, C.; Lu, Y.; Salavati, R.; Basu, N.; Xia, J. MetaboAnalystR 4.0: A Unified LC-MS Workflow for Global Metabolomics. Nat. Commun. 2024, 15, 3675. [Google Scholar] [CrossRef]
- De Mendiburu, F.; Simon, R. Agricolae—Ten Years of an Open Source Statistical Tool for Experiments in Breeding, Agriculture and Biology. PeerJ PrePrints 2015, e1404v1. [Google Scholar]
- Rohlf, F.J. NTSYS-Pc Numerical Taxonomy and Multivariate Analysis System; Version 2.0; Applied Biostatistics Inc.: Setauket, NY, USA, 1992. [Google Scholar]
- Girgel, U. Principle Component Analysis (PCA) of Bean Genotypes (Phaseolus vulgaris L.) Concerning Agronomic, Morphological and Biochemical Characteristics. Appl. Ecol. Environ. Res. 2021, 19, 1999–2011. [Google Scholar] [CrossRef]
- Sharma, R.S.; Vaidya, N.; Maloo, S.R.; Kumar, A.; Sharma, S.; Ramkrishnan, R.S.; Kumari, V. Application of Molecular Markers in Assessment of Genetic Diversity of Medicinal Plants. In Molecular Marker Techniques; Springer Nature: Singapore, 2023; pp. 103–116. [Google Scholar]
- Sharma, S.; Sharma, Y.P.; Thakur, P. Quantification of Colchicine in Different Parts of Gloriosa superba L. Int. J. Chem. Stud. 2017, 5, 147–149. [Google Scholar]
- Uchimahali, J.; Jebamalar, A.; Duraikannu, G.; Thirumal, S. Phytochemical Analysis and Evaluation of Antimicrobial Activity in the Whole Plant Extracts of Gloriosa superba. Asian J. Pharm. Clin. Res. 2019, 12, 245–249. [Google Scholar] [CrossRef]
- Tilwari, A.; Sharma, R. Random Amplified Polymorphic DNA and Inter Simple Sequence Repeat Markers Reveals Genetic Diversity between Micro Propagated, Wild and Field Cultivated Genotypes of Gloriosa superba: An Endangered Medicinal Plant. Mol. Biol. Rep. 2021, 48, 2437–2452. [Google Scholar] [CrossRef]
- Parkash, C.; Kumar, S.; Singh, R.; Kumar, A.; Kumar, S.; Dey, S.S.; Bhatia, R.; Kumar, R. ‘Ogura’-Based ‘CMS’ Lines with Different Nuclear Backgrounds of Cabbage Revealed Substantial Diversity at Morphological and Molecular Levels. 3 Biotech 2018, 8, 27. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Nie, W.; Zhu, P.; Liang, C.; Deng, S. Genetic Diversity in Callicarpa Kwangtungensis Chun. Based on Morphological, Biochemical and ISSR Markers. J. Appl. Res. Med. Aromat. Plants 2018, 10, 41–48. [Google Scholar] [CrossRef]
- Harish; Gupta, A.K.; Phulwaria, M.; Rai, M.K.; Shekhawat, N.S. Conservation Genetics of Endangered Medicinal Plant Commiphora Wightii in Indian Thar Desert. Gene 2014, 535, 266–272. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chen, W.; Shen, X.; Yang, Y.; Qi, F.; Liu, Y.; Meng, H. Analysis of the Genetic Diversity of Garlic (Allium sativum L.) by Simple Sequence Repeat and Inter Simple Sequence Repeat Analysis and Agro-Morphological Traits. Biochem. Syst. Ecol. 2014, 55, 260–267. [Google Scholar] [CrossRef]
- Sahana, K.S.; Gnanam, R.; Rajesh, S.; Rajamani, K. Evaluation of Genetic Diversity in Gloriosa superba L, an Endangered Medicinal Plant Using Molecular Marker. Int. J. Curr. Microbiol. Appl. Sci. 2019, 8, 2125–2134. [Google Scholar] [CrossRef]



| S. No | Collection Site | District | State | Country | Altitude (Meters from Sea Level) | Longitude | Latitude | Site Description |
|---|---|---|---|---|---|---|---|---|
| 1 | Darjeeling 1 | Darjeeling | West Bengal | India | 2042 | 88.2627° E | 27.0360° N | Cultivated |
| 2 | Darjeeling 2 | Darjeeling | West Bengal | India | 2042 | 88.2627° E | 27.0360° N | Cultivated |
| 3 | Darjeeling 3 | Darjeeling | West Bengal | India | 2042 | 88.2627° E | 27.0360° N | Cultivated |
| 4 | Salyan | Salyan | Karnali | Nepal | 1530 | 82.1278° E | 28.3525° N | Cultivated |
| 5 | Sumbuk | South Sikkim | Sikkim | India | 457 | 88.3811° E | 27.0986° N | Cultivated |
| 6 | Chhatarpur | Chhatarpur | Madhya Pradesh | India | 305 | 79.5812° E | 24.9164° N | Wild |
| 7 | Mandla | Mandla | Madhya Pradesh | India | 445 | 80.3714° E | 22.5979° N | Wild |
| 8 | Chhindwara | Chhindwara | Madhya Pradesh | India | 675 | 79.5812° E | 24.9164° N | Wild |
| 9 | Kesla | Hoshangabad | Madhya Pradesh | India | 278 | 77.7370° E | 22.7441° N | Wild |
| 10 | Pachmarhi | Hoshangabad | Madhya Pradesh | India | 1069 | 78.4346° E | 22.4674° N | Wild |
| 11 | Amarkantak | Anuppur | Madhya Pradesh | India | 1048 | 81.7532° E | 22.6822° N | Wild |
| 12 | Jabalpur | Jabalpur | Madhya Pradesh | India | 412 | 79.9864° E | 23.1815° N | Cultivated |
| 13 | Nellore | Nellore | Andhra Pradesh | India | 18 | 79.9865° E | 14.4426° N | Cultivated |
| 14 | Tenkasi | Tirunelveli | Tamil Nadu | India | 143 | 77.3129° E | 8.9590° N | Cultivated |
| 15 | Aruppukottai | Virudhunagar | Tamil Nadu | India | 97 | 78.1000° E | 9.5156° N | Cultivated |
| 16 | Vallioor | Tirunelveli | Tamil Nadu | India | 92 | 77.6236° E | 8.4127° N | Cultivated |
| 17 | Kallimandayam | Dindigul | Tamil Nadu | India | 301 | 77.6870° E | 10.5894° N | Cultivated |
| 18 | Markampatti | Dindigul | Tamil Nadu | India | 156 | 77.8074° E | 10.6731° N | Cultivated |
| 19 | Mulanur | Tirupur | Tamil Nadu | India | 238 | 77.7080° E | 10.7930° N | Cultivated |
| Location | PH | NLPP | NBPP | DF | DFF | NFPP | NPP | NSPP | FPW | FSWPP |
|---|---|---|---|---|---|---|---|---|---|---|
| Amarkantak | 96.4 ± 29.64 | 27.8 ± 9.7 | 1.4 ± 0.54 | 78 ± 2.4 | 85.8 ± 1.1 | 4 ± 0.04 | 3.6 ± 0.55 | 22.8 ± 1.09 | 6.18 ± 0.11 | 2.62 ± 1.64 |
| Arruppukotai | 63 ± 7.5 | 37.2 ± 7.0 | 2.2 ± 0.83 | 82.25 ± 4.5 | 90.25 ± 2.63 | 4.5 ± 0.57 | 2.5 ± 0.58 | 30 ± 3.65 | 5.32 ± 0.38 | 2.97 ± 0.3 |
| Chhatarpur | 132 ± 11.2 | 59.4 ± 6.8 | 2 ± 1.4 | 61.8 ± 0.44 | 81.6 ± 0.89 | 9 ± 2.24 | 7.2 ± 1.8 | 33.6 ± 3.57 | 6.84 ± 0.76 | 3.38 ± 0.40 |
| Chindwara | 108.8 ± 50.9 | 37.6 ± 15.4 | 1.6 ± 0.89 | 58.4 ± 0.54 | 64.2 ± 1.64 | 4.6 ± 0.55 | 4 ± 0.02 | 26.4 ± 2.19 | 3.96 ± 0.33 | 1.24 ± 0.02 |
| Darjeeling1 | 116 ± 6.5 | 35.4 ± 3.36 | 1.8 ± 1.09 | 50.75 ± 0.95 | 56.5 ± 1.29 | 4 ± 0.82 | 2.75 ± 1.25 | 16.5 ± 5.44 | 4.41 ± 1.47 | 1.68 ± 0.42 |
| Darjeeling2 | 132 ± 46.7 | 38.6 ± 16.6 | 1.4 ± 0.54 | 50 ± 1.63 | 52.5 ± 3.69 | 2.5 ± 0.57 | 1.5 ± 0.57 | 29.25 ± 10.24 | 6.46 ± 0.62 | 2.53 ± 0.6 |
| Darjeeling3 | 92.6 ± 13.8 | 34 ± 6.5 | 1.4 ± 0.89 | 49.2 ± 1.78 | 57 ± 1.41 | 3.2 ± 0.45 | 2.4 ± 0.89 | 18.4 ± 4.33 | 4.09 ± 1.18 | 1.8 ± 0.4 |
| Kallimanthayam | 91.5 ± 34 | 38.25 ± 19.0 | 1.75 ± 0.83 | 103.2 ± 1.7 | 113.2 ± 1.8 | 3.7 ± 0.45 | 3 ± 0.02 | 31.5 ± 2.23 | 7.08 ± 0.17 | 4.08 ± 0.18 |
| Kesla | 140 ± 32.8 | 39.2 ± 12.7 | 1.8 ± 0.83 | 70.2 ± 1.64 | 75 ± 2.7 | 4.6 ± 0.55 | 3.2 ± 1.09 | 27.6 ± 2.19 | 4.08 ± 0.38 | 1.23 ± 0.016 |
| Mandla | 119.2 ± 32.9 | 23.4 ± 11.3 | 2.2 ± 1.64 | 72 ± 10.1 | 78.2 ± 9.3 | 7.2 ± 1.09 | 5.2 ± 1.09 | 36 ± 5.5 | 6.972 ± 0.2 | 2.84 ± 0.33 |
| Markampatti | 158.2 ± 33.4 | 67.2 ± 18.8 | 3.2 ± 2.2 | 115.5 ± 3 | 124.5 ± 1.91 | 5.25 ± 0.5 | 3.75 ± 0.95 | 31.5 ± 9 | 6.275 ± 0.9 | 3.32 ± 0.67 |
| Mullanoor | 95 ± 21.7 | 40.4 ± 10.2 | 3.6 ± 2.4 | 67.2 ± 5.21 | 72.2 ± 6.2 | 5.4 ± 0.89 | 4.6 ± 0.54 | 35.8 ± 3.7 | 6.26 ± 0.33 | 3.04 ± 0.26 |
| Nellore | 57 ± 11 | 28.4 ± 8.8 | 1.4 ± 0.54 | 94 ± 4 | 98.5 ± 7 | 3.25 ± 0.5 | 3 ± 0.04 | 19 ± 2 | 4.17 ± 0.75 | 1.96 ± 0.56 |
| Nepal | 83 ± 13 | 27.6 ± 17.3 | 1.4 ± 0.89 | 65.6 ± 3.6 | 72.8 ± 1.8 | 5 ± 1.4 | 4.2 ± 1.3 | 28.2 ± 5.7 | 6.43 ± 0.36 | 2.88 ± 0.46 |
| Pachmarhi | 99.8 ± 26.5 | 33.2 ± 14.6 | 1.2 ± 0.44 | 65 ± 2.2 | 72 ± 1.2 | 4 ± 0.02 | 3 ± 0.02 | 33 ± 0.05 | 6.5 ± 0.3 | 3.6 ± 0.23 |
| SFRI_Jabalpur | 72.6 ± 13.2 | 31.6 ± 8.08 | 2 ± 1 | 76.8 ± 1.1 | 85 ± 2.12 | 3.2 ± 0.45 | 2.4 ± 0.55 | 26 ± 1.4 | 6.32 ± 0.18 | 3.28 ± 0.11 |
| Sikkim | 96.8 ± 33.4 | 22 ± 8.2 | 1.8 ± 1 | 66.2 ± 1.1 | 74.8 ± 1.64 | 4.8 ± 1.09 | 3.4 ± 0.55 | 39.6 ± 3.3 | 6.8 ± 0.27 | 4.04 ± 0.22 |
| Tenkashi | 94.6 ± 28.7 | 45.8 ± 20.6 | 2 ± 1.4 | 59 ± 2.7 | 66.2 ± 1.64 | 5.6 ± 0.55 | 4 ± 0.55 | 33 ± 2.7 | 4.76 ± 0.76 | 3.26 ± 0.49 |
| Valliour | 44.2 ± 6.6 | 24 ± 2.34 | 1.6 ± 0.89 | 90.8 ± 1.8 | 98.8 ± 1.8 | 5.2 ± 0.45 | 4 ± 0.55 | 40.8 ± 2.7 | 6.45 ± 0.14 | 3.12 ± 0.18 |
| CV | 32.99647 | 35.43879 | 63.53912 | 4.75557 | 4.19714 | 18.23983 | 22.45335 | 14.85898 | 10.60104 | 12.77759 |
| LSD | 41.44136 | 16.25074 | 1.50731 | 4.49835 | 4.38345 | 1.11455 | 1.04310 | 5.70651 | 0.79717 | 0.46453 |
| Location | FPYPP | FSYPP | DSWPP | DSYPP | LL | LW | IL | TS | DT | Colchicine |
| Amarkantak | 21.96 ± 3.06 | 9.4 ± 1.1 | 1.38 ± 0.054 | 5.08 ± 0.71 | 11.7 ± 4.18 | 2.9 ± 0.94 | 4.3 ± 2.02 | 8 ± 3.46 | 1.22 ± 0.18 | 0.17 ± 0.19 |
| Arruppukotai | 13.15 ± 3.32 | 7.375 ± 1.68 | 1.84 ± 0.3 | 3.69 ± 1.05 | 11.15 ± 1.51 | 3.72 ± 0.55 | 2.73 ± 0.22 | 9.4 ± 1.34 | 1.47 ± 0.167 | 0.53 ± 0.0036 |
| Chhatarpur | 47.46 ± 8.36 | 23.64 ± 1.51 | 1.7 ± 0.22 | 12.9 ± 2.86 | 12.2 ± 0.67 | 3.34 ± 0.36 | 3.67 ± 1.32 | 12 ± 2.23 | 1.67 ± 0.1 | 0.19 ± 0.0032 |
| Chindwara | 15.52 ± 1.2 | 4.98 ± 0.164 | 0.874 ± 0.02 | 3.49 ± 0.08 | 12.4 ± 0.65 | 2.83 ± 0.24 | 5.05 ± 2.4 | 11.2 ± 1.48 | 1.65 ± 0.09 | 0.18 ± 0.016 |
| Darjeeling1 | 12.75 ± 7.9 | 4.75 ± 2.57 | 1.085 ± 0.26 | 1.93 ± 1.2 | 16.56 ± 1.48 | 3.03 ± 1.49 | 5.52 ± 1.02 | 7.72 ± 1.42 | 1.37 ± 0.09 | 0.21 ± 0.029 |
| Darjeeling2 | 9.26 ± 3.42 | 3.695 ± 1.8 | 1.69 ± 0.32 | 2.08 ± 1.07 | 14.3 ± 4.12 | 4.35 ± 1.40 | 5.4 ± 0.65 | 6.8 ± 1.35 | 1.31 ± 0.08 | 0.23 ± 0.0023 |
| Darjeeling3 | 10.52 ± 7.65 | 4.5 ± 2.85 | 1.15 ± 0.10 | 1.824 ± 0.77 | 13.82 ± 2.13 | 3.09 ± 1.88 | 5.34 ± 1.09 | 8.48 ± 1.26 | 1.66 ± 0.134 | 0.22 ± 0.0228 |
| Kallimanthayam | 21.01 ± 0.58 | 12.32 ± 0.71 | 2.78 ± 0.62 | 7.08 ± 0.17 | 12.4 ± 1.45 | 1.7 ± 0.51 | 3.16 ± 0.61 | 13.02 ± 1.1 | 1.75 ± 0.147 | 0.55 ± 0.0089 |
| Kesla | 12.76 ± 3.6 | 3.916 ± 1.29 | 0.87 ± 0.02 | 2.76 ± 0.95 | 13.85 ± 0.89 | 2.62 ± 0.32 | 6.55 ± 1.76 | 14 ± 2.23 | 1.75 ± 0.08 | 0.049 ± 0.018 |
| Mandla | 35.9 ± 6.84 | 16.84 ± 1.97 | 1.31 ± 0.15 | 7.4 ± 0.82 | 13.3 ± 3.18 | 4.26 ± 1.16 | 4.75 ± 1.87 | 9.6 ± 3.43 | 1.73 ± 0.29 | 0.062 ± 0.0007 |
| Markampatti | 23.9 ± 8.86 | 12.55 ± 4.8 | 1.87 ± 0.42 | 7.05 ± 2.84 | 10.6 ± 1.19 | 2.2 ± 0.24 | 2.1 ± 0.26 | 16.58 ± 1.78 | 2.3 ± 0.1 | 0.58 ± 0.019 |
| Mullanoor | 28.7 ± 3.41 | 13.76 ± 2.2 | 1.30 ± 0.14 | 8.34 ± 1.36 | 13.04 ± 1.27 | 2.26 ± 0.75 | 6.7 ± 3.17 | 18.54 ± 2.35 | 2.284 ± 0.15 | 0.37 ± 0.01 |
| Nellore | 12.725 ± 2.25 | 5.72 ± 1.85 | 1.23 ± 0.065 | 2.85 ± 1.5 | 12.1 ± 2.94 | 3.13 ± 0.72 | 1.8 ± 0.30 | 11.9 ± 2.63 | 1.81 ± 0.21 | 0.62 ± 0.00017 |
| Nepal | 26.72 ± 9.47 | 12.28 ± 5.04 | 1.54 ± 0.13 | 6.2 ± 2.75 | 10.8 ± 2.1 | 4 ± 0.40 | 3.72 ± 0.89 | 9.9 ± 1.47 | 1.56 ± 0.15 | 0.352 ± 0.0106 |
| Pachmarhi | 19.2 ± 0.58 | 10.3 ± 0.65 | 1.62 ± 0.03 | 6.8 ± 0.05 | 11.3 ± 2.34 | 2.93 ± 0.99 | 3.91 ± 0.85 | 10.2 ± 2.16 | 1.37 ± 0.17 | 0.225 ± 0.0075 |
| SFRI_Jabalpur | 14.98 ± 3.56 | 8.38 ± 2.93 | 1.27 ± 0.18 | 5.08 ± 2.62 | 11.65 ± 2.17 | 2.9 ± 0.76 | 2.72 ± 0.44 | 5.8 ± 0.90 | 1.17 ± 0.22 | 0.33 ± 0.0106 |
| Sikkim | 23.14 ± 2.79 | 13.48 ± 1.47 | 1.84 ± 0.05 | 7.8 ± 0.27 | 13.3 ± 3.3 | 5.76 ± 0.77 | 5.6 ± 1.96 | 13.54 ± 0.84 | 1.75 ± 0.15 | 0.374 ± 0.0014 |
| Tenkashi | 18.94 ± 3.34 | 12.74 ± 1.69 | 1.932 ± 0.15 | 7.08 ± 0.38 | 11.08 ± 1.67 | 2.84 ± 0.88 | 2.42 ± 0.49 | 15.6 ± 1.81 | 2.14 ± 0.24 | 0.64 ± 0.0102 |
| Valliour | 25.76 ± 0.76 | 11.92 ± 0.62 | 1.68 ± 0.06 | 7.5 ± 0.66 | 10.15 ± 0.83 | 3.83 ± 0.31 | 3.11 ± 0.66 | 7.9 ± 0.74 | 1.45 ± 0.178 | 0.505 ± 0.0015 |
| CV | 24.34162 | 23.50275 | 14.88319 | 25.38813 | 18.75123 | 27.74781 | 34.93603 | 17.80573 | 10.054402 | 4.687235141 |
| LSD | 6.58590 | 3.10355 | 0.29703 | 1.86165 | 2.93259 | 1.13620 | 1.82232 | 2.48339 | 0.2093201 | 0.022386397 |
| (a) | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Source of Variation | DF | Plant Height (cm) | Number of Leaves per Plant | Number of Branches per Plant | Leaf Length | Leaf Width | Internodal Length | Tuber Size | Diameter of Tuber | Days to Flowering | Days to Fifty Percent Flowering |
| Replication | 4 | 132.6421 | 42.5993 | 1.7599 | 1.0838 | 0.6528 | 0.3505 | 2.5808 | 0.0228 | 20.7039 | 27.1842 |
| Ecotypes/Genotypes | 19 | 4252.868 ** | 662.7018 ** | 1.8767 | 12.1177 ** | 4.2471 ** | 11.0872 ** | 60.3448 ** | 0.5209 ** | 1453.968 ** | 1580.364 ** |
| Error | 76 | 1080.417 | 166.1382 | 1.4293 | 5.4104 | 0.8121 | 2.0892 | 3.8798 | 0.0276 | 11.858 | 11.2599 |
| (b) | |||||||||||
| Source of Variation | DF | Number of flowers per plant | Number of pods per plant | Number of seeds per pod | Fresh pod weight | Fresh seed weight per pod | Fresh pod yield per plant | Fresh seed yield per plant | Dry seed weight per pod | Dry seed yield per plant | Colcichine content |
| Replication | 4 | 1.0819 | 0.995 | 6.8488 | 0.0863 | 0.0303 | 35.1351 | 12.1019 | 0.0434 | 4.4547 | 0.0061 |
| Ecotypes/Genotypes | 19 | 10.9201 ** | 7.4051 ** | 217.6051 ** | 6.0549 ** | 3.4837 ** | 437.8888 ** | 126.9486 ** | 0.9718 ** | 38.4499 ** | 0.1406 ** |
| Error | 76 | 0.7279 | 0.6376 | 19.0829 | 0.3724 | 0.1265 | 25.4175 | 5.6445 | 0.0517 | 2.031 | 0.0002 |
| SSR Markers | H | PIC | E | H. av | MI | D |
|---|---|---|---|---|---|---|
| RGM-56194 | 0.432133 | 0.405246 | 2.052632 | 0.007581 | 0.015562 | 0.535714 |
| RGM-59440 | 0.49771 | 0.374757 | 3.263158 | 0.003742 | 0.012211 | 0.784575 |
| RGM-60741 | 0.487535 | 0.37977 | 2.105263 | 0.005132 | 0.010804 | 0.825308 |
| RGM-56691 | 0.458102 | 0.393686 | 2.578947 | 0.006028 | 0.015545 | 0.587368 |
| RGM-56692 | 0.491343 | 0.332698 | 4.526316 | 0.003233 | 0.014631 | 0.681509 |
| RGM-24219 | 0 | 0.453407 | 1 | 0 | 0 | 0 |
| RGM-24910 | 0.496153 | 0.330323 | 1.631579 | 0.008704 | 0.014202 | 0.708647 |
| RGM-52632 | 0.361496 | 0.388068 | 4.578947 | 0.003171 | 0.01452 | 0.41919 |
| RGM-51956 | 0.487535 | 0.334562 | 0.421053 | 0.02566 | 0.010804 | 0.836257 |
| RGM-49006 | 0.188366 | 0.435666 | 0.894737 | 0.009914 | 0.00887 | 0.204678 |
| RGM-49355 | 0.188366 | 0.367257 | 0.894737 | 0.009914 | 0.00887 | 0.204678 |
| RGM-47736 | 0.49446 | 0.262753 | 0.894737 | 0.013012 | 0.011642 | 0.806543 |
| RGM-51635 | 0.498615 | 0.26069 | 0.526316 | 0.026243 | 0.013812 | 0.736842 |
| RGM-48348 | 0.33241 | 0.32975 | 1.578947 | 0.008748 | 0.013812 | 0.381223 |
| RGM-69599 | 0.145429 | 0.374423 | 1.842105 | 0.003827 | 0.00705 | 0.153627 |
| RGM-66027 | 0.352108 | 0.323008 | 2.315789 | 0.006177 | 0.014305 | 0.407268 |
| RGM-66661 | 0.425516 | 0.294467 | 4.157895 | 0.003733 | 0.01552 | 0.521658 |
| RGM-67617 | 0.099723 | 0.380026 | 1.894737 | 0.002624 | 0.004972 | 0.103841 |
| RGM-69875 | 0.208795 | 0.363201 | 3.526316 | 0.002747 | 0.009688 | 0.224211 |
| RGM-70748 | 0.33241 | 0.32975 | 2.368421 | 0.005832 | 0.013812 | 0.379699 |
| RGM-73459 | 0.241305 | 0.355884 | 5.157895 | 0.002117 | 0.010918 | 0.262071 |
| RGM-69870 | 0.361496 | 0.319659 | 1.526316 | 0.009513 | 0.01452 | 0.422475 |
| RGM-56853 | 0.483033 | 0.268338 | 1.631579 | 0.006356 | 0.01037 | 0.836842 |
| RGM-57783 | 0.455525 | 0.281247 | 1.947368 | 0.007992 | 0.015563 | 0.582707 |
| RGM-25035 | 0.487535 | 0.266153 | 1.736842 | 0.008553 | 0.014856 | 0.669173 |
| RGM-26631 | 0.188366 | 0.367257 | 1.789474 | 0.004957 | 0.00887 | 0.201991 |
| RGM-27384 | 0.465374 | 0.276712 | 1.894737 | 0.008164 | 0.015469 | 0.605263 |
| RGM-58197 | 0 | 0.384998 | 1 | 0 | 0 | 0 |
| RGM-58198 | 0.432133 | 0.291629 | 0.315789 | 0.022744 | 0.007182 | 0.912281 |
| RGM-58412 | 0.49446 | 0.262753 | 1.105263 | 0.013012 | 0.014382 | 0.70128 |
| RGM-58791 | 0.496153 | 0.261915 | 1.631579 | 0.008704 | 0.014202 | 0.708647 |
| RGM-59946 | 0.498615 | 0.26069 | 0.526316 | 0.026243 | 0.013812 | 0.736842 |
| RGM-60707 | 0.473992 | 0.272664 | 1.842105 | 0.008316 | 0.015318 | 0.627193 |
| RGM-60731 | 0.487535 | 0.266153 | 1.263158 | 0.008553 | 0.010804 | 0.827068 |
| RGM-59023 | 0.498615 | 0.26069 | 1.578947 | 0.008748 | 0.013812 | 0.727444 |
| RGM-59025 | 0 | 0.384998 | 2 | 0 | 0 | 0 |
| RGM-51958 | 0.496153 | 0.261915 | 1.368421 | 0.008704 | 0.011911 | 0.796366 |
| RGM-52500 | 0.496153 | 0.261915 | 1.631579 | 0.008704 | 0.014202 | 0.708647 |
| RGM-51196 | 0.099723 | 0.380026 | 1.894737 | 0.002624 | 0.004972 | 0.103841 |
| RGM-51200 | 0 | 0.384998 | 2 | 0 | 0 | 0 |
| RGM-25491 | 0.247576 | 0.354351 | 3.421053 | 0.003258 | 0.011144 | 0.270175 |
| RGM-43597 | 0.215451 | 0.361789 | 2.631579 | 0.00378 | 0.009947 | 0.232456 |
| RGM-44449 | 0 | 0.384998 | 3 | 0 | 0 | 0 |
| RGM-39722 | 0.387812 | 0.309799 | 2.210526 | 0.006804 | 0.01504 | 0.460526 |
| RGM-40085 | 0.188366 | 0.367257 | 1.789474 | 0.004957 | 0.00887 | 0.201991 |
| RGM-46426 | 0.300554 | 0.339832 | 1.631579 | 0.007909 | 0.012905 | 0.338549 |
| RGM-47507 | 0.432133 | 0.291629 | 2.052632 | 0.007581 | 0.015562 | 0.535714 |
| RGM-47202 | 0.33241 | 0.32975 | 1.578947 | 0.008748 | 0.013812 | 0.381223 |
| RGM-47204 | 0 | 0.384998 | 3 | 0 | 0 | 0 |
| RGM-47147 | 0 | 0.384998 | 4 | 0 | 0 | 0 |
| RGM-47164 | 0 | 0.391169 | 1 | 0 | 0 | 0 |
| RGM-41552 | 0.265928 | 0.355811 | 1 | 0.265928 | 0.265928 | 0.293233 |
| RGM-44907 | 0.589252 | 0.317107 | 1 | 0.589252 | 0.589252 | 0.507602 |
| RGM-45128 | 0.31148 | 0.342659 | 1 | 0.31148 | 0.31148 | 0.351504 |
| RGM-44513 | 0.473992 | 0.278835 | 1 | 0.473992 | 0.473992 | 0.625058 |
| RGM-42854 | 0 | 0.391169 | 1 | 0 | 0 | 0 |
| RGM-40092 | 0.498615 | 0.266861 | 1 | 0.498615 | 0.498615 | 0.782361 |
| RGM-40096 | 0.473992 | 0.278835 | 1 | 0.473992 | 0.473992 | 0.627193 |
| RGM-86857 | 0 | 0.391169 | 1 | 0 | 0 | 0 |
| RGM-87387 | 0 | 0.391169 | 1 | 0 | 0 | 0 |
| RGM-85888 | 0 | 0.391169 | 1 | 0 | 0 | 0 |
| RGM-85889 | 0.188366 | 0.373429 | 1 | 0.188366 | 0.188366 | 0.200702 |
| RGM-39603 | 0 | 0.391169 | 1 | 0 | 0 | 0 |
| RGM-39606 | 0.130502 | 0.382654 | 1 | 0.130502 | 0.130502 | 0.136591 |
| RGM-39698 | 0.215451 | 0.36796 | 1 | 0.215451 | 0.215451 | 0.232456 |
| RGM-41186 | 0.432133 | 0.2978 | 1 | 0.432133 | 0.432133 | 0.534737 |
| RGM-39500 | 0 | 0.391169 | 1 | 0 | 0 | 0 |
| RGM-4743 | 0.265928 | 0.355811 | 1 | 0.265928 | 0.265928 | 0.298246 |
| RGM-7349 | 0.099723 | 0.386197 | 1 | 0.099723 | 0.099723 | 0.105263 |
| RGM-8423 | 0.188366 | 0.373429 | 1 | 0.188366 | 0.188366 | 0.201991 |
| RGM-8583 | 0 | 0.391169 | 1 | 0 | 0 | 0 |
| RGM-13552 | 0.31148 | 0.342659 | 1 | 0.31148 | 0.31148 | 0.351504 |
| RGM-4194 | 0.41859 | 0.30356 | 1 | 0.41859 | 0.41859 | 0.511278 |
| RGM-17502 | 0.265928 | 0.355811 | 1 | 0.265928 | 0.265928 | 0.298246 |
| RGM-18069 | 0.496153 | 0.268086 | 1 | 0.496153 | 0.496153 | 0.708647 |
| RGM-18938 | 0.099723 | 0.386197 | 1 | 0.099723 | 0.099723 | 0.105263 |
| RGM-18896 | 0.387812 | 0.31597 | 1 | 0.387812 | 0.387812 | 0.467836 |
| RGM-14067 | 0.265928 | 0.355811 | 1 | 0.265928 | 0.265928 | 0.298246 |
| RGM-64349 | 0.160049 | 0.378362 | 1 | 0.160049 | 0.160049 | 0.169173 |
| RGM-64355 | 0.228532 | 0.365056 | 1 | 0.228532 | 0.228532 | 0.248933 |
| RGM-76857 | 0.411357 | 0.306562 | 1 | 0.411357 | 0.411357 | 0.500711 |
| RGM-77245 | 0.265928 | 0.355811 | 1 | 0.265928 | 0.265928 | 0.298246 |
| RGM-71542 | 0.41859 | 0.30356 | 1 | 0.41859 | 0.41859 | 0.511278 |
| RGM-71641 | 0.099723 | 0.386197 | 1 | 0.099723 | 0.099723 | 0.105263 |
| RGM-80230 | 0.411357 | 0.306562 | 1 | 0.411357 | 0.411357 | 0.500711 |
| RGM-73616 | 0.465374 | 0.282883 | 1 | 0.465374 | 0.465374 | 0.605263 |
| RGM-79522 | 0.265928 | 0.355811 | 1 | 0.265928 | 0.265928 | 0.298246 |
| RGM-79523 | 0.265928 | 0.355811 | 1 | 0.265928 | 0.265928 | 0.298246 |
| RGM-74641 | 0 | 0.391169 | 1 | 0 | 0 | 0 |
| RGM-88771 | 0.453407 | 0.28838 | 1 | 0.453407 | 0.453407 | 0.576484 |
| RGM-81186 | 0 | 0.396271 | 1 | 0 | 0 | 0 |
| RGM-81897 | 0 | 0.396271 | 1 | 0 | 0 | 0 |
| RGM-38349 | 0.188366 | 0.37853 | 1 | 0.188366 | 0.188366 | 0.201991 |
| RGM-34224 | 0.188366 | 0.37853 | 1 | 0.188366 | 0.188366 | 0.204678 |
| RGM-34165 | 0.498615 | 0.271963 | 1 | 0.498615 | 0.498615 | 0.736842 |
| RGM-34358 | 0.265928 | 0.360912 | 1 | 0.265928 | 0.265928 | 0.298246 |
| RGM-34357 | 0.387812 | 0.321072 | 1 | 0.387812 | 0.387812 | 0.467836 |
| RGM-38344 | 0.188366 | 0.37853 | 1 | 0.188366 | 0.188366 | 0.204678 |
| RGM-38348 | 0 | 0.396271 | 1 | 0 | 0 | 0 |
| RGM-7400 | 0 | 0.396271 | 1 | 0 | 0 | 0 |
| RGM-7527 | 0.099723 | 0.391299 | 1 | 0.099723 | 0.099723 | 0.105263 |
| RGM-39279 | 0.188366 | 0.37853 | 1 | 0.188366 | 0.188366 | 0.201991 |
| RGM-36147 | 0.487535 | 0.277426 | 1 | 0.487535 | 0.487535 | 0.671408 |
| RGM-36361 | 0.265928 | 0.360912 | 1 | 0.265928 | 0.265928 | 0.298246 |
| RGM-36788 | 0 | 0.396271 | 1 | 0 | 0 | 0 |
| RGM-29406 | 0.487535 | 0.277426 | 1 | 0.487535 | 0.487535 | 0.678363 |
| RGM-30615 | 0.498615 | 0.313951 | 1 | 0.498615 | 0.498615 | 0.526316 |
| RGM-36974 | 0.300554 | 0.351105 | 1 | 0.300554 | 0.300554 | 0.338549 |
| RGM-34372 | 0.099723 | 0.391299 | 1 | 0.099723 | 0.099723 | 0.105263 |
| RGM-34368 | 0 | 0.396271 | 1 | 0 | 0 | 0 |
| RGM-35987 | 0.361496 | 0.330932 | 1 | 0.361496 | 0.361496 | 0.422475 |
| RGM-34705 | 0.432133 | 0.302902 | 1 | 0.432133 | 0.432133 | 0.54386 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Majumdar, M.; Arya, R.; Sahu, S.P.; Tiwari, A.; Kim, J.-J. Assessment of Genetic Diversity by Morphological, Biochemical, and Molecular Markers in Gloriosa superba Ecotypes Collected from Different Agro-Climatic Zones in India. Horticulturae 2025, 11, 723. https://doi.org/10.3390/horticulturae11070723
Majumdar M, Arya R, Sahu SP, Tiwari A, Kim J-J. Assessment of Genetic Diversity by Morphological, Biochemical, and Molecular Markers in Gloriosa superba Ecotypes Collected from Different Agro-Climatic Zones in India. Horticulturae. 2025; 11(7):723. https://doi.org/10.3390/horticulturae11070723
Chicago/Turabian StyleMajumdar, Moumita, Rakesh Arya, Soumya Prakash Sahu, Archana Tiwari, and Jong-Joo Kim. 2025. "Assessment of Genetic Diversity by Morphological, Biochemical, and Molecular Markers in Gloriosa superba Ecotypes Collected from Different Agro-Climatic Zones in India" Horticulturae 11, no. 7: 723. https://doi.org/10.3390/horticulturae11070723
APA StyleMajumdar, M., Arya, R., Sahu, S. P., Tiwari, A., & Kim, J.-J. (2025). Assessment of Genetic Diversity by Morphological, Biochemical, and Molecular Markers in Gloriosa superba Ecotypes Collected from Different Agro-Climatic Zones in India. Horticulturae, 11(7), 723. https://doi.org/10.3390/horticulturae11070723






