Comprehensive Evaluation and Screening for Salt Tolerance Germplasms at Seedling Stage in Eggplant
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Determination of Optimal Salt Stress Levels
2.3. Salt Tolerance Screening of Germplasm
2.4. Descriptor Measurements
2.5. Evaluation of Seedling Physiological Indices
2.6. Statistical Analysis
3. Results
3.1. Optimal Salt Concentration for Screening
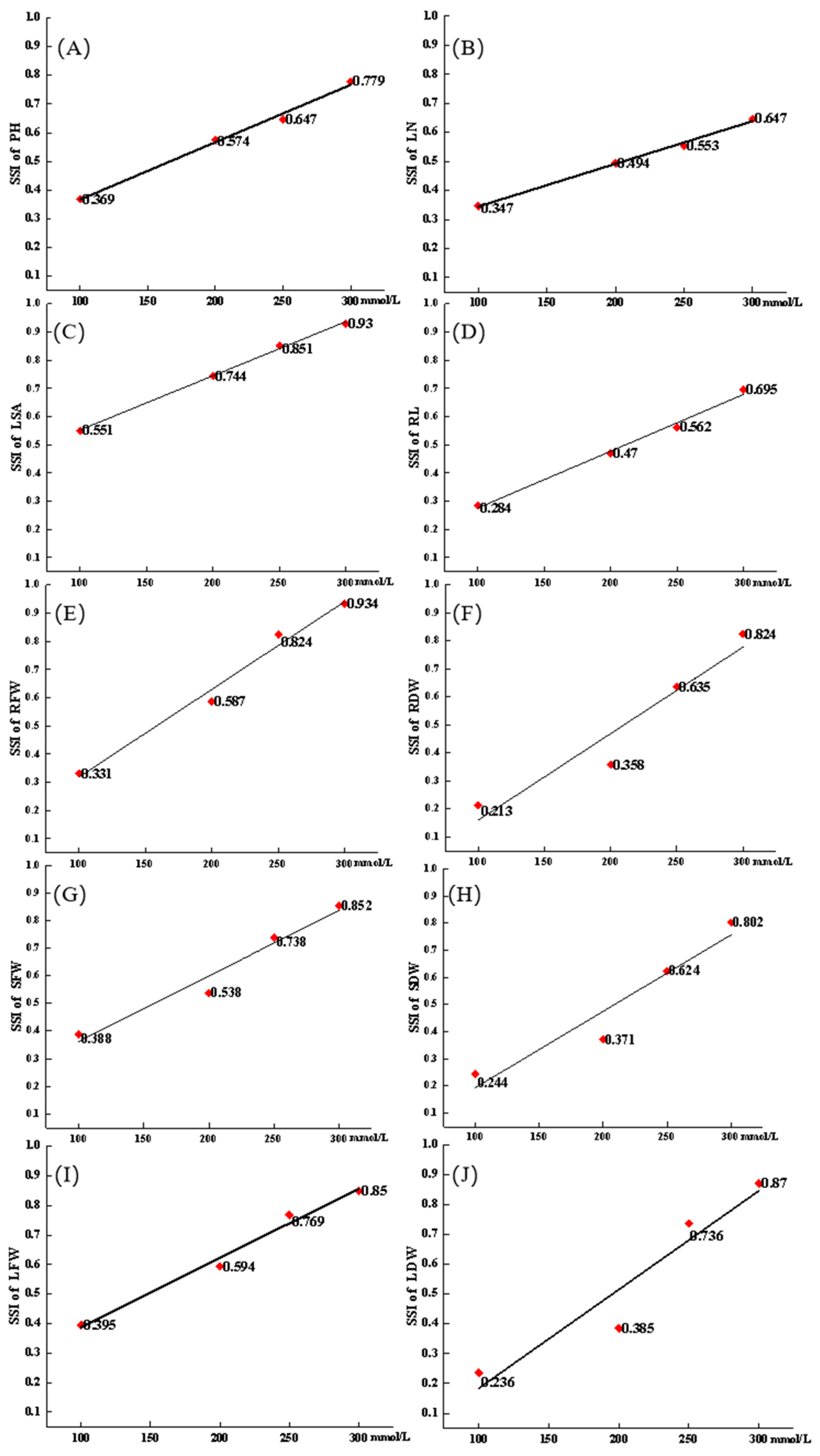
3.2. Variations in Salt Tolerance Indices
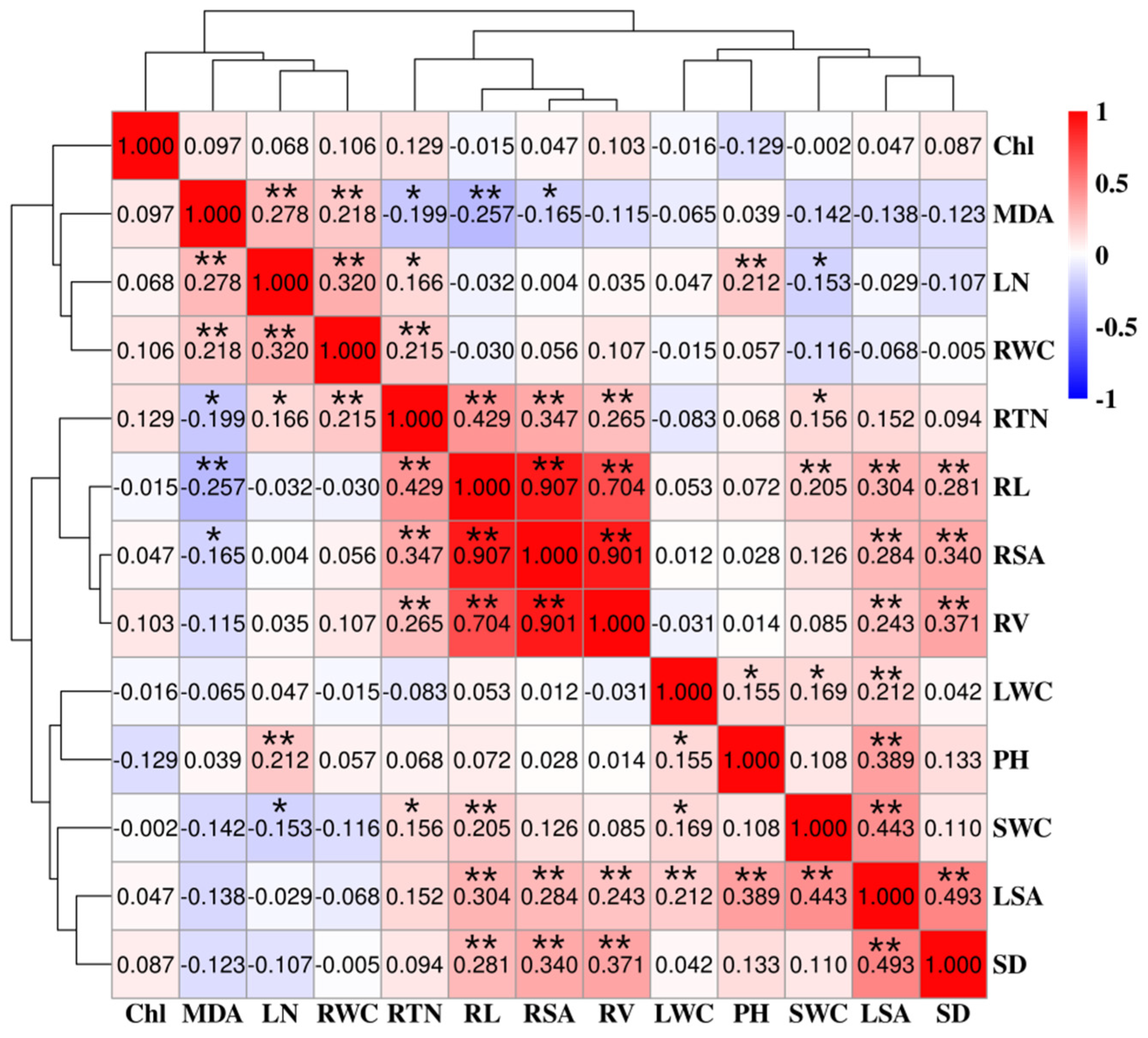
3.3. Correlation Analysis of STI Using Different Indices in Seedlings
3.4. Principal Component Analysis of STI
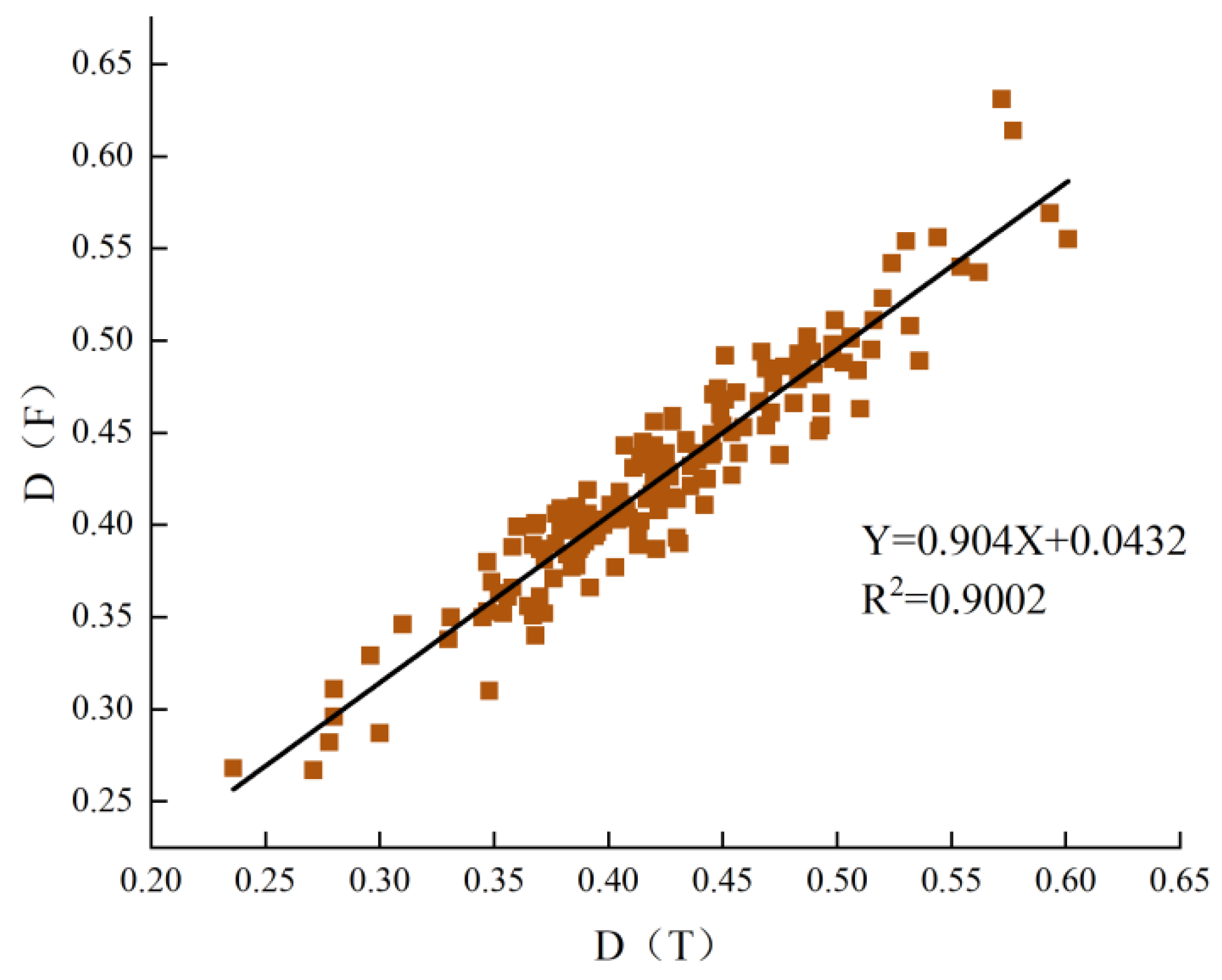
3.5. Membership Function Evaluation
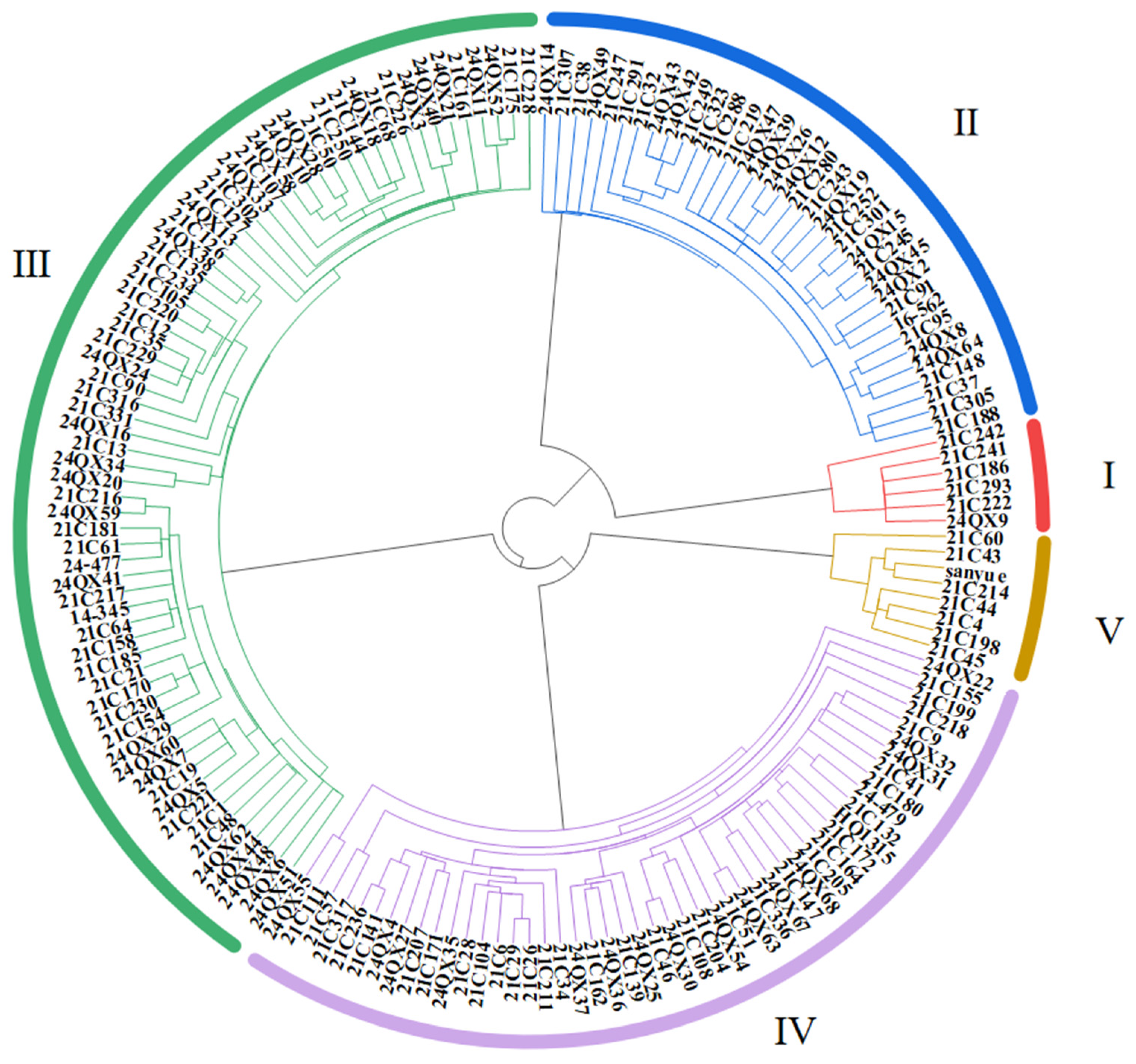
3.6. Germplasm Classification by Salt Tolerance
3.7. Stepwise Regression Analysis of STIs Using Different Indices
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zelm, E.V.; Zhang, Y.; Testerink, C. Salt-tolerance mechanisms of plants. Annu. Rev. Plant Biol. 2020, 71, 403–433. [Google Scholar] [CrossRef] [PubMed]
- Hopmans, J.W.; Qureshi, A.S.; Kisekka, I.; Munns, R.; Grattan, S.R.; Rengasamy, P.; Ben-Gal, A.; Assouline, S.; Javaux, M.; Minhas, P.S.; et al. Critical knowledge gaps and research priorities in global soil salinity. Adv. Agron. 2021, 169, 1–191. [Google Scholar]
- Tian, F.; Hou, M.; Qiu, Y.; Zhang, T.; Yuan, Y. Salinity stress effects on transpiration and plant growth under different salinity soil levels based on thermal infrared remote (TIR) technique. Geoderma 2020, 357, 113961. [Google Scholar] [CrossRef]
- Liu, X.; Guo, K.; Feng, X.; Sun, H. Discussion on the agricultural efficient utilization of saline-alkali land resources. China J. Ecol. Agric. 2023, 31, 345–353. [Google Scholar]
- Zhang, M.X.; Bai, R.; Nan, M.; Ren, W.; Wang, C.M.; Shabala, S.; Zhang, J.L. Evaluation of salt-tolerance of oat cultivars and the mechanism of adaptation to salinity. J. Plant Physiol. 2022, 273, 153708. [Google Scholar] [CrossRef] [PubMed]
- Yu, R.; Wang, G.; Yu, X.; Li, L.; Li, C.; Song, Y.; Xu, Z.; Zhang, J.; Guan, C. Assessing alfalfa (Medicago sativa L.) tolerance to salinity at seedling stage and screening of the salinity tolerance traits. Plant Biol. 2021, 23, 664–674. [Google Scholar] [CrossRef]
- Zhao, S.; Zhang, Q.; Liu, M.; Zhou, H.; Ma, C.; Wang, P. Regulation of plant responses to salt stress. Int. J. Mol. Sci. 2021, 22, 4609. [Google Scholar] [CrossRef]
- de Freitas, P.A.F.; de Carvalho, H.H.; Costa, J.H.; Miranda, R.D.S.; Saraiva, K.D.D.C.; de Oliveira, F.D.B.; Coelho, D.G.; Prisco, J.T.; Gomes-Filho, E. Salt acclimation in sorghum plants by exogenous proline: Physiological and biochemical changes and regulation of proline metabolismp. Plant Cell Rep. 2019, 38, 403–416. [Google Scholar] [CrossRef] [PubMed]
- Zörb, C.; Geilfus, C.M.; Dietz, K.J. Salinity and crop yield. Plant Biol. 2019, 21, 31–38. [Google Scholar] [CrossRef]
- Yichie, Y.; Brien, C.; Berger, B.; Roberts, T.H.; Atwell, B.J. Salinity tolerance in Australian wild Oryza species varies widely and matches that observed in O. sativa. .Rice 2018, 11, 66. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, Z.; Chen, T.; Zhao, Q.; Yao, S.; Zhou, L.; Zhao, L.; Zhao, C.; Liang, W.; Lu, K.; et al. Breeding and Characteristics of the New Salt-tolerant Rice Variety Nangengyan No.1 with Fine Quality. Barley Cereal Sci. 2023, 40, 67–70. [Google Scholar] [CrossRef]
- Chang, Q. Let the white-flowering saline land produce golden yields. People’s Daily, 6 November 2024. [Google Scholar]
- Feng, L.F. The ability of technology and science to construct a “black soil granary”. Sci. News 2023, 25, 18–21. [Google Scholar]
- Chartzoulakis, K.; Klapaki, G. Response of two greenhouse pepper hybrids to NaCl salinity during different growth stages. Sci. Hortic. 2000, 86, 247–260. [Google Scholar] [CrossRef]
- Tabassum, R.; Tahjib-Ul-Arif, M.; Hasanuzzaman, M.; Sohag, A.A.M.; Islam, M.S.; Shafi, S.S.H.; Islam, M.M.; Hassan, L. Screening salt-tolerant rice at the seedling and reproductive stages: An effective and reliable approach. Environ. Exp. Bot. 2021, 192, 104629. [Google Scholar] [CrossRef]
- Zhang, F.L. Screening and Identification of Saline Tolerant Germplasms in Rice; Huazhong Agricultural University: Wuhan, China, 2022. [Google Scholar]
- Benabderrahim, M.A.; Guiza, M.; Haddad, M. Genetic diversity of salt-tolerance in tetraploid alfalfa (Medicago sativa L.). Acta Physiol. Plant. 2020, 42, 5. [Google Scholar] [CrossRef]
- Masuda, M.S.; Azad, M.A.K.; Hasanuzzaman, M.; Arifuzzaman, M. Evaluation of salt-tolerance in maize (Zea mays L.) at seedling stage through morphological characters and salt-tolerance index. Plant Physiol. Rep. 2021, 26, 419–427. [Google Scholar] [CrossRef]
- Sultana, R.; Uddin, M.N.; Rahim, M.A.; Fakir, M.S.A. Effect of salt stress on germination and seedling growth in four maize genotypes. Pak. J. Biol. Sci. 2016, 4, 359–360. [Google Scholar]
- Heuer, B.; Meiri, A.; Shalhevet, J. Salt-tolerance of eggplant. Plant Soil 1986, 95, 9–13. [Google Scholar] [CrossRef]
- Savvas, D.; Lenz, F. Influence of NaCl concentration in the nutrient solution on mineral composition of eggplants grown in sand culture. Angew. Bot. 1996, 70, 124–127. [Google Scholar]
- Akinci, I.E.; Akinci, S.; Yilmaz, K.; Dikici, H. Response of eggplant varieties (Solanum melongena) to salinity in germination and seedling stages. N. Z. J. Crop Hortic. Sci. 2004, 32, 193–200. [Google Scholar] [CrossRef]
- Wu, H.; Lin, B.; Li, C.; Liu, X.; Zhang, Z.; Zhong, L. Evaluation of Salt-tolerance of Different Eggplant Varieties at Seedling Stage; Henan Agricultural Science: Zhengzhou, China, 2020. [Google Scholar]
- Brenes, M.; Solana, A.; Boscaiu, M.; Fita, A.; Vicente, O.; Calatayud, Á.; Plazas, M. Physiological and Biochemical Responses to Salt Stress in Cultivated Eggplant (Solanum melongena L.) and in S. insanum L., a Close Wild Relative. Agronomy 2020, 10, 651. [Google Scholar] [CrossRef]
- Hannachi, S.; Van Labeke, M.C. Salt stress affects germination, seedling growth and physiological responses differentially in eggplant cultivars (Solanum melongena L.). Sci. Hortic. 2018, 228, 56–65. [Google Scholar] [CrossRef]
- Xi, J.Y. Evaluation of Maize Germplasm for Salt-Alkali Tolerance at Seedling Stage and Association Analysis; Shandong Agricultural University: Tai’an, China, 2024. [Google Scholar]
- Li, M. Screening and GWAS Analysis of Salt Tolerant Soybean Germplasm Resources; Jilin Agricultural University: Changchun, China, 2024. [Google Scholar]
- Ashraf, M.; Foolad, M.R. Crop breeding for salt-tolerance in the era of molecular markers and marker-assisted selection. Plant Breed. 2013, 132, 10–20. [Google Scholar] [CrossRef]
- Li, Y. Principal Component Analysis and Comprehensive Evaluation of Saline-alkaline Tolerance Related Traits in Rice in Cold Regio; Heilongjiang Bayi Agricultural University: Daqing, China, 2022. [Google Scholar]
- Fernandez George, C.J. Effective selection criteria for assessing stress tolerance. In Proceedings of the International Symposium on Adaptation of Vegetables and Other Food Crops in Temperature and Water Stress; Kuo, C.G., Ed.; Asian Vegetable Research and Development Center: Shanhua, Taiwan, 1992; pp. 257–270. [Google Scholar]
- Zhang, R.; Hussain, S.; Wang, Y.; Liu, Y.; Li, Q.; Chen, Y.; Wei, H.; Gao, P.; Dai, Q. Comprehensive Evaluation of Salt-tolerance in Rice (Oryza sativa L.) Germplasm at the Germination Stage. Agronomy 2021, 11, 1569. [Google Scholar] [CrossRef]
- Choudhary, A.; Kaur, N.; Sharma, A.; Kumar, A. Evaluation and screening of elite wheat germplasm for salinity stress at the seedling phase. Physiol. Plant. 2021, 173, 2207–2215. [Google Scholar] [CrossRef]
- Geng, L.Y. Linkage Analysis and GWAS Reveal QTL ControllingSalt-tolerance in Rice and Candidate Genes Discovery; Chinese Academy of Agricultural Sciences: Beijing, China, 2020. [Google Scholar]
- Cebeci, E.; Boyaci, H.F.; Kiran, S.; Ellialtioglu, S.S. Comprehensive assessment to reveal the salt-tolerance potential of cultivated eggplants and their wild relatives. Front. Plant Sci. 2025, 16, 1483409. [Google Scholar] [CrossRef] [PubMed]
- Tavakoli, M.; Poustini, K.; Besharati, H.; Ali, S. Variable salinity responses of 25 alfalfa genotypes and comparative salt-response ion distribution. Russ. J. Plant Physiol. 2019, 66, 231–239. [Google Scholar] [CrossRef]
- Tian, L.; Chen, Y.; Liu, J.; Ma, X.; Wang, N.; Yang, B.; Li, Y.; Guo, H.; Li, J.; Hu, H.; et al. Comprehensive evaluation and selection of rice (Oryza sativa japonica) germplasm for saline tolerance at germination stage. Chin. J. Rice Sci. 2017, 31, 631. [Google Scholar]
- Chunthaburee, S.; Dongsansuk, A.; Sanitchon, J.; Pattanagul, W.; Theerakulpisut, P. Physiological and biochemical parameters for evaluation and clustering of rice cultivars differing in salt-tolerance at seedling stage. Saudi J. Biol. Sci. 2016, 23, 467–477. [Google Scholar] [CrossRef]
- Zhang, G.W.; Lu, H.L.; Zhang, L.; Chang, B.L.; Zhou, Z.G. Salt-tolerance evaluation of cotton (Gossypium hirsutum) at its germinating and seedling stages and selection of related indices. J. Appl. Ecol. 2011, 22, 2045. [Google Scholar]

| Sequence | Number | Variety Name | Source |
|---|---|---|---|
| 1 | 24QX2 | Shenqie two | Shenyang Fengnong Agriculture |
| 2 | 24QX3 | Shenqie one | Shenyang Fengnong Agriculture |
| 3 | 24QX4 | Heibawang | Shenyang Fengnong Agriculture |
| 4 | 24QX5 | Huakui | Shenyang Jinming Hang Agricultural Technology |
| 5 | 24QX6 | Jinmingchang | Shenyang Jinming Hang Agricultural Technology |
| 6 | 24QX7 | Daguolv | Shenyang Jinming Hang Agricultural Technology |
| 7 | 24QX8 | Ziyue | Jinan Tianren Seed |
| 8 | 24QX9 | Yuanqieguanjun | Jinan Tianren Seed |
| 9 | 24QX10 | Jindunlvguan | Xi’an Kington Seed Technology |
| 10 | 24QX11 | New dahongbao | Xi’an Kington Seed Technology |
| 11 | 24QX12 | Zixianqie | Shenyang Fengnong Agriculture |
| 12 | 24QX13 | Yuanqie307 | Unknown |
| 13 | 24QX14 | Yuanqieheiqiang | Unknown |
| 14 | 24QX15 | Yuanqieheijie | Unknown |
| 15 | 24QX16 | Gaoke189 | Unknown |
| 16 | 24QX18 | Zihong two | Unknown |
| 17 | 24QX19 | Zihong three | Unknown |
| 18 | 24QX20 | Zihong four | Unknown |
| 19 | 24QX21 | Zihong five | Unknown |
| 20 | 24QX22 | Lvqie one | Unknown |
| 21 | 24QX24 | Nanhanheijiaolong | Xi’an Hangfeng Seed Industry |
| 22 | 24QX25 | Changjiangzilong | Xi’an Hangfeng Seed Industry |
| 23 | 24QX26 | Baimawangzi | Xi’an Hangfeng Seed Industry |
| 24 | 24QX27 | Nanhanbaiyuqie | Xi’an Hangfeng Seed Industry |
| 25 | 24QX28 | Heipangzi | Xi’an Hangfeng Seed Industry |
| 26 | 24QX29 | Meijiaozi six | Xi’an Hangfeng Seed Industry |
| 27 | 24QX30 | Zhusiqie | Xi’an Hangfeng Seed Industry |
| 28 | 24QX31 | Zirong six | Guangdong Gannong Agricultural Technology |
| 29 | 24QX32 | Meigui | Guangdong Gannong Agricultural Technology |
| 30 | 24QX33 | Zirong eight | Guangdong Gannong Agricultural Technology |
| 31 | 24QX34 | Feicui | Guangdong Gannong Agricultural Technology |
| 32 | 24QX35 | A116 | Unknown |
| 33 | 24QX36 | Laolunsi | Beijing Zhongnong Kunchuang Seed |
| 34 | 24QX37 | AGC6317 | Unknown |
| 35 | 24QX38 | Fengyuanheichangie | Xi’an Golden Seed Fengyuan Seed Industry |
| 36 | 24QX39 | Fengyuandaminqie | Xi’an Golden Seed Fengyuan Seed Industry |
| 37 | 24QX40 | Fengyuankuaiqie | Xi’an Golden Seed Fengyuan Seed Industry |
| 38 | 24QX41 | Fengyuanlvguanqie | Xi’an Golden Seed Fengyuan Seed Industry |
| 39 | 24QX42 | Fengyuanziguan | Xi’an Golden Seed Fengyuan Seed Industry |
| 40 | 24QX43 | Fengyuanziyuanqie | Xi’an Golden Seed Fengyuan Seed Industry |
| 41 | 24QX44 | Caoqingqie | Henan Dazheng Runhe Seed Industry |
| 42 | 24QX45 | Bailong332 | Fangzhen Agriculture |
| 43 | 24QX47 | Hangqie one | Shouguang Xinxinran Horticulture |
| 44 | 24QX48 | Heibao two | Shouguang Xinxinran Horticulture |
| 45 | 24QX49 | Fenyanhua | Shouguang Xinxinran Horticulture |
| 46 | 24QX51 | Zidaichangqie | Shouguang Xinxinran Horticulture |
| 47 | 24QX52 | Guangshenxianqie | Shouguang Xinxinran Horticulture |
| 48 | 24QX54 | Heiliangzai | Jing Yan Yi Agricultural Seed Technology |
| 49 | 24QX55 | Yuhang | Shouguang Xinxinran Horticulture |
| 50 | 24QX58 | Zili | Xi’an Jinsheng Seed Industry |
| 51 | 24QX59 | Jingqieheiba | Jing Yan Yi Agricultural Seed Technology |
| 52 | 24QX60 | Jingqie130 | Jing Yan Yi Agricultural Seed Technology |
| 53 | 24QX62 | Rongyaochangqie | Shouguang Jinpeng Seed |
| 54 | 24QX63 | Nongda601 | Hebei Agricultural University |
| 55 | 24QX64 | Nongda604 | Hebei Agricultural University |
| 56 | 24QX67 | Nongda702 | Hebei Agricultural University |
| 57 | 24QX68 | Nongda703 | Hebei Agricultural University |
| Sequence | Variety Name | Source | Sequence | Variety Name | Source |
|---|---|---|---|---|---|
| 1 | 24-477 | China | 55 | 21C158 | France |
| 2 | 24-479 | China | 56 | 21C161 | United States of America |
| 3 | 14-345 | China | 57 | 21C162 | United States of America |
| 4 | 16-562 | China | 58 | 21C164 | India |
| 5 | Sanyue | China | 59 | 21C170 | China |
| 6 | HQ1315 | China | 60 | 21C171 | China |
| 7 | 21C1 | Turkey | 61 | 21C172 | China |
| 8 | 21C4 | Russian Federation | 62 | 21C175 | India |
| 9 | 21C6 | India | 63 | 21C180 | China |
| 10 | 21C9 | United Kingdom | 64 | 21C181 | Union of Soviet Soc. Rep. |
| 11 | 21C12 | India | 65 | 21C185 | Unknown |
| 12 | 21C13 | Union of Soviet Soc. Rep. | 66 | 21C186 | Italy |
| 13 | 21C19 | India | 67 | 21C188 | Ukraine |
| 14 | 21C21 | Turkey | 68 | 21C198 | China |
| 15 | 21C26 | Netherlands | 69 | 21C199 | Unknown |
| 16 | 21C28 | Japan | 70 | 21C204 | Turkey |
| 17 | 21C29 | Turkey | 71 | 21C205 | Turkey |
| 18 | 21C32 | Unknown | 72 | 21C207 | Turkey |
| 19 | 21C34 | Unknown | 73 | 21C211 | Turkey |
| 20 | 21C35 | United Kingdom | 74 | 21C214 | Turkey |
| 21 | 21C37 | Turkey | 75 | 21C216 | Turkey |
| 22 | 21C38 | Turkey | 76 | 21C217 | Turkey |
| 23 | 21C41 | Germany | 77 | 21C218 | Turkey |
| 24 | 21C43 | Japan | 78 | 21C219 | Turkey |
| 25 | 21C44 | Nepal | 79 | 21C220 | Turkey |
| 26 | 21C45 | India | 80 | 21C221 | Turkey |
| 27 | 21C46 | India | 81 | 21C222 | Turkey |
| 28 | 21C48 | Malaysia | 82 | 21C226 | Japan |
| 29 | 21C50 | India | 83 | 21C228 | United States of America |
| 30 | 21C51 | Japan | 84 | 21C229 | United States of America |
| 31 | 21C57 | Unknown | 85 | 21C230 | Japan |
| 32 | 21C60 | India | 86 | 21C234 | India |
| 33 | 21C61 | United States of America | 87 | 21C236 | China |
| 34 | 21C64 | India | 88 | 21C241 | Iran (Islamic Republic of) |
| 35 | 21C68 | Turkey | 89 | 21C242 | Iran (Islamic Republic of) |
| 36 | 21C90 | Turkey | 90 | 21C243 | India |
| 37 | 21C91 | Spain | 91 | 21C245 | India |
| 38 | 21C95 | Turkey | 92 | 21C247 | India |
| 39 | 21C104 | Japan | 93 | 21C249 | Turkey |
| 40 | 21C105 | Japan | 94 | 21C250 | Turkey |
| 41 | 21C107 | India | 95 | 21C252 | China |
| 42 | 21C108 | China | 96 | 21C280 | Maldives |
| 43 | 21C111 | Hungary | 97 | 21C288 | Ukraine |
| 44 | 21C126 | India | 98 | 21C291 | Syrian Arab Republic |
| 45 | 21C127 | India | 99 | 21C293 | Syrian Arab Republic |
| 46 | 21C132 | China | 100 | 21C301 | China |
| 47 | 21C135 | Iran (Islamic Republic of) | 101 | 21C302 | China |
| 48 | 21C139 | India | 102 | 21C305 | China |
| 49 | 21C141 | India | 103 | 21C307 | China |
| 50 | 21C144 | Turkey | 104 | 21C316 | China |
| 51 | 21C147 | Turkey | 105 | 21C317 | China |
| 52 | 21C148 | Turkey | 106 | 21C323 | China |
| 53 | 21C154 | India | 107 | 21C331 | China |
| 54 | 21C155 | Unknown | 108 | 21C336 | China |
| Trait | Maximum Value | Minimum Value | Average Value | Standard Deviation | Coefficient of Variation |
|---|---|---|---|---|---|
| PH | 0.74 | 0.24 | 0.456 | 0.092 | 20.12% |
| SD | 0.73 | 0.34 | 0.492 | 0.079 | 15.96% |
| Chl | 1.73 | 0.79 | 1.261 | 0.143 | 11.32% |
| LN | 0.79 | 0.33 | 0.546 | 0.090 | 16.45% |
| LSA | 0.76 | 0.09 | 0.234 | 0.116 | 49.44% |
| RWC | 1.28 | 0.72 | 0.961 | 0.045 | 4.67% |
| SWC | 1.09 | 0.59 | 0.880 | 0.065 | 7.38% |
| LWC | 1.10 | 0.71 | 0.934 | 0.058 | 6.17% |
| MDA | 2.23 | 0.35 | 1.030 | 0.404 | 39.28% |
| RL | 2.05 | 0.25 | 0.663 | 0.220 | 33.16% |
| RTN | 4.12 | 0.21 | 0.968 | 0.424 | 43.80% |
| RSA | 1.61 | 0.12 | 0.504 | 0.216 | 42.79% |
| RV | 0.89 | 0.06 | 0.415 | 0.188 | 45.25% |
| KMO and Bartlett Ball Tests | ||
|---|---|---|
| Kaiser–Meyer–Olkin sampling adequacy measurement | 0.644 | |
| Approx. chi-squared | 807.515 | |
| Bartlett ball test | df | 78 |
| sig | 0.00 | |
| Trait | Principal Component | ||||||
|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | |
| RSA | 0.963 | 0.076 | −0.022 | −0.024 | −0.001 | 0.001 | 0.062 |
| RL | 0.918 | 0.026 | 0.069 | −0.179 | 0.008 | −0.115 | 0.162 |
| RV | 0.892 | 0.172 | −0.107 | 0.069 | 0.026 | 0.149 | −0.1 |
| RTN | 0.551 | −0.042 | 0.407 | −0.277 | −0.386 | −0.067 | 0.34 |
| LSA | 0.142 | 0.762 | −0.04 | −0.094 | 0.17 | 0.113 | 0.225 |
| PH | 0.001 | 0.68 | 0.318 | −0.024 | 0.106 | −0.294 | 0.04 |
| SD | 0.282 | 0.577 | −0.464 | −0.085 | −0.03 | 0.304 | −0.12 |
| LN | 0.020 | 0.132 | 0.813 | 0.208 | 0.031 | 0.165 | −0.175 |
| MDA | −0.108 | −0.123 | 0.15 | 0.909 | −0.024 | 0.073 | −0.006 |
| LWC | 0.007 | 0.272 | 0.098 | 0.02 | 0.813 | −0.033 | 0.084 |
| RWC | 0.003 | 0.45 | 0.204 | 0.406 | −0.538 | −0.055 | −0.03 |
| Chl | −0.006 | −0.011 | 0.1 | 0.066 | −0.001 | 0.936 | 0.042 |
| SWC | 0.096 | 0.156 | −0.12 | −0.001 | 0.083 | 0.04 | 0.925 |
| Eigenvalue | 2.99 | 1.746 | 1.259 | 1.17 | 1.149 | 1.146 | 1.119 |
| Contribution rate | 22.998 | 13.432 | 9.688 | 9.000 | 8.839 | 8.816 | 8.606 |
| Cumulative contribution rate | 22.998 | 36.430 | 46.118 | 55.118 | 63.956 | 72.772 | 81.378 |
| Variety Name | Comprehensive Index | Membership Function Value | D Value | Rank | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PCA1 | PCA2 | PCA3 | PCA4 | PCA5 | PCA6 | PCA7 | UX1 | UX2 | UX3 | UX4 | UX5 | UX6 | UX7 | |||
| 24QX2 | −0.025 | 0.214 | 1.529 | 1.026 | −0.222 | 1.357 | −0.277 | 0.274 | 0.459 | 0.779 | 0.647 | 0.409 | 0.750 | 0.528 | 0.499 | 20 |
| 24QX3 | −0.157 | 0.652 | −0.639 | 1.158 | −0.437 | 0.645 | 0.225 | 0.253 | 0.521 | 0.357 | 0.672 | 0.381 | 0.644 | 0.600 | 0.449 | 50 |
| 24QX4 | −0.915 | 0.387 | 0.466 | 0.225 | −0.490 | −0.206 | −0.487 | 0.137 | 0.483 | 0.572 | 0.495 | 0.374 | 0.518 | 0.498 | 0.391 | 115 |
| 24QX5 | −0.185 | 0.518 | −0.169 | 0.038 | −0.238 | −0.357 | −0.788 | 0.249 | 0.502 | 0.448 | 0.459 | 0.407 | 0.495 | 0.454 | 0.404 | 105 |
| 24QX6 | −0.650 | 0.089 | 0.786 | 0.835 | −0.255 | −0.783 | −0.056 | 0.178 | 0.442 | 0.634 | 0.610 | 0.405 | 0.432 | 0.560 | 0.417 | 91 |
| 24QX7 | −0.948 | −0.379 | 1.685 | −0.265 | 0.484 | −0.208 | −0.387 | 0.132 | 0.376 | 0.809 | 0.402 | 0.502 | 0.518 | 0.512 | 0.405 | 103 |
| 24QX8 | 0.162 | −0.239 | 1.834 | 1.758 | 0.331 | 0.460 | 0.206 | 0.302 | 0.396 | 0.838 | 0.785 | 0.482 | 0.617 | 0.597 | 0.520 | 12 |
| 24QX9 | 1.479 | 1.091 | 1.263 | 1.150 | 0.150 | 2.189 | −0.192 | 0.504 | 0.582 | 0.727 | 0.670 | 0.458 | 0.874 | 0.540 | 0.601 | 1 |
| 24QX10 | −1.338 | −0.567 | 2.184 | 0.194 | −0.042 | 1.600 | 0.403 | 0.073 | 0.350 | 0.907 | 0.489 | 0.433 | 0.786 | 0.626 | 0.439 | 61 |
| 24QX11 | −0.312 | −0.094 | 0.616 | 1.059 | 0.060 | 0.988 | −0.498 | 0.230 | 0.416 | 0.601 | 0.653 | 0.447 | 0.695 | 0.496 | 0.454 | 45 |
| 24QX12 | −0.118 | −0.958 | 0.638 | 2.319 | 0.293 | 1.307 | −0.162 | 0.259 | 0.295 | 0.605 | 0.892 | 0.477 | 0.743 | 0.545 | 0.483 | 31 |
| 24QX13 | 0.930 | −0.012 | 1.004 | 0.060 | 0.205 | 0.440 | −2.925 | 0.420 | 0.428 | 0.677 | 0.464 | 0.466 | 0.614 | 0.147 | 0.454 | 46 |
| 24QX14 | 0.739 | 0.092 | −0.055 | 1.616 | 0.088 | 0.755 | −0.080 | 0.391 | 0.442 | 0.470 | 0.759 | 0.450 | 0.661 | 0.556 | 0.503 | 19 |
| 24QX15 | 0.094 | −0.037 | −0.402 | 1.398 | 0.267 | 2.715 | −0.127 | 0.292 | 0.424 | 0.403 | 0.717 | 0.474 | 0.951 | 0.549 | 0.493 | 24 |
| 24QX16 | −1.125 | 0.429 | 0.864 | 0.475 | −0.296 | 0.832 | −0.178 | 0.105 | 0.489 | 0.649 | 0.542 | 0.400 | 0.672 | 0.542 | 0.422 | 79 |
| 24QX18 | −1.400 | −0.443 | 1.583 | 2.103 | −0.009 | 0.507 | 0.250 | 0.063 | 0.367 | 0.789 | 0.851 | 0.437 | 0.624 | 0.604 | 0.446 | 54 |
| 24QX19 | 0.445 | −1.271 | 2.232 | 0.390 | 0.909 | 0.622 | −0.686 | 0.346 | 0.252 | 0.916 | 0.526 | 0.558 | 0.641 | 0.469 | 0.487 | 28 |
| 24QX20 | 0.066 | −0.050 | 1.038 | −0.140 | −0.448 | −0.785 | −0.535 | 0.287 | 0.422 | 0.683 | 0.426 | 0.380 | 0.432 | 0.491 | 0.420 | 87 |
| 24QX21 | 1.011 | 0.131 | −0.184 | 1.426 | −1.203 | −1.108 | −0.702 | 0.432 | 0.448 | 0.445 | 0.722 | 0.280 | 0.384 | 0.467 | 0.451 | 48 |
| 24QX22 | −0.912 | −0.696 | 1.507 | −0.193 | −0.890 | 0.158 | −0.325 | 0.138 | 0.332 | 0.775 | 0.416 | 0.321 | 0.572 | 0.521 | 0.384 | 124 |
| 24QX24 | −0.632 | 0.073 | 1.352 | −0.027 | −0.428 | −0.123 | 0.269 | 0.181 | 0.440 | 0.745 | 0.447 | 0.382 | 0.530 | 0.607 | 0.425 | 75 |
| 24QX25 | −0.158 | −0.693 | 0.059 | −0.071 | −0.124 | −0.869 | −0.855 | 0.253 | 0.332 | 0.493 | 0.439 | 0.422 | 0.420 | 0.445 | 0.372 | 133 |
| 24QX26 | 1.199 | 1.435 | 0.431 | −0.602 | −0.790 | 0.677 | −1.516 | 0.461 | 0.630 | 0.565 | 0.338 | 0.335 | 0.649 | 0.350 | 0.483 | 32 |
| 24QX27 | −0.312 | −0.289 | −0.462 | 0.040 | −0.290 | 0.807 | −0.806 | 0.230 | 0.389 | 0.391 | 0.460 | 0.400 | 0.668 | 0.452 | 0.390 | 117 |
| 24QX28 | 0.131 | −0.505 | 1.324 | −0.161 | −0.663 | 0.698 | −0.429 | 0.297 | 0.359 | 0.739 | 0.422 | 0.351 | 0.652 | 0.506 | 0.440 | 60 |
| 24QX29 | 0.033 | 0.202 | −0.947 | 0.758 | −0.226 | 0.838 | −1.697 | 0.282 | 0.458 | 0.297 | 0.596 | 0.409 | 0.673 | 0.323 | 0.408 | 99 |
| 24QX30 | −0.170 | −0.941 | 0.918 | −0.765 | 0.612 | 0.319 | −2.858 | 0.251 | 0.298 | 0.660 | 0.307 | 0.519 | 0.596 | 0.156 | 0.370 | 135 |
| 24QX31 | −1.086 | −0.327 | 0.448 | 0.807 | −0.680 | −1.020 | −1.555 | 0.111 | 0.384 | 0.568 | 0.605 | 0.349 | 0.397 | 0.344 | 0.347 | 154 |
| 24QX32 | −0.753 | −0.362 | 0.416 | −0.480 | −0.216 | −0.457 | −2.676 | 0.162 | 0.379 | 0.562 | 0.361 | 0.410 | 0.481 | 0.183 | 0.331 | 156 |
| 24QX33 | 0.430 | 0.456 | 1.452 | 0.316 | −0.751 | −1.374 | −0.797 | 0.343 | 0.493 | 0.764 | 0.512 | 0.340 | 0.345 | 0.453 | 0.449 | 51 |
| 24QX34 | −0.764 | −0.819 | 1.142 | 0.940 | 0.185 | 0.697 | −0.795 | 0.160 | 0.315 | 0.703 | 0.630 | 0.463 | 0.652 | 0.453 | 0.420 | 85 |
| 24QX35 | −0.307 | −0.506 | 0.588 | 0.015 | −0.380 | −0.538 | −0.721 | 0.230 | 0.359 | 0.596 | 0.455 | 0.389 | 0.469 | 0.464 | 0.388 | 120 |
| 24QX36 | −1.013 | 0.593 | 0.061 | 0.700 | −0.698 | −0.880 | −0.559 | 0.122 | 0.512 | 0.493 | 0.585 | 0.347 | 0.418 | 0.487 | 0.377 | 131 |
| 24QX37 | −0.995 | −0.480 | −0.176 | 1.736 | −0.091 | −1.197 | −0.197 | 0.125 | 0.362 | 0.447 | 0.781 | 0.427 | 0.371 | 0.539 | 0.379 | 129 |
| 24QX38 | −0.133 | −0.284 | 0.484 | 1.668 | 0.051 | −0.835 | −0.754 | 0.257 | 0.390 | 0.575 | 0.768 | 0.445 | 0.425 | 0.459 | 0.434 | 65 |
| 24QX39 | 1.354 | 0.301 | 0.723 | 0.203 | −0.146 | −0.464 | −1.597 | 0.485 | 0.471 | 0.622 | 0.491 | 0.419 | 0.480 | 0.338 | 0.477 | 34 |
| 24QX40 | −0.715 | −1.405 | 1.505 | 1.267 | 0.854 | 0.769 | 0.184 | 0.168 | 0.233 | 0.774 | 0.692 | 0.551 | 0.663 | 0.594 | 0.450 | 49 |
| 24QX41 | −1.702 | −0.490 | 1.688 | 1.412 | 0.494 | −0.139 | 0.481 | 0.017 | 0.361 | 0.810 | 0.720 | 0.504 | 0.528 | 0.637 | 0.420 | 84 |
| 24QX42 | −0.196 | 0.378 | 0.830 | 0.678 | 0.254 | 0.326 | 0.132 | 0.247 | 0.482 | 0.643 | 0.581 | 0.472 | 0.597 | 0.587 | 0.469 | 39 |
| 24QX43 | −0.105 | −0.668 | 1.471 | 1.271 | 0.651 | −0.837 | 0.394 | 0.261 | 0.336 | 0.768 | 0.693 | 0.524 | 0.424 | 0.625 | 0.467 | 40 |
| 24QX44 | −0.557 | −0.235 | 0.478 | 0.667 | −0.024 | −1.369 | 0.619 | 0.192 | 0.397 | 0.574 | 0.579 | 0.435 | 0.345 | 0.657 | 0.407 | 100 |
| 24QX45 | 0.631 | −1.052 | 0.283 | 2.607 | 0.087 | 0.513 | 0.028 | 0.374 | 0.282 | 0.536 | 0.946 | 0.450 | 0.625 | 0.572 | 0.498 | 21 |
| 24QX47 | 0.209 | −0.709 | −0.277 | 2.374 | 0.109 | 1.258 | 0.447 | 0.309 | 0.330 | 0.427 | 0.902 | 0.453 | 0.735 | 0.632 | 0.489 | 27 |
| 24QX48 | −0.334 | 0.297 | −0.645 | 0.858 | 0.090 | −0.998 | −0.255 | 0.226 | 0.471 | 0.355 | 0.615 | 0.450 | 0.400 | 0.531 | 0.401 | 108 |
| 24QX49 | 0.509 | −0.353 | −1.274 | 0.845 | −0.229 | 3.042 | 0.246 | 0.355 | 0.380 | 0.233 | 0.613 | 0.408 | 1.000 | 0.603 | 0.475 | 35 |
| 24QX51 | −0.586 | 0.809 | −0.220 | 0.066 | −1.107 | 0.453 | 0.603 | 0.188 | 0.543 | 0.438 | 0.465 | 0.293 | 0.616 | 0.655 | 0.414 | 93 |
| 24QX52 | 0.927 | −0.249 | −0.687 | 0.385 | 0.411 | 0.046 | −0.060 | 0.419 | 0.395 | 0.347 | 0.525 | 0.493 | 0.555 | 0.559 | 0.456 | 44 |
| 24QX54 | −1.216 | −1.128 | −0.090 | 0.498 | 0.764 | −0.657 | 0.936 | 0.091 | 0.272 | 0.463 | 0.547 | 0.539 | 0.451 | 0.703 | 0.368 | 138 |
| 24QX55 | −0.519 | −0.793 | 0.098 | 0.864 | −0.862 | −0.398 | 0.857 | 0.198 | 0.318 | 0.500 | 0.616 | 0.325 | 0.489 | 0.691 | 0.398 | 109 |
| 24QX58 | −0.170 | 0.391 | 0.101 | 2.184 | −1.933 | −0.531 | −0.093 | 0.251 | 0.484 | 0.501 | 0.866 | 0.184 | 0.470 | 0.554 | 0.436 | 62 |
| 24QX59 | −1.014 | 0.960 | −0.190 | 1.219 | −0.658 | −0.189 | 0.779 | 0.122 | 0.564 | 0.444 | 0.683 | 0.352 | 0.520 | 0.680 | 0.423 | 78 |
| 24QX60 | 0.024 | −0.212 | −0.091 | 1.196 | −1.254 | 0.022 | −1.285 | 0.281 | 0.400 | 0.463 | 0.679 | 0.273 | 0.552 | 0.383 | 0.406 | 102 |
| 24QX62 | 0.746 | 0.391 | 0.092 | −0.204 | −2.214 | −0.295 | −1.337 | 0.392 | 0.484 | 0.499 | 0.414 | 0.147 | 0.505 | 0.375 | 0.406 | 101 |
| 24QX63 | −0.646 | −1.055 | −1.135 | 1.107 | −1.980 | 0.574 | 1.184 | 0.179 | 0.282 | 0.260 | 0.662 | 0.178 | 0.634 | 0.738 | 0.367 | 141 |
| 24QX64 | 1.119 | −0.688 | 0.518 | 2.890 | −1.388 | −0.827 | 1.301 | 0.449 | 0.333 | 0.582 | 1.000 | 0.256 | 0.426 | 0.755 | 0.516 | 13 |
| 24QX67 | −0.955 | −0.171 | −1.217 | 0.100 | −0.988 | 0.441 | 1.290 | 0.131 | 0.405 | 0.244 | 0.471 | 0.308 | 0.614 | 0.754 | 0.365 | 143 |
| 24QX68 | −0.814 | −0.380 | −0.504 | −0.215 | −0.759 | 0.126 | 0.237 | 0.153 | 0.376 | 0.383 | 0.412 | 0.338 | 0.567 | 0.602 | 0.358 | 145 |
| 24-477 | −0.557 | −0.455 | 1.549 | 0.025 | 0.426 | −0.293 | −0.381 | 0.192 | 0.366 | 0.783 | 0.457 | 0.495 | 0.505 | 0.513 | 0.421 | 82 |
| 24-479 | −0.339 | −1.073 | 0.081 | −0.314 | 0.673 | −1.223 | −1.611 | 0.225 | 0.279 | 0.497 | 0.393 | 0.527 | 0.367 | 0.336 | 0.345 | 155 |
| 14-345 | 0.470 | −0.510 | −0.549 | 0.751 | 0.059 | −0.088 | −1.040 | 0.349 | 0.358 | 0.374 | 0.595 | 0.446 | 0.535 | 0.418 | 0.419 | 88 |
| 16-562 | 1.638 | 0.916 | −0.127 | 0.657 | −0.305 | 0.195 | −1.118 | 0.528 | 0.557 | 0.456 | 0.577 | 0.398 | 0.577 | 0.407 | 0.509 | 16 |
| Sanyue | −0.747 | −0.910 | −0.725 | −1.861 | −3.328 | 0.545 | −0.320 | 0.163 | 0.302 | 0.340 | 0.100 | 0.000 | 0.629 | 0.522 | 0.271 | 164 |
| HQ1315 | −0.284 | −0.309 | 0.078 | −1.252 | −1.287 | −0.139 | −0.655 | 0.234 | 0.386 | 0.496 | 0.215 | 0.269 | 0.528 | 0.474 | 0.349 | 151 |
| 21C1 | −0.488 | 1.621 | −1.616 | −0.488 | −0.388 | 0.954 | 0.067 | 0.203 | 0.656 | 0.166 | 0.360 | 0.387 | 0.690 | 0.577 | 0.403 | 106 |
| 21C4 | −1.600 | −0.301 | 0.554 | −1.361 | −0.010 | −2.174 | −1.105 | 0.032 | 0.387 | 0.589 | 0.194 | 0.437 | 0.226 | 0.409 | 0.280 | 161 |
| 21C6 | 0.826 | −0.111 | −2.469 | −0.841 | −0.421 | 1.434 | −1.007 | 0.404 | 0.414 | 0.000 | 0.293 | 0.383 | 0.761 | 0.423 | 0.384 | 125 |
| 21C9 | 0.165 | −0.262 | −0.660 | −0.977 | −0.395 | −1.295 | −2.132 | 0.303 | 0.393 | 0.352 | 0.267 | 0.386 | 0.356 | 0.261 | 0.330 | 157 |
| 21C12 | 1.046 | −1.084 | 1.074 | −1.029 | −0.584 | 1.242 | −2.100 | 0.438 | 0.278 | 0.690 | 0.257 | 0.362 | 0.733 | 0.266 | 0.427 | 72 |
| 21C13 | −0.912 | 1.400 | 1.355 | −1.439 | −0.345 | 0.742 | −0.279 | 0.138 | 0.625 | 0.745 | 0.180 | 0.393 | 0.659 | 0.528 | 0.421 | 83 |
| 21C19 | −1.053 | −0.162 | −0.275 | −0.288 | 0.496 | 1.033 | 1.231 | 0.116 | 0.407 | 0.427 | 0.398 | 0.504 | 0.702 | 0.745 | 0.405 | 104 |
| 21C21 | −0.141 | −0.180 | −0.112 | −0.471 | 0.060 | 0.660 | 0.069 | 0.256 | 0.404 | 0.459 | 0.363 | 0.446 | 0.647 | 0.578 | 0.414 | 94 |
| 21C26 | −0.010 | −0.119 | −0.808 | −0.598 | −0.294 | −1.066 | 0.692 | 0.276 | 0.413 | 0.324 | 0.339 | 0.400 | 0.390 | 0.667 | 0.379 | 128 |
| 21C28 | −0.128 | 0.044 | −0.951 | −0.456 | −0.711 | 0.698 | −0.182 | 0.258 | 0.435 | 0.296 | 0.366 | 0.345 | 0.652 | 0.542 | 0.386 | 122 |
| 21C29 | −0.553 | 0.674 | −0.532 | −1.066 | −0.693 | 0.815 | −0.142 | 0.193 | 0.524 | 0.377 | 0.250 | 0.347 | 0.670 | 0.547 | 0.382 | 127 |
| 21C32 | 1.575 | −0.660 | −0.728 | 0.282 | −0.644 | 0.639 | −0.064 | 0.519 | 0.337 | 0.339 | 0.506 | 0.354 | 0.643 | 0.559 | 0.466 | 41 |
| 21C34 | −0.618 | −0.061 | −0.515 | −0.167 | −0.442 | 0.058 | 0.804 | 0.183 | 0.421 | 0.381 | 0.421 | 0.380 | 0.557 | 0.684 | 0.387 | 121 |
| 21C35 | 0.860 | −0.711 | −0.247 | −1.131 | −0.014 | 0.030 | 0.666 | 0.409 | 0.330 | 0.433 | 0.238 | 0.437 | 0.553 | 0.664 | 0.426 | 73 |
| 21C37 | 2.134 | 1.015 | −0.077 | −1.024 | 0.292 | 0.850 | −0.175 | 0.604 | 0.571 | 0.466 | 0.258 | 0.477 | 0.675 | 0.543 | 0.532 | 9 |
| 21C38 | 3.279 | 0.519 | −0.471 | −0.294 | −0.882 | −1.697 | 0.562 | 0.779 | 0.502 | 0.389 | 0.397 | 0.322 | 0.297 | 0.649 | 0.530 | 10 |
| 21C41 | −1.269 | 0.271 | −0.429 | −1.290 | −1.189 | 1.786 | −0.108 | 0.083 | 0.467 | 0.397 | 0.208 | 0.282 | 0.814 | 0.552 | 0.348 | 152 |
| 21C43 | −1.812 | −0.333 | −1.158 | −0.727 | −1.717 | 0.177 | 0.259 | 0.000 | 0.383 | 0.255 | 0.315 | 0.212 | 0.575 | 0.605 | 0.278 | 163 |
| 21C44 | −1.775 | −1.415 | 0.270 | −1.407 | 0.497 | −0.684 | 0.593 | 0.006 | 0.231 | 0.534 | 0.186 | 0.504 | 0.447 | 0.653 | 0.296 | 160 |
| 21C45 | −0.690 | −0.863 | −1.566 | −0.580 | 0.565 | −0.710 | −0.817 | 0.172 | 0.309 | 0.176 | 0.342 | 0.513 | 0.443 | 0.450 | 0.310 | 158 |
| 21C46 | −1.214 | −0.465 | 0.245 | −0.670 | −0.565 | 1.944 | −0.214 | 0.091 | 0.364 | 0.529 | 0.325 | 0.364 | 0.837 | 0.537 | 0.372 | 134 |
| 21C48 | 0.849 | 0.432 | 0.174 | −0.943 | −1.297 | −0.981 | −0.789 | 0.407 | 0.490 | 0.515 | 0.274 | 0.268 | 0.403 | 0.454 | 0.409 | 98 |
| 21C50 | 0.692 | 0.648 | −1.893 | −0.252 | −0.169 | 1.785 | −0.341 | 0.383 | 0.520 | 0.112 | 0.405 | 0.416 | 0.814 | 0.519 | 0.441 | 59 |
| 21C51 | −0.335 | 0.244 | −1.210 | −0.906 | −0.689 | 1.079 | −0.513 | 0.226 | 0.463 | 0.245 | 0.281 | 0.348 | 0.709 | 0.494 | 0.368 | 140 |
| 21C57 | 0.485 | −1.641 | 0.349 | −0.552 | −1.836 | 0.152 | 1.063 | 0.352 | 0.200 | 0.549 | 0.348 | 0.197 | 0.571 | 0.721 | 0.396 | 110 |
| 21C60 | −1.507 | 0.252 | −1.726 | −0.101 | −2.653 | −3.695 | 0.743 | 0.047 | 0.465 | 0.145 | 0.433 | 0.089 | 0.000 | 0.675 | 0.236 | 165 |
| 21C61 | −0.703 | 0.126 | 0.292 | −0.314 | 0.079 | 0.036 | 1.521 | 0.170 | 0.447 | 0.538 | 0.393 | 0.449 | 0.554 | 0.787 | 0.422 | 81 |
| 21C64 | −0.191 | 0.502 | 0.433 | −0.215 | 0.644 | −0.803 | −0.709 | 0.248 | 0.500 | 0.565 | 0.412 | 0.523 | 0.429 | 0.466 | 0.418 | 89 |
| 21C68 | 1.279 | −0.825 | −0.466 | 0.200 | 0.171 | −0.914 | 0.295 | 0.473 | 0.314 | 0.390 | 0.490 | 0.461 | 0.413 | 0.610 | 0.446 | 53 |
| 21C90 | 0.814 | 0.116 | −0.176 | −0.605 | −0.060 | −1.149 | −0.142 | 0.402 | 0.446 | 0.447 | 0.338 | 0.431 | 0.378 | 0.547 | 0.424 | 76 |
| 21C91 | 0.639 | 0.092 | 1.532 | −0.549 | 0.700 | 0.667 | 0.460 | 0.375 | 0.442 | 0.780 | 0.348 | 0.531 | 0.648 | 0.634 | 0.506 | 18 |
| 21C95 | 0.347 | 0.966 | 0.148 | 1.261 | 0.828 | 0.321 | 0.451 | 0.331 | 0.564 | 0.510 | 0.691 | 0.548 | 0.596 | 0.633 | 0.515 | 14 |
| 21C104 | 0.300 | −1.730 | −0.760 | −1.132 | 1.856 | 0.086 | 0.135 | 0.323 | 0.187 | 0.333 | 0.238 | 0.683 | 0.561 | 0.587 | 0.386 | 123 |
| 21C105 | 0.695 | −2.140 | 0.457 | −0.755 | 1.563 | 0.294 | 0.117 | 0.384 | 0.130 | 0.570 | 0.309 | 0.645 | 0.592 | 0.585 | 0.428 | 69 |
| 21C107 | 0.254 | 1.002 | −0.557 | −0.916 | 0.194 | 0.826 | 0.215 | 0.316 | 0.569 | 0.372 | 0.279 | 0.464 | 0.671 | 0.599 | 0.445 | 56 |
| 21C108 | −1.153 | −0.680 | 0.918 | −1.348 | 0.096 | 0.480 | 0.640 | 0.101 | 0.334 | 0.660 | 0.197 | 0.451 | 0.620 | 0.660 | 0.370 | 136 |
| 21C111 | −1.059 | −0.480 | 0.776 | −1.053 | 0.861 | 0.623 | 0.633 | 0.115 | 0.362 | 0.632 | 0.253 | 0.552 | 0.641 | 0.659 | 0.395 | 111 |
| 21C126 | 0.656 | 0.671 | 0.710 | −1.282 | 0.461 | −1.478 | −0.286 | 0.378 | 0.523 | 0.619 | 0.209 | 0.499 | 0.329 | 0.527 | 0.436 | 63 |
| 21C127 | 0.096 | 0.489 | 0.506 | −0.966 | 0.914 | −0.301 | −0.336 | 0.292 | 0.498 | 0.580 | 0.269 | 0.559 | 0.504 | 0.519 | 0.434 | 64 |
| 21C132 | −0.304 | −0.076 | −1.310 | −0.319 | 0.204 | −1.848 | 0.131 | 0.231 | 0.419 | 0.226 | 0.392 | 0.466 | 0.274 | 0.587 | 0.347 | 153 |
| 21C135 | 0.176 | 0.107 | −0.936 | −0.251 | −0.048 | 2.327 | −0.840 | 0.304 | 0.444 | 0.299 | 0.405 | 0.432 | 0.894 | 0.447 | 0.431 | 66 |
| 21C139 | 0.171 | −0.845 | −0.839 | −0.788 | −0.105 | −0.124 | 0.248 | 0.304 | 0.311 | 0.317 | 0.303 | 0.425 | 0.530 | 0.603 | 0.376 | 132 |
| 21C141 | −0.560 | 1.244 | −0.327 | −1.044 | −0.644 | 0.611 | −0.464 | 0.192 | 0.603 | 0.417 | 0.254 | 0.354 | 0.639 | 0.501 | 0.392 | 114 |
| 21C144 | −0.476 | 1.307 | −0.310 | −0.239 | 0.793 | 0.395 | 0.420 | 0.205 | 0.612 | 0.421 | 0.407 | 0.543 | 0.607 | 0.628 | 0.445 | 55 |
| 21C147 | −0.814 | −2.426 | −0.077 | −1.944 | 4.260 | 0.528 | 0.038 | 0.153 | 0.090 | 0.466 | 0.084 | 1.000 | 0.627 | 0.573 | 0.360 | 144 |
| 21C148 | 1.504 | −0.356 | 0.335 | 0.351 | 1.418 | −0.049 | 0.548 | 0.508 | 0.380 | 0.546 | 0.519 | 0.625 | 0.541 | 0.647 | 0.524 | 11 |
| 21C154 | −0.603 | −0.226 | −1.065 | 0.093 | 1.242 | 0.095 | 1.448 | 0.185 | 0.398 | 0.273 | 0.470 | 0.602 | 0.563 | 0.776 | 0.411 | 97 |
| 21C155 | −0.178 | −0.997 | −0.130 | −1.091 | 2.555 | −0.723 | −0.112 | 0.250 | 0.290 | 0.456 | 0.246 | 0.775 | 0.441 | 0.552 | 0.391 | 116 |
| 21C158 | −0.506 | −0.429 | −0.110 | −0.840 | 0.774 | 1.408 | 0.742 | 0.200 | 0.369 | 0.460 | 0.293 | 0.541 | 0.757 | 0.675 | 0.417 | 90 |
| 21C161 | 0.198 | −1.027 | 0.633 | 0.437 | 0.426 | −0.129 | 0.945 | 0.308 | 0.286 | 0.604 | 0.535 | 0.495 | 0.529 | 0.704 | 0.451 | 47 |
| 21C162 | −0.080 | −0.460 | −1.531 | −0.303 | −0.259 | 0.008 | 0.853 | 0.265 | 0.365 | 0.183 | 0.395 | 0.404 | 0.550 | 0.691 | 0.377 | 130 |
| 21C164 | −1.027 | −0.944 | −0.135 | −1.091 | −0.477 | −0.081 | 1.976 | 0.120 | 0.297 | 0.455 | 0.246 | 0.376 | 0.537 | 0.852 | 0.354 | 148 |
| 21C170 | −0.546 | 0.332 | 0.805 | −1.232 | −1.270 | 0.582 | 1.377 | 0.194 | 0.476 | 0.638 | 0.219 | 0.271 | 0.635 | 0.766 | 0.413 | 95 |
| 21C171 | 0.279 | −1.546 | −0.754 | −0.161 | −2.014 | 1.168 | 1.253 | 0.320 | 0.213 | 0.334 | 0.422 | 0.173 | 0.722 | 0.748 | 0.388 | 119 |
| 21C172 | −0.087 | −0.686 | −0.847 | −1.023 | −1.970 | 0.418 | 0.783 | 0.264 | 0.333 | 0.316 | 0.258 | 0.179 | 0.611 | 0.681 | 0.354 | 149 |
| 21C175 | 1.390 | −0.151 | 0.192 | −0.929 | −0.826 | 0.658 | −0.490 | 0.490 | 0.408 | 0.518 | 0.276 | 0.330 | 0.646 | 0.497 | 0.457 | 43 |
| 21C180 | −1.219 | −0.396 | 0.207 | −0.978 | −0.490 | −0.078 | 0.943 | 0.091 | 0.374 | 0.521 | 0.267 | 0.374 | 0.537 | 0.704 | 0.352 | 150 |
| 21C181 | 0.361 | −0.739 | −0.833 | 0.238 | −0.869 | 0.881 | 0.764 | 0.333 | 0.326 | 0.319 | 0.497 | 0.324 | 0.679 | 0.678 | 0.422 | 80 |
| 21C185 | 0.467 | −1.840 | −1.392 | 2.246 | −0.006 | −0.814 | 0.720 | 0.349 | 0.172 | 0.210 | 0.878 | 0.438 | 0.428 | 0.671 | 0.415 | 92 |
| 21C186 | 3.769 | −2.441 | 0.945 | 0.532 | −1.200 | 0.191 | 1.418 | 0.854 | 0.088 | 0.665 | 0.553 | 0.280 | 0.577 | 0.772 | 0.572 | 4 |
| 21C188 | 4.719 | −3.070 | 2.664 | −2.387 | −1.208 | −2.768 | 2.380 | 1.000 | 0.000 | 1.000 | 0.000 | 0.279 | 0.138 | 0.910 | 0.544 | 7 |
| 21C198 | −0.398 | −0.808 | −0.057 | −0.964 | 0.058 | −0.459 | −3.944 | 0.216 | 0.316 | 0.470 | 0.270 | 0.446 | 0.480 | 0.000 | 0.300 | 159 |
| 21C199 | −0.134 | −1.406 | −2.103 | −0.049 | −0.055 | 1.041 | 0.252 | 0.257 | 0.233 | 0.071 | 0.443 | 0.431 | 0.703 | 0.604 | 0.356 | 147 |
| 21C204 | 0.263 | −0.786 | −0.870 | −0.823 | 0.408 | −0.585 | −0.452 | 0.318 | 0.319 | 0.311 | 0.296 | 0.492 | 0.462 | 0.503 | 0.369 | 137 |
| 21C205 | 0.376 | −0.447 | −1.127 | −0.217 | 1.051 | −1.473 | −2.137 | 0.335 | 0.367 | 0.261 | 0.411 | 0.577 | 0.330 | 0.260 | 0.358 | 146 |
| 21C207 | −0.061 | −0.506 | −1.406 | 1.254 | 0.098 | −0.089 | −0.949 | 0.268 | 0.359 | 0.207 | 0.690 | 0.451 | 0.535 | 0.431 | 0.389 | 118 |
| 21C211 | −0.295 | −1.038 | −0.560 | 0.504 | 0.920 | 0.056 | −1.116 | 0.232 | 0.284 | 0.372 | 0.548 | 0.560 | 0.557 | 0.407 | 0.382 | 126 |
| 21C214 | −1.014 | −0.751 | −1.991 | −0.479 | 0.612 | −1.166 | −1.089 | 0.122 | 0.324 | 0.093 | 0.362 | 0.519 | 0.376 | 0.411 | 0.280 | 162 |
| 21C216 | −0.542 | 0.981 | −0.631 | 0.427 | 1.194 | −0.589 | −0.141 | 0.194 | 0.566 | 0.358 | 0.533 | 0.596 | 0.461 | 0.548 | 0.423 | 77 |
| 21C217 | 0.079 | −0.184 | −1.801 | 1.497 | 1.045 | −1.063 | 0.583 | 0.290 | 0.404 | 0.130 | 0.736 | 0.576 | 0.391 | 0.652 | 0.420 | 86 |
| 21C218 | −0.617 | −0.091 | −1.504 | 0.440 | 1.639 | −1.743 | 0.182 | 0.183 | 0.417 | 0.188 | 0.536 | 0.655 | 0.290 | 0.594 | 0.368 | 139 |
| 21C219 | 0.761 | 1.908 | −1.790 | 0.448 | 1.371 | 0.895 | −0.629 | 0.394 | 0.696 | 0.132 | 0.537 | 0.619 | 0.681 | 0.477 | 0.493 | 23 |
| 21C220 | −0.069 | −0.192 | −0.696 | 0.738 | 1.855 | −1.880 | 1.036 | 0.267 | 0.402 | 0.345 | 0.592 | 0.683 | 0.269 | 0.717 | 0.428 | 70 |
| 21C221 | −1.207 | 0.524 | −0.593 | 0.802 | 1.815 | −1.261 | 0.612 | 0.093 | 0.503 | 0.365 | 0.604 | 0.678 | 0.361 | 0.656 | 0.402 | 107 |
| 21C222 | 2.012 | 1.522 | −1.012 | 1.856 | 2.612 | 0.031 | −0.454 | 0.585 | 0.642 | 0.284 | 0.804 | 0.783 | 0.553 | 0.503 | 0.593 | 2 |
| 21C226 | −0.593 | 0.861 | 0.040 | 0.444 | 2.021 | −1.113 | 0.537 | 0.187 | 0.550 | 0.489 | 0.536 | 0.705 | 0.383 | 0.645 | 0.448 | 52 |
| 21C228 | −0.280 | 1.188 | −0.258 | 0.678 | 0.830 | −0.588 | 0.616 | 0.235 | 0.595 | 0.431 | 0.581 | 0.548 | 0.461 | 0.657 | 0.459 | 42 |
| 21C229 | −0.223 | 0.225 | −0.137 | −0.374 | 0.400 | −0.043 | 0.767 | 0.243 | 0.461 | 0.454 | 0.381 | 0.491 | 0.542 | 0.678 | 0.425 | 74 |
| 21C230 | 0.541 | 1.709 | −0.238 | −0.603 | 0.032 | −0.710 | −2.945 | 0.360 | 0.668 | 0.435 | 0.338 | 0.443 | 0.443 | 0.144 | 0.413 | 96 |
| 21C234 | −0.101 | 1.284 | −1.158 | −0.669 | 0.475 | −0.178 | 1.166 | 0.262 | 0.609 | 0.255 | 0.325 | 0.501 | 0.522 | 0.736 | 0.430 | 67 |
| 21C236 | 0.070 | −0.738 | 0.030 | −0.942 | −0.146 | −0.945 | 1.268 | 0.288 | 0.326 | 0.487 | 0.274 | 0.419 | 0.408 | 0.750 | 0.393 | 113 |
| 21C241 | 1.359 | 2.410 | 2.640 | −1.419 | 0.161 | −0.650 | 0.008 | 0.486 | 0.766 | 0.995 | 0.183 | 0.460 | 0.452 | 0.569 | 0.562 | 5 |
| 21C242 | 2.268 | 2.183 | −1.014 | −0.741 | 0.657 | 0.049 | 0.676 | 0.625 | 0.735 | 0.283 | 0.312 | 0.525 | 0.556 | 0.665 | 0.554 | 6 |
| 21C243 | 0.111 | 2.005 | −0.571 | 0.442 | 0.435 | −0.309 | 0.716 | 0.294 | 0.710 | 0.370 | 0.536 | 0.496 | 0.503 | 0.671 | 0.483 | 30 |
| 21C245 | 0.560 | 0.791 | −0.239 | −0.817 | 0.043 | 0.488 | 3.002 | 0.363 | 0.540 | 0.434 | 0.297 | 0.444 | 0.621 | 1.000 | 0.498 | 22 |
| 21C247 | 0.366 | 1.088 | 0.391 | −0.414 | 0.087 | 0.801 | 0.113 | 0.333 | 0.582 | 0.557 | 0.374 | 0.450 | 0.667 | 0.584 | 0.481 | 33 |
| 21C249 | 0.103 | 1.222 | 0.533 | −0.311 | 0.128 | −0.028 | 0.356 | 0.293 | 0.600 | 0.585 | 0.393 | 0.455 | 0.544 | 0.619 | 0.469 | 38 |
| 21C250 | −0.060 | 1.044 | −0.875 | −0.550 | −0.023 | 0.779 | 1.148 | 0.268 | 0.575 | 0.310 | 0.348 | 0.436 | 0.664 | 0.733 | 0.443 | 57 |
| 21C252 | 1.473 | 0.360 | −0.500 | −0.809 | 0.703 | 0.325 | 0.479 | 0.503 | 0.480 | 0.384 | 0.299 | 0.531 | 0.597 | 0.637 | 0.490 | 26 |
| 21C280 | 0.754 | 0.063 | −0.233 | 0.212 | 0.958 | −0.016 | 0.962 | 0.393 | 0.438 | 0.436 | 0.492 | 0.565 | 0.546 | 0.706 | 0.485 | 29 |
| 21C288 | 0.924 | 1.852 | 0.006 | −0.762 | 0.182 | −0.345 | 1.155 | 0.419 | 0.688 | 0.482 | 0.308 | 0.463 | 0.497 | 0.734 | 0.506 | 17 |
| 21C291 | 1.452 | −0.297 | −1.180 | 0.022 | 0.737 | 0.281 | 0.170 | 0.500 | 0.388 | 0.251 | 0.456 | 0.536 | 0.590 | 0.592 | 0.471 | 37 |
| 21C293 | 2.493 | 1.270 | −0.495 | 0.290 | 1.064 | −0.043 | 0.424 | 0.659 | 0.607 | 0.385 | 0.507 | 0.579 | 0.542 | 0.629 | 0.577 | 3 |
| 21C301 | −0.912 | 1.894 | 2.346 | −1.869 | 0.874 | 1.360 | 0.937 | 0.138 | 0.694 | 0.938 | 0.098 | 0.554 | 0.750 | 0.703 | 0.492 | 25 |
| 21C302 | −1.410 | 1.776 | 0.525 | −0.055 | 1.440 | −0.195 | 0.637 | 0.061 | 0.678 | 0.583 | 0.442 | 0.628 | 0.520 | 0.660 | 0.442 | 58 |
| 21C305 | 0.233 | 1.849 | 2.067 | −1.049 | 0.921 | 0.562 | 0.700 | 0.313 | 0.688 | 0.884 | 0.254 | 0.560 | 0.632 | 0.669 | 0.536 | 8 |
| 21C307 | −0.507 | 4.080 | 1.368 | 2.107 | −2.553 | −2.217 | 0.665 | 0.200 | 1.000 | 0.748 | 0.852 | 0.102 | 0.219 | 0.664 | 0.510 | 15 |
| 21C316 | 0.105 | 0.935 | −1.045 | −0.425 | 0.259 | 0.520 | −0.050 | 0.293 | 0.560 | 0.277 | 0.372 | 0.473 | 0.626 | 0.561 | 0.428 | 71 |
| 21C317 | −0.784 | 1.553 | 0.074 | −0.890 | −0.057 | −1.609 | 0.765 | 0.157 | 0.647 | 0.495 | 0.284 | 0.431 | 0.310 | 0.678 | 0.394 | 112 |
| 21C323 | −0.552 | 0.245 | 1.135 | −0.036 | 1.281 | 0.237 | 1.238 | 0.193 | 0.464 | 0.702 | 0.446 | 0.607 | 0.584 | 0.746 | 0.472 | 36 |
| 21C331 | −0.485 | 0.954 | 0.953 | −0.899 | −0.695 | 0.215 | 0.548 | 0.203 | 0.563 | 0.667 | 0.282 | 0.347 | 0.580 | 0.647 | 0.430 | 68 |
| 21C336 | −0.953 | 0.262 | −0.225 | −0.603 | 0.417 | −1.202 | 0.630 | 0.131 | 0.466 | 0.437 | 0.338 | 0.494 | 0.370 | 0.659 | 0.367 | 142 |
| Classification of Salt Tolerance | Variety Name | Quantity |
|---|---|---|
| Highly salt-tolerant (HST) | 24QX9, 21C222, 21C293, 21C186, 21C241, 21C242 | 6 |
| Salt-tolerant (ST) | 21C188, 21C305, 21C37, 21C38, 21C148, 24QX8, 24QX64, 21C95, 21C307, 16-562, 21C288, 21C91, 24QX14, 24QX2, 24QX45, 21C245, 21C219, 24QX15, 21C301, 21C252, 24QX47, 24QX19, 21C280, 21C243, 24QX12, 24QX26, 21C247, 24QX39, 24QX49, 21C323, 21C291, 21C249, 24QX42, 24QX43, 21C32 | 35 |
| Moderately salt-tolerant (MST) | 21C228, 21C175, 24QX52, 24QX11, 24QX13, 21C161, 24QX21, 24QX40, 24QX3, 24QX33, 21C226, 21C68, 24QX18, 21C144, 21C107, 21C250, 21C302, 21C50, 24QX28, 24QX10, 24QX58, 21C126, 21C127, 24QX38, 21C135, 21C234, 21C331, 21C105, 21C220, 21C316, 21C12, 21C35, 21C229, 24QX24, 21C90, 21C216, 24QX59, 24QX16, 21C181, 21C61, 24-477, 21C13, 24QX41, 24QX34, 21C217, 24QX20, 14-345, 21C64, 21C158, 24QX6, 21C185, 24QX51, 21C21, 21C170, 21C230, 21C154, 21C48, 24QX29, 24QX44, 24QX62, 24QX60, 24QX7, 21C19, 24QX5, 21C1, 21C221, 24QX48 | 67 |
| Salt-sensitive (SS) | 24QX55, 21C57, 21C111, 21C317, 21C236, 21C141, 24QX4, 21C155, 24QX27, 21C207, 21C171, 24QX35, 21C34, 21C28, 21C104, 24QX22, 21C6, 21C211, 21C29, 21C26, 24QX37, 21C162, 24QX36, 21C139, 24QX25, 21C46, 24QX30, 21C108, 21C204, 24QX54, 21C218, 21C51, 24QX63, 21C336, 24QX67, 21C147, 24QX68, 21C205, 21C199, 21C164, 21C172, 21C180, HQ1315, 21C41, 21C132, 24QX31, 24-479, 24QX32, 21C9 | 49 |
| Highly salt-sensitive (HSS) | 21C45, 21C198, 21C44, 21C4, 21C214, 21C43, Sanyue, 21C60 | 8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, Y.; Wang, Z.; Du, Y.; Di, S.; Gao, Z.; Chen, X.; Zhang, W.; Song, L.; Luo, S.; Li, Q. Comprehensive Evaluation and Screening for Salt Tolerance Germplasms at Seedling Stage in Eggplant. Horticulturae 2025, 11, 697. https://doi.org/10.3390/horticulturae11060697
Fang Y, Wang Z, Du Y, Di S, Gao Z, Chen X, Zhang W, Song L, Luo S, Li Q. Comprehensive Evaluation and Screening for Salt Tolerance Germplasms at Seedling Stage in Eggplant. Horticulturae. 2025; 11(6):697. https://doi.org/10.3390/horticulturae11060697
Chicago/Turabian StyleFang, Yu, Zhiguo Wang, Yingnan Du, Shuaitao Di, Zhenwei Gao, Xueping Chen, Weiwei Zhang, Lijun Song, Shuangxia Luo, and Qiang Li. 2025. "Comprehensive Evaluation and Screening for Salt Tolerance Germplasms at Seedling Stage in Eggplant" Horticulturae 11, no. 6: 697. https://doi.org/10.3390/horticulturae11060697
APA StyleFang, Y., Wang, Z., Du, Y., Di, S., Gao, Z., Chen, X., Zhang, W., Song, L., Luo, S., & Li, Q. (2025). Comprehensive Evaluation and Screening for Salt Tolerance Germplasms at Seedling Stage in Eggplant. Horticulturae, 11(6), 697. https://doi.org/10.3390/horticulturae11060697





