Identification of Potential Key Genes for Stem Polysaccharide Synthesis Based on Transcriptome Analysis of Different Developmental Stages of Dendrobium officinale
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Determination of Water-Soluble Polysaccharide Content
2.3. Transcriptome Sequencing and Analysis
2.3.1. Ribonucleic Acid Extraction and Complementary DNA Library Development
2.3.2. Sequencing and Raw Data Filtering
2.3.3. Reference Genome Alignment
2.3.4. Functional Annotation and GO, KEGG Analysis of Differentially Expressed Genes
2.3.5. Expression Trend and Weighted Gene Co-Expression Network Analysis
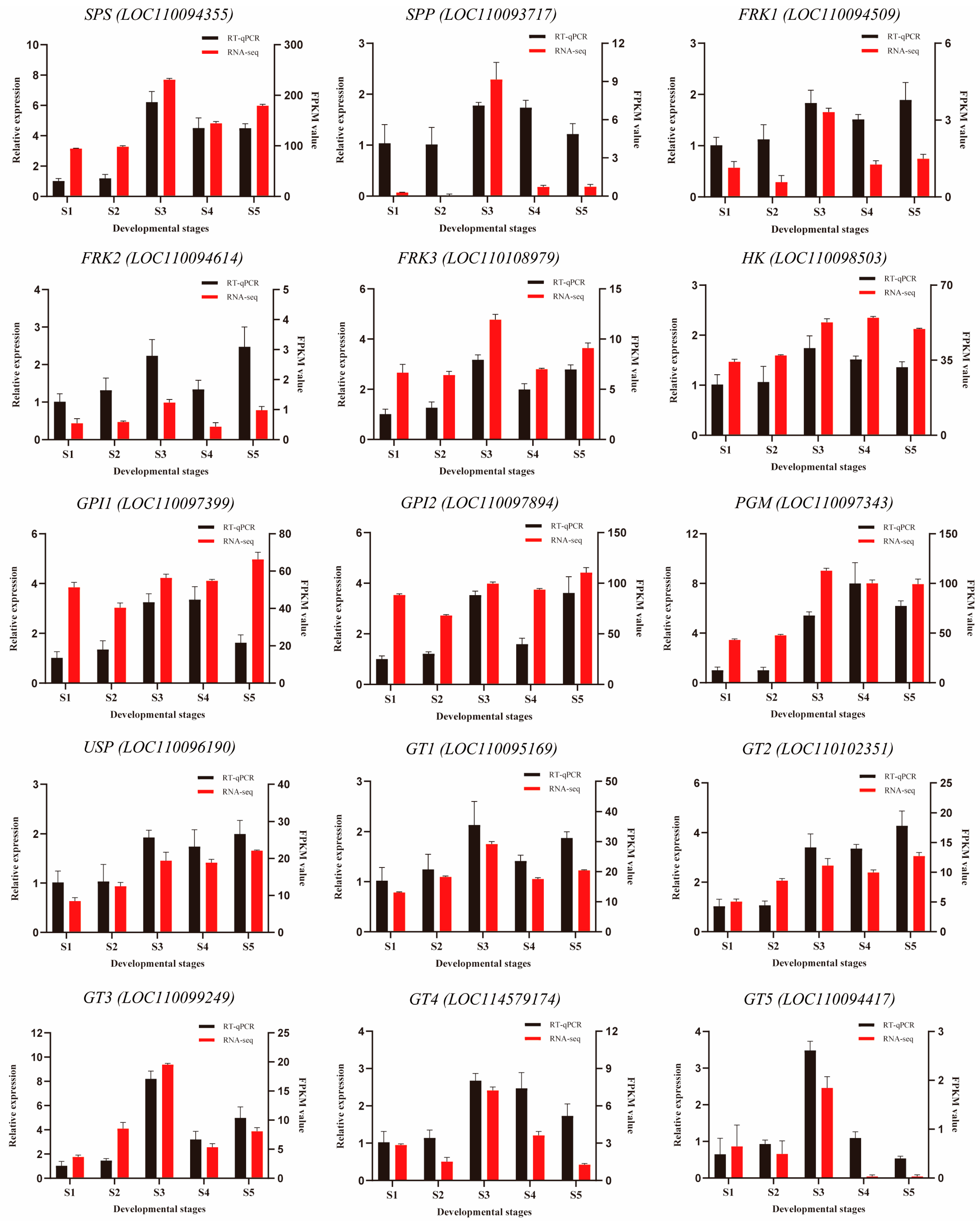
2.3.6. Sugar Metabolism Genes with qRT-PCR Analysis
3. Results
3.1. Phenotypic Changes and Water-Soluble Polysaccharide Content Variations in D. officinale at Different Developmental Stages
3.2. Transcriptome Quality Examination and Statistics
3.3. Expression Pattern Analysis
3.4. Analysis of DEGs
3.4.1. Statistics of DEGs
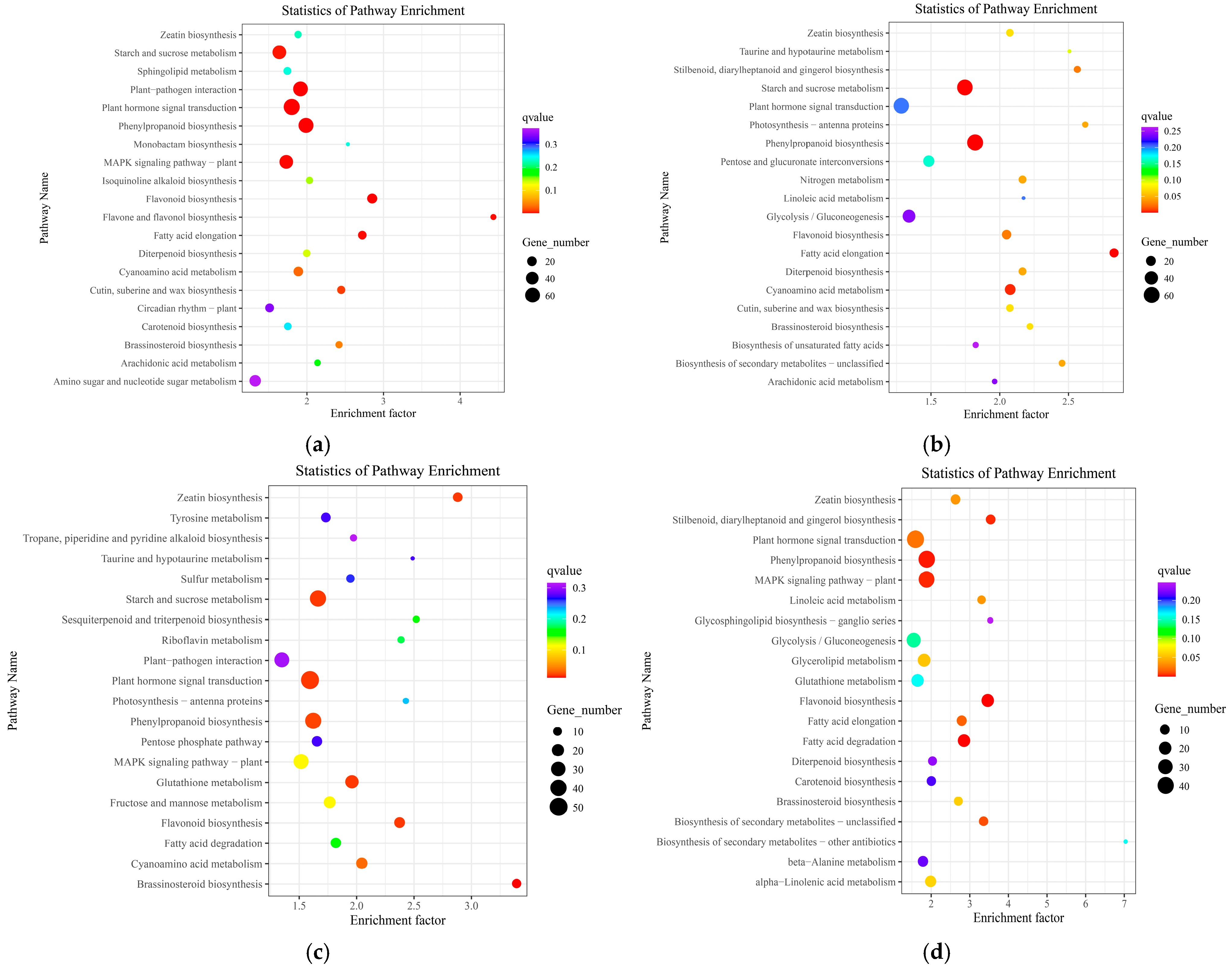
3.4.2. KEGG Pathway Analysis of DEGs
3.5. Analysis of the Biosynthetic Pathway of D. officinale Stem Polysaccharides
3.6. Weighted Gene Co-Expression Network Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Li, X.; Ding, X.; Chu, B.; Zhou, Q.; Ding, G.; Gu, S. Genetic diversity analysis and conservation of the endangered Chinese endemic herb Dendrobium officinale Kimura et Migo (Orchidaceae) based on AFLP. Genetica 2008, 133, 159–166. [Google Scholar] [CrossRef]
- Shen, C.; Guo, H.; Chen, H.; Shi, Y.; Meng, Y.; Lu, J.; Feng, S.; Wang, H. Identification and analysis of genes associated with the synthesis of bioactive constituents in Dendrobium officinale using RNA-Seq. Sci. Rep. 2017, 7, 187. [Google Scholar] [CrossRef]
- Ng, T.; Liu, J.; Wong, J.; Ye, X.; Sze, S.; Tong, Y.; Zhang, K. Review of research on Dendrobium, a prized folk medicine. Appl. Microbiol. Biotechnol. 2012, 93, 1795–1803. [Google Scholar] [CrossRef]
- Wang, Z.; Zhao, M.; Cui, H.; Li, J.; Wang, M. Transcriptomic landscape of medicinal Dendrobium reveals genes associated with the biosynthesis of bioactive components. Front. Plant Sci. 2020, 11, 391. [Google Scholar] [CrossRef]
- Yuan, Y.; Zhang, J.; Liu, X.; Meng, M.; Wang, J.; Lin, J. Tissue-specific transcriptome for Dendrobium officinale reveals genes involved in flavonoid biosynthesis. Genomics 2020, 112, 1781–1794. [Google Scholar] [CrossRef]
- Zhang, W.; Liu, X.; Sun, X.; Han, R.; Yu, N.; Liang, J.; Zhou, A. Comparison of the antioxidant activities and polysaccharide characterization of fresh and dry Dendrobium officinale Kimura et Migo. Molecules 2022, 27, 6654. [Google Scholar] [CrossRef]
- Luo, Z.; Liu, L.; Nie, Q.; Huang, M.; Luo, C.; Sun, Y.; Ma, Y.; Yu, J.; Du, F. HPLC-based metabolomics of Dendrobium officinale revealing its antioxidant ability. Front. Plant Sci. 2023, 14, 1060242. [Google Scholar] [CrossRef]
- Wang, H.; Ge, J.; Zhang, F.; Zha, X.; Liu, J.; Li, Q.; Luo, J. Dendrobium officinale polysaccharide promotes M1 polarization of TAMs to inhibit tumor growth by targeting TLR2. Carbohydr. Polym. 2022, 292, 119683. [Google Scholar] [CrossRef]
- Sun, C.; Zhang, N.; Xu, G.; Jiang, P.; Huang, S.; Zhao, Q.; He, Y. Anti-tumor and immunomodulation activity of polysaccharides from Dendrobium officinale in S180 tumor-bearing mice. J. Funct. Foods 2022, 94, 105105. [Google Scholar] [CrossRef]
- Xu, L.; Zeng, X.; Liu, Y.; Wu, Z.; Zheng, X.; Zhang, X. Inhibitory effect of Dendrobium officinale polysaccharide on oxidative damage of glial cells in aging mice by regulating gut microbiota. Int. J. Biol. Macromol. 2023, 247, 125787. [Google Scholar] [CrossRef]
- Huang, H.; Lee, W.; Zou, H.; Li, H.; Zhang, S.; Li, H.; Lin, J. Antimicrobial peptides in Dendrobium officinale: Genomic parameters, peptide structures, and gene expression patterns. Plant Genome 2023, 16, e20348. [Google Scholar] [CrossRef]
- Cai, M.; Zhu, H.; Xu, L.; Wang, J.; Xu, J.; Li, Z.; Yang, K.; Wu, J.; Sun, P. Structure, anti-fatigue activity and regulation on gut microflora in vivo of ethanol-fractional polysaccharides from Dendrobium officinale. Int. J. Biol. Macromol. 2023, 234, 123572. [Google Scholar] [CrossRef]
- Tao, S.; Lei, Z.; Huang, K.; Li, Y.; Ren, Z.; Zhang, X.; Wei, G.; Chen, H. Structural characterization and immunomodulatory activity of two novel polysaccharides derived from the stem of Dendrobium officinale Kimura et Migo. J. Funct. Foods 2019, 57, 121–134. [Google Scholar] [CrossRef]
- Guo, X.; Yang, M.; Wang, C.; Nie, S.; Cui, S.; Guo, Q. Acetyl-glucomannan from Dendrobium officinale: Structural modification and immunomodulatory activities. Front. Nutr. 2022, 9, 1016961. [Google Scholar] [CrossRef]
- Fang, J.; Lin, Y.; Xie, H.; Farag, M.; Feng, S.; Li, J.; Shao, P. Dendrobium officinale leaf polysaccharides ameliorated hyperglycemia and promoted gut bacterial associated SCFAs to alleviate type 2 diabetes in adult mice. Food Chem.-X 2022, 13, 100207. [Google Scholar] [CrossRef]
- Zhao, W.; Chen, Y.; Tian, Y.; Wang, Y.; Du, J.; Ye, X.; Lu, L.; Sun, C. Dietary supplementation with Dendrobium officinale leaves improves growth, antioxidant status, immune function, and gut health in broilers. Front. Microbiol. 2023, 14, 1255894. [Google Scholar] [CrossRef]
- Wang, L.; Li, C.; Hu, C.; Gong, P.; Zhao, S. Purification and structural characterization of Dendrobium officinale polysaccharides and its activities. Chem. Biodivers. 2021, 18, e2001023. [Google Scholar] [CrossRef]
- Tian, W.; Dai, L.; Lu, S.; Luo, Z.; Qiu, Z.; Li, J.; Li, P.; Du, B. Effect of Bacillus sp. DU-106 fermentation on Dendrobium officinale chock for polysaccharide: Structure and immunoregulatory activities. Int. J. Biol. Macromol. 2019, 135, 1034–1042. [Google Scholar] [CrossRef]
- Pharmacopoeia Commission of the People’s Republic of China. Pharmacopoeia of the People’s Republic of China, 2020 ed.; China Medical Science Press: Beijing, China, 2020; Volume I. [Google Scholar]
- Wang, Y.; Tong, Y.; Adejobi, O.; Wang, Y.; Liu, A. Research advances in Multi-Omics on the traditional chinese herb Dendrobium officinale. Front. Plant Sci. 2022, 12, 808228. [Google Scholar] [CrossRef]
- Jiang, M.; Li, S.; Zhao, C.; Zhao, M.; Xu, S.; Wen, G. Identification and analysis of sucrose synthase gene family associated with polysaccharide biosynthesis in Dendrobium catenatum by transcriptomic analysis. PeerJ 2022, 10, e13222. [Google Scholar] [CrossRef]
- He, C.; Zhang, J.; Liu, X.; Zeng, S.; Wu, K.; Yu, Z.; Wang, X.; da Silva, J.; Lin, Z.; Duan, J. Identification of genes involved in biosynthesis of mannan polysaccharides in Dendrobium officinale by RNA-seq analysis. Plant Mol. Biol. 2015, 88, 219–231. [Google Scholar] [CrossRef]
- Yu, Z.; He, C.; da Silva, J.; Luo, J.; Yang, Z.; Duan, J. The GDP-mannose transporter gene (DoGMT) from Dendrobium officinale is critical for mannan biosynthesis in plant growth and development. Plant Sci. 2018, 277, 43–54. [Google Scholar] [CrossRef]
- Li, D.; Ye, G.; Li, J.; Lai, Z.; Ruan, S.; Qi, Q.; Wang, Z.; Duan, S.; Jin, H.; Wang, H. High light triggers flavonoid and polysaccharide synthesis through DoHY5-dependent signaling in Dendrobium officinale. Plant J. 2023, 115, 1114–1133. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; He, C.; Wu, K.; da Silva, J.; Zeng, S.; Zhang, X.; Yu, Z.; Xia, H.; Duan, J. Transcriptome analysis of Dendrobium officinale and its application to the identification of genes associated with polysaccharide synthesis. Front. Plant Sci. 2016, 7, 5. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zhang, T.; Wang, L.; Dong, S.; Wang, D.; Li, B.; Cao, X. The dynamic changes in the main substances in Codonopsis pilosula root provide insights into the carbon flux between primary and secondary metabolism during different growth stages. Metabolites 2023, 13, 456. [Google Scholar] [CrossRef]
- Wang, J.; Su, H.; Wu, Z.; Wang, W.; Zhou, Y.; Li, M. Integrated metabolites and transcriptomics at different growth stages reveal polysaccharide and flavonoid biosynthesis in Cynomorium songaricum. Int. J. Mol. Sci. 2022, 23, 10675. [Google Scholar] [CrossRef]
- Chen, S.; Zhou, Y.; Chen, Y.; Gu, J. Fastp: An ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 2018, 34, 884–890. [Google Scholar] [CrossRef]
- Kim, D.; Pertea, G.; Trapnell, C.; Pimentel, H.; Kelley, R.; Salzberg, S. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013, 14, R36. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.; Park, C.; Bennett, C.; Salzberg, S. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Anders, S.; Huber, W. Differential expression analysis for sequence count data. Genome Biol. 2010, 11, R106. [Google Scholar] [CrossRef]
- Apweiler, R.; Bairoch, A.; Wu, C.; Barker, W.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The universal protein knowledgebase. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef] [PubMed]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.; Sonnhammer, E.; Tosatto, S.; Paladin, L.; Raj, S.; Richardson, L.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.; Galperin, M.; Natale, D.; Koonin, E. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Klopfenstein, D.; Zhang, L.; Pedersen, B.; Ramirez, F.; Vesztrocy, A.; Naldi, A.; Mungall, C.; Yunes, J.; Botvinnik, O.; Weigel, M.; et al. GOATOOLS: A Python library for Gene Ontology analyses. Sci. Rep. 2018, 8, 10872. [Google Scholar] [CrossRef]
- Xie, C.; Mao, X.; Huang, J.; Ding, Y.; Wu, J.; Dong, S.; Kong, L.; Gao, G.; Li, C.; Wei, L. KOBAS 2.0: A web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011, 39, W316–W322. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Wang, B.; Hua, W.; Niu, J.; Dang, K.; Qiang, Y.; Wang, Z. De Novo assembly and analysis of polygonatum sibiricum transcriptome and identification of genes involved in polysaccharide biosynthesis. Int. J. Mol. Sci. 2017, 18, 1950. [Google Scholar] [CrossRef]
- Niu, J.; Zhao, G.; Mi, Z.; Chen, L.; Liu, S.; Wang, S.; Wang, D.; Wang, Z. De novo sequencing of Bletilla striata (Orchidaceae) transcriptome and identification of genes involved in polysaccharide biosynthesis. Genet. Mol. Biol. 2020, 43, e20190417. [Google Scholar] [CrossRef]
- Zhang, S.; Shi, Y.; Huang, L.; Wang, C.; Zhao, D.; Ma, K.; Wu, J.; Peng, D. Comparative transcriptomic analysis of rhizomes, stems, and leaves of Polygonatum odoratum (Mill.) Druce reveals candidate genes associated with polysaccharide synthesis. Gene 2020, 744, 144626. [Google Scholar] [CrossRef]
- Zhang, G.; Xu, Q.; Bian, C.; Tsai, W.; Yeh, C.; Liu, K.; Yoshida, K.; Zhang, L.; Chang, S.; Chen, F.; et al. The Dendrobium catenatum Lindl. genome sequence provides insights into polysaccharide synthase, floral development and adaptive evolution. Sci. Rep. 2016, 6, 19029. [Google Scholar] [CrossRef]
- Misra, V.; Mall, A.; Ansari, S.; Ansari, M.I. Sugar transporters, sugar-metabolizing enzymes, and their interaction with phytohormones in sugarcane. J. Plant Growth Regul. 2023, 42, 4975–4988. [Google Scholar] [CrossRef]
- Yoon, J.; Cho, L.; Tun, W.; Jeon, J.; An, G. Sucrose signaling in higher plants. Plant Sci. 2021, 302, 110703. [Google Scholar] [CrossRef] [PubMed]
- Shen, Q.; Wu, X.; Tao, Y.; Yan, G.; Wang, X.; Cao, S.; Wang, C.; He, W. Mining candidate genes related to heavy metals in Mature Melon (Cucumis melo L.) peel and pulp using WGCNA. Genes 2022, 13, 1767. [Google Scholar] [CrossRef] [PubMed]
- Ma, P.; Zhang, X.; Chen, L.; Zhao, Q.; Zhang, Q.; Hua, X.; Wang, Z.; Tang, H.; Yu, Q.; Zhang, M.; et al. Comparative analysis of sucrose phosphate synthase (SPS) gene family between Saccharum officinarum and Saccharum spontaneum. BMC Plant Biol. 2020, 20, 422. [Google Scholar] [CrossRef]
- Huang, D.; Qin, C.; Gui, Y.; Zhao, L.; Chen, Z.; Wang, M.; Sun, Y.; Liao, Q.; Li, Y.; Lakshmanan, P. Role of the SPS Gene Families in the Regulation of Sucrose Accumulation in Sugarcane. Sugar Tech 2017, 19, 117–124. [Google Scholar] [CrossRef]
- Chen, S.; Hajirezaei, M.; Peisker, M.; Tschiersch, H.; Sonnewald, U.; Börnke, F. Decreased sucrose-6-phosphate phosphatase level in transgenic tobacco inhibits photosynthesis, alters carbohydrate partitioning, and reduces growth. Planta 2005, 221, 479–492. [Google Scholar] [CrossRef]
- Vanderwall, M.; Gendron, J.M. HEXOKINASE1 and glucose-6-phosphate fuel plant growth and development. Development 2023, 150, dev202346. [Google Scholar] [CrossRef] [PubMed]
- Aguilera-Alvarado, G.; Sanchez-Nieto, S. Plant hexokinases are multifaceted proteins. Plant Cell Physiol. 2017, 58, 1151–1160. [Google Scholar] [CrossRef]
- Zabotina, O.A.; Zhang, N.; Weerts, R. Polysaccharide biosynthesis: Glycosyltransferases and their complexes. Front. Plant Sci. 2021, 12, 625307. [Google Scholar] [CrossRef]
- He, C.; Wu, K.; Zhang, J.; Liu, X.; Zeng, S.; Yu, Z.; Zhang, X.; da Silva, J.; Deng, R.; Tan, J.; et al. Cytochemical localization of polysaccharides in Dendrobium officinale and the involvement of DoCSLA6 in the synthesis of mannan polysaccharides. Front. Plant Sci. 2017, 8, 173. [Google Scholar] [CrossRef]
- Zhang, S.; Li, J.; Shen, Y.; Korkor, L.; Pu, Q.; Lu, J.; Shakeela, B.; Kong, D.; Li, O.; Zeng, G.; et al. Physiological responses of Dendrobium officinale under exposure to cold stress with two cultivars. Phyton 2020, 89, 599–617. [Google Scholar] [CrossRef]








| S1 | S2 | S3 | S4 | S5 | |
|---|---|---|---|---|---|
| Content of water-soluble polysaccharide (%) | 15.37 ± 0.41% e | 21.18 ± 0.22% d | 40.44 ± 0.75% a | 35.62 ± 0.23% b | 32.61 ± 0.52% c |
| Sample | ReadSum | BaseSum | GC (%) | Q20 (%) | Q30 (%) | Uniquely Mapped (%) |
|---|---|---|---|---|---|---|
| S1-1 | 24,600,661 | 7,380,198,300 | 45.74 | 97.96 | 94.49 | 91.57 |
| S1-2 | 28,335,191 | 8,500,557,300 | 45.65 | 97.87 | 94.3 | 91.27 |
| S1-3 | 29,071,046 | 8,721,313,800 | 45.82 | 98.07 | 94.72 | 91.90 |
| S2-1 | 26,070,164 | 7,821,049,200 | 45.95 | 98.02 | 94.66 | 91.73 |
| S2-2 | 23,229,828 | 6,968,948,400 | 45.84 | 98.02 | 94.57 | 91.71 |
| S2-3 | 23,112,220 | 6,933,666,000 | 46.07 | 97.84 | 94.28 | 91.76 |
| S3-1 | 23,947,748 | 7,184,324,400 | 45.35 | 97.87 | 94.37 | 90.23 |
| S3-2 | 31,578,561 | 9,473,568,300 | 45.22 | 97.97 | 94.55 | 90.11 |
| S3-3 | 25,886,613 | 7,765,983,900 | 45.42 | 97.55 | 93.41 | 90.25 |
| S4-1 | 22,837,990 | 6,851,397,000 | 45.61 | 97.96 | 94.52 | 92.30 |
| S4-2 | 23,227,517 | 6,968,255,100 | 45.65 | 98.19 | 94.97 | 92.60 |
| S4-3 | 21,288,991 | 6,404,681,386 | 45.58 | 98.03 | 94.78 | 90.89 |
| S5-1 | 24,748,756 | 7,474,124,312 | 45.49 | 98.16 | 95.19 | 88.68 |
| S5-2 | 25,158,085 | 7,597,741,670 | 45.43 | 98.31 | 95.48 | 89.21 |
| S5-3 | 24,674,333 | 7,451,648,566 | 45.46 | 98.39 | 95.65 | 89.52 |
| Type | Total | Up | Down |
|---|---|---|---|
| S1 vs. S2 | 5764 | 2898 | 2866 |
| S2 vs. S3 | 6408 | 2735 | 3673 |
| S3 vs. S4 | 4477 | 2217 | 2260 |
| S4 vs. S5 | 3809 | 1958 | 1851 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, T.; Huang, S.; Tian, S.; Gao, M.; Zhang, X.; He, L.; Zhang, S. Identification of Potential Key Genes for Stem Polysaccharide Synthesis Based on Transcriptome Analysis of Different Developmental Stages of Dendrobium officinale. Horticulturae 2025, 11, 679. https://doi.org/10.3390/horticulturae11060679
Yang T, Huang S, Tian S, Gao M, Zhang X, He L, Zhang S. Identification of Potential Key Genes for Stem Polysaccharide Synthesis Based on Transcriptome Analysis of Different Developmental Stages of Dendrobium officinale. Horticulturae. 2025; 11(6):679. https://doi.org/10.3390/horticulturae11060679
Chicago/Turabian StyleYang, Tianwei, Shiyu Huang, Shanshan Tian, Manrong Gao, Xiangjun Zhang, Longfei He, and Shangwen Zhang. 2025. "Identification of Potential Key Genes for Stem Polysaccharide Synthesis Based on Transcriptome Analysis of Different Developmental Stages of Dendrobium officinale" Horticulturae 11, no. 6: 679. https://doi.org/10.3390/horticulturae11060679
APA StyleYang, T., Huang, S., Tian, S., Gao, M., Zhang, X., He, L., & Zhang, S. (2025). Identification of Potential Key Genes for Stem Polysaccharide Synthesis Based on Transcriptome Analysis of Different Developmental Stages of Dendrobium officinale. Horticulturae, 11(6), 679. https://doi.org/10.3390/horticulturae11060679





