Abstract
Chinese Cymbidium are prized for their ornamental beauty, ecological significance, and economic value. However, genomic resources crucial for breeding studies within this genus remain scarce, which has hindered the identification of key genes controlling economically important traits and posed challenges for conservation efforts. We performed a comprehensive identification of whole-genome simple sequence repeats (SSRs) and single-nucleotide polymorphism (SNP) markers using the restriction-site associated DNA sequencing (RADseq) on C. goeringii and C. faberi. A total of 49,640 SSR loci were identified across both species, with an average density of 12.7 SSRs/Mb. Among these, 17,637 SSRs were common to both C. goeringii and C. faber, while 17,676 and 14,329 SSRs were uniquely identified in C. goeringii and C. faberi, respectively. Additionally, we identified 405,416 SNPs and 26,870 InDels, with average densities of 105.2/Mb and 6.5/Mb. Furthermore, we validated two SSRs (located at Chr01:78857480-78860461 and Chr01:93382182-93384869) and developed an efficient method for identifying hybrids among the progeny resulting from crosses between C. goeringii and C. faberi. We also validated two SNP markers that showed a close association with the petal and lip length using Sanger sequencing. Our findings revealed that the Chr01_99657375 SNP achieved 73% predictive accuracy for identifying long-petal/lip phenotypes. The results are expected to greatly benefit marker-assisted breeding efforts in Cymbidium orchids and lay a solid foundation for the molecular breeding process of improving flower shape traits in orchid plants.
1. Introduction
Cymbidium spp. (Orchidaceae) are highly valued ornamental potted flowers with centuries of cultivation history. Key floral traits, including petal length and lip length, exhibit strong positive correlations with overall floral dimensions, profoundly influencing aesthetic appeal and ornamental value. While the ABCDE model posits primarily regulation of petal and lip development by class A and E genes [1,2], interspecific variability in these traits suggests co-regulation by additional genetic factors [3]. However, the intricate regulatory networks governing these processes remain incompletely understood, and further investigation is necessary to elucidate their precise roles.
Marker-assisted selection (MAS) technology has proven effective in dissecting genetic pathways underlying ornamental traits, facilitating identification of linked markers in species such as C. sinense [4], Prunus meme [5], Matthiola incana [6]. MAS relies on the identification of molecular markers significantly associated with target traits [7]. Among available markers, SNPs offer high-density genomic distribution and stability, while SSRs provide co-dominance, repeatability, and polymorphism [8]. Though SNPs and SSRs are widely used in orchids for genetic diversity and phylogenetic studies [9,10], their applications in floral trait-associated marker development remain limited.
Next-generation sequencing (NGS) enables high-throughput SNPs/SSRs discovery in plants [11]. However, whole-genome resequencing remains economically impractical for species with large, complex genomes such as C. ensifolium (3.62 Gb) and C.sinense (3.53 Gb). To address this challenge, restriction-site associated DNA sequencing (RADseq) provides an elegant solution by selectively targeting genomic regions flanking restriction enzyme sites, enabling cost-effective genotyping, marker development, and high-resolution genetic mapping across diverse species [12].
Recent advancements in Cymbidium genomics have established critical foundational resources. Multiple high-quality genome assemblies for this orchid genus have now been completed [13,14,15], creating unprecedented opportunities for RADseq-based marker discovery pipelines. These genomic blueprints have enabled the development of innovative tools like the 51 K Axiom Cymbidium SNP array, which was generated through resequencing 195 C. sinense accessions. This array has already demonstrated utility in identifying trait-associated molecular markers and candidate genes via genome-wide association studies (GWAS), particularly for floral trait dissection [16]. Notably, RADseq offers distinct advantages over array-based genotyping platforms. By leveraging NGS to sequence reduced-representation libraries focused on restriction-associated sites, RADseq provides a more flexible and scalable solution with significantly lower per-sample costs compared to SNP arrays [17]. This cost-effectiveness, combined with comprehensive genome coverage and transferability across diverse germplasm, positions RADseq as a transformative technology for accelerating marker-assisted selection and genomic-informed breeding programs in Cymbidium and related ornamentals.
C. goeringii and C. faberi are traditional terrestrial orchids with different flowering times, floral structures, and leaf morphology. C. faberi usually exhibits taller growth, more flowers per inflorescence, and more variation in lip shape and color compared to the typically shorter C. goeringii. These phenotypic differences make them valuable for exploring the genetic basis of floral trait diversity. Therefore, we employed the RADseq method to sequence C. goeringii ‘Huanghemei’ and C. faberi ‘Laojiping’, aiming to uncover the underlying genetic variation between these two species. Additionally, SSRs and SNPs derived from RADseq sequences, which were previously identified as closely linked to the locus influencing petal and lip length [18], were validated using both DNA analyzer and Sanger sequencing. Subsequently, two reliable SSRs and SNPs were selected to establish a robust and efficient approach for hybrid identification and marker-assisted selection, specifically targeting traits associated with petal and lip length in the progeny resulting from crosses between C. goeringii and C. faberi. This study not only corroborates the findings of our prior QTL analysis but also facilitates the development of markers that significantly contribute to flower shape breeding in Chinese Cymbidium, thereby advancing our understanding and manipulation of floral traits in this genus.
2. Materials and Methods
2.1. Plant Materials
In this study, genomic DNA extracted from C. goeringii ‘Huanghemei’ and C. faberi ‘Laojiping’ was utilized for the development of SSR markers. To assess the SSRs’ efficiency, genotyping was conducted on 30 F1 progenies resulting from a cross between C. goeringii ‘Huanghemei’ (female parent) and C. faberi ‘Laojiping’ (male parent). This genotyping process was carried out using a DNA analyzer (ABI 3730 XL) at Sangon Biotech (Shanghai, China). Furthermore, to identify SNP markers associated with petal length and lip length traits, Sanger sequencing was performed on a selected group of 15 F1 progenies.
2.2. Measurement of Petal and Lip Length
To obtain precise measurements of the petal length and lip length, the flowers were carefully dissected and subsequently scanned to generate digital images. Following this, ImageJ software (version 1.53k) was employed to measure the lengths of both the petals and the lip.
2.3. DNA Isolation and Quality Control
Genomic DNA was extracted from fresh leaf tissue using the Rapid Non-Toxic DNA Extraction Kit (High Yield Version, Fuzhou MELISSA Biotechnology Co., Ltd., Fuzhou, China). DNA quality was examined using a UV (ultraviolet) spectrophotometer (260/280 nm absorbance ratio) and 1.5% agarose gel electrophoresis. Only samples meeting stringent quality criteria (DNA concentration > 50 ng/μL, 260/280 ratio of 1.8–2.0) were selected for downstream applications, including RAD library construction, PCR amplification, and SNP genotyping array analysis.
2.4. RAD Library Preparation, Sequencing, Assembly
The RAD library was prepared by Genedenovo Biotechnology Co., Ltd. (Guangzhou, China) following the protocol established by Baird et al. [19]. Briefly, genomic DNA was subjected to double digestion using the restriction enzyme EcoRI (NEB, Ipswich, MA, USA) at 37 °C for 60 min (recognition motif: G|AATTC), and the reaction was terminated at 65 °C for 20 min. The resulting fragments were ligated with P1 barcode adapters, pooled across samples, and sheared to a 300–700 bp size range using sonication. After end repair and A-tailing, P2 adapters were incorporated via ligation. PCR amplification generated 150 bp paired-end (PE) sequencing libraries for C. goeringii and C. faberi, which were sequenced on the Illumina Hiseq 4000 platform (San Diego, CA, USA).
Raw reads underwent adapter trimming and quality filtering (Q ≥ 30) using Trimmomatic v0.39 [20]. High-quality clean reads were de novo assembled into LongRead contigs with Velvet v1.2.10 [21], employing optimized k-mer parameters. The assembly was screened against the Repbase repeat database using CENSOR v4.2.29 [22] with default parameter settings to identify and mask repetitive elements. After stringent quality control filtering (Phred score ≥ 30), we obtained 4.35 Gb (94.90% of raw reads) and 4.36 Gb (95.23% of raw reads) of high-quality clean data for C. faberi and C. goeringii, respectively. This corresponds to 31.38 million and 31.39 million clean reads per species, demonstrating consistent sequencing quality across both orchid species.
2.5. SSR Marker Identification and Genomic Distribution Analysis
The genome of C. goeringii was used as the reference genome in this study [15]. SSR motifs were computationally identified using MISA v1.0 software [23] with optimized search parameters: minimum repeat units of 10 for monomers, 6 for dimers, and 5 for trimers, tetramers, pentamers, and hexamers. To visualize genomic SSR distribution, we generated a density plot using the R package “LinkageMapView” [24], mapping SSR loci across chromosomes. Additionally, Pearson correlation analysis was performed in R to assess the relationship between chromosome length and SSR abundance, providing insights into SSR distribution patterns in relation to genome architecture.
2.6. SNP and InDel Detection Pipeline
High-quality clean reads from C. goeringii and C. faberi were aligned to the C. goeringii reference genome using BWA-MEM v0.7.17 with default parameters [25]. Variant calling was performed using GATK v4.2.6.1 software [26]. Specifically, HaplotypeCaller was run in ERC GVCF mode to generate per-sample gVCF files, followed by joint genotyping with GenotypeGVCFs to produce a multi-sample VCF file containing SNPs and small insertions/deletions (InDels). To ensure variant reliability, stringent filtering criteria were applied to both SNPs and InDels using VCFtools v4.2 [27], retaining biallelic sites with minor allele frequency (MAF) ≥ 0.2, read depth (DP) ≥ 6, and Phred-scaled quality score (QUAL) ≥ 20. This multi-step process enabled the identification of high-confidence markers suitable for genetic analyses.
2.7. Validation of SSR Polymorphisms
Building upon a previously mapped QTL (Qzhbc1-1) controlling petal length and lip length, located at 31.56 cM (Chr01_93439794) on linkage group LG01 and spanning 78.60–100.51 Mb [physical position] [18], we selected eight SSR markers within this interval for polymorphism validation across 30 individuals from a cross between C. goeringii and C. faberi.
SSR-specific primers were designed using Primer Premier 5.0 (PREMIER Biosoft, Palo Alto, CA, USA) with default parameters. Primer design sequences and genomic coordinates are provided in Supplementary File S1. The primers were synthesized by Sangon Biotech (Shanghai, China), and their properties are summarized in Table S1.
Amplifications were conducted in a 25 μL reaction containing: 2 μL 50 ng/μL genomic DNA, 12.5 μL 2× Pro Taq Master Mix (Accurate Biotechnology, Hunan, Changsha, China), 1.0 μL each of forward/reverse primers (10 μM), and 8.5 μL nuclease-free H2O. Thermal cycling parameters were: initial denaturation at 94 °C for 2 min; 35 cycles of 94 °C (30 s), locus-specific annealing temperature (30 s), and 72 °C (1 min); final extension at 72 °C for 5 min using an Eppendorf Mastercycler (Eppendorf, Shanghai, China). Amplicons were examined using 2% agarose gel electrophoresis stained with GelRed™ (Biotium, Fremont, CA, USA) and visualized under UV light. Markers producing clear, species-specific bands of expected size were fluorescently labeled (FAM and HEX) and subjected to capillary electrophoresis on an ABI 3730xl DNA Analyzer (Applied Biosystems, Foster City, CA, USA). Fragment sizing was performed using GeneMarker v1.5 (SoftGenetics, State College, PA, USA).
2.8. SNP Validation and Genotyping
Six candidate SNPs identified within the RADseq dataset were prioritized for validation. These markers reside within the Qzhbc1-1 QTL interval (79.26–100.05 Mb) on chromosome 1, a region previously associated with petal length variation. To enable targeted sequencing, we extracted 800 bp flanking sequences around each SNP and locus-specific primers using Primer 5.0 software (PREMIER Biosoft, Palo Alto, CA, USA). Primer sequences with amplification parameters and genomic coordinates are detailed in Table S2 and Supplementary File S2.
The PCR reactions were prepared as described for SSR markers, using the same reagent concentrations and thermal cycling profile. Amplicons were visualized on 2% agarose gels to confirm successful amplification. Prior to sequencing, PCR products were purified using the QIAquick Gel Extraction Kit (Qiagen, Hilden, Germany) following the manufacturer’s protocol. PCR products were sequenced using Sanger sequencing technology by Sango Biotech Co., Ltd. Sequence traces were analyzed using Geneious Prime v2023.1.1 (Biomatters Ltd., Auckland, New Zealand) to confirm SNP identities and genotype calls.
3. Results
3.1. Genomic Distribution and Characteristics of SSR Markers
A total of 49,640 SSR loci were identified across the genomes of C. goeringii and C. faberi, with 17,676 species-specific loci in C. goeringii, 14,329 in C. faberi, and 17,635 conserved between both species (Table S3). Classification of SSR motifs revealed a predominance of dinucleotide (P2) repeats, comprising 28.96% of all SSRs, followed by trinucleotide (20.52%) and mononucleotide (16.92%) repeats (Figure 1A).
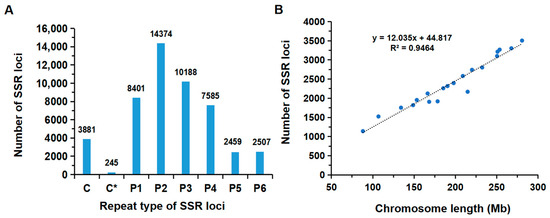
Figure 1.
The characteristics of SSR markers in the Chinese Cymbidium. (A) The number of different SSR motif types. (B) The Pearson correlation analysis between chromosome length and the number of SSR loci. The “C*” indicates SSR type with specific repeat motifs, positions, or functional characteristics.
SSR density varied substantially across chromosomes, with chromosome 1 (Chr01) harboring the highest number of SSR loci (3506), followed by Chr02 and Chr03 (Figure 1B). The overall SSR density ranged from 10.1 to 14.3 SSR/Mb across chromosomes (Table S4), with Chr19 exhibiting the highest density (14.3 SSR/Mb) and Chr08 the lowest (10.1 SSR/Mb). Heatmap visualization revealed a non-uniform distribution pattern, with elevated SSR densities observed in telomeric and centromeric regions (Figure 2A). A linear regression analysis demonstrated a strong positive correlation (r2 = 0.946) between chromosome length and SSR abundance, suggesting co-evolution between genome expansion and SSR proliferation.
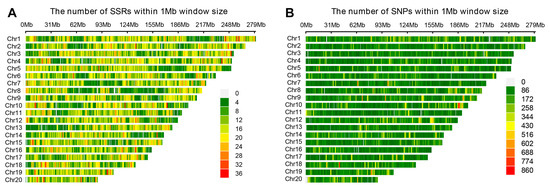
Figure 2.
Genomic distribution of SSR and SNP markers across 20 chromosomes of Cymbidium goeringii and C. faberi based on RADseq analysis. The figure visualizes the comparative density patterns of simple sequence repeat (SSR) loci (A) and single-nucleotide polymorphism (SNP) loci (B) across all 20 chromosomes (Chr1-Chr20) using the C. goeringii reference genome. The X-axis represents chromosome positions within 1 Mb sliding windows (spanning 0 to 279 Mb). Heatmap color gradients (as indicated in the legend) quantify the number of markers identified within each window.
3.2. Identification and Characteristics of SNPs and InDels in C. goeringii and C. faberi
After stringent filtering to remove missing data across all samples, we identified 432,286 high-quality variant loci, comprising 405,416 SNPs and 26,870 InDels (Table S5). These variants exhibited a non-uniform distribution pattern, with average genomic densities of 105.2 SNPs/Mb and 6.5 InDels/Mb, respectively, reflecting moderate polymorphism levels in Cymbidium genomes (Figure 2B).
Transition mutations constituted the majority of SNPs in both species: 64.23% in C. goeringii and 65.09% in C. faberi (Figure 3A,B). The transition/transversion (Ts/Tv) ratio, a proxy for genetic divergence, was higher in C. faberi (1.84) than in C. goeringii (1.79), suggesting closer phylogenetic relatedness within C. faberi populations.
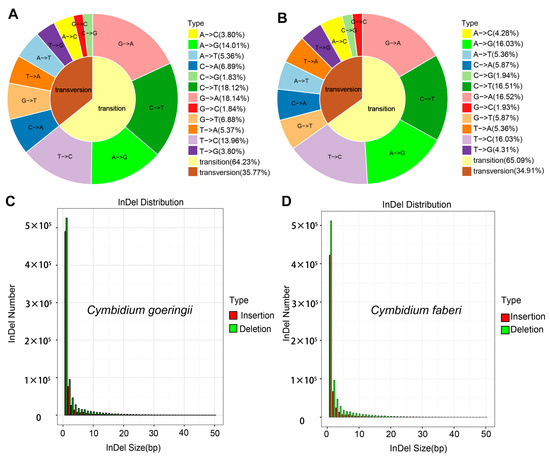
Figure 3.
The characteristics of genomic variation in Cymbidium goeringii and C. faberi. (A,B). The type of SNP variation in (A) C. goeringii and (B) C. faberi. (C,D). The distribution of InDel size in (C) C. goeringii and (D) C. faberi.
Consistent with plant genome characteristics, small InDels (1–2 bp) were predominant, accounting for >90% of all InDel events in both species (Figure 3C,D). These micro-insertions/deletions primarily occurred in non-coding regions, highlighting their potential regulatory roles in shaping phenotypic diversity. The comprehensive SNP/InDel catalog provides a valuable resource for marker-assisted selection (MAS) of ornamental traits in Cymbidium. Particularly, species-specific variants identified here could facilitate targeted introgression of desirable floral characteristics between C. goeringii and C. faberi.
Beyond the observed genetic differences, C. goeringii and C. faberi also exhibited notable similarities at the genomic level. Both species displayed comparable SNP and InDel size distributions, particularly the predominance of 1–2 bp InDels, and a similar proportion of transition mutations. These parallels suggest a conserved genomic architecture and potential overlap in regulatory elements, which may underlie shared floral traits and support the feasibility of trait introgression through interspecific hybridization.
3.3. SSR Marker Validation and Polymorphism Assessment
Based on prior QTL mapping studies [18], five SSR markers within a genomic region associated with petal and lip length were prioritized for validation. Eight primer pairs were initially designed to amplify these loci. Although all primer pairs produced amplification products, P3 and P4 were excluded due to weak amplification with no visible bands on agarose gel electrophoresis. The remaining six primer pairs demonstrated robust and specific amplification, generating amplicons of expected sizes ranging from 114 to 144 bp, as verified by agarose gel electrophoresis (Figure S1).
Target fragments from three SSR markers (P1, P7, P8) were successfully sequenced using capillary electrophoresis. The results showed distinct polymorphism patterns between C. goeringii and C. faberi (Figure 4). Specifically, markers P1 and P8 exhibited high cross-species polymorphism, with multiple alleles detected in both species. In contrast, P7 showed limited allelic variation, indicating potential species-specific evolution at this locus.
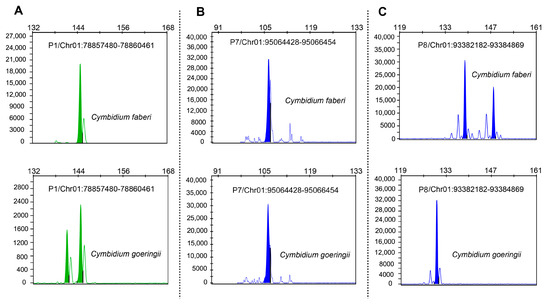
Figure 4.
The genotyping results of capillary electrophoresis. (A–C) Three SSR markers, P1 (A), P7 (B), and P8 (C), were successfully genotyped using capillary electrophoresis.
3.4. SNP Marker Validation and Polymorphism Analysis
Six SNPs within the Qzhbc1-1 QTL interval (previously associated with petal and lip length by Jiang [18]) were selected for validation via Sanger sequencing (Table S2). The sequencing traces confirmed two polymorphic loci in both C. goeringii and C. faberi: Chr01_99657201 (T/A polymorphism) and Chr01_99657375 (A/G polymorphism) (Figure 5). The remaining four candidates either failed to amplify or exhibited monomorphic patterns across species and were subsequently excluded from further analysis. These validated SNPs demonstrate utility for dissecting quantitative trait variation in Cymbidium ornamental traits.
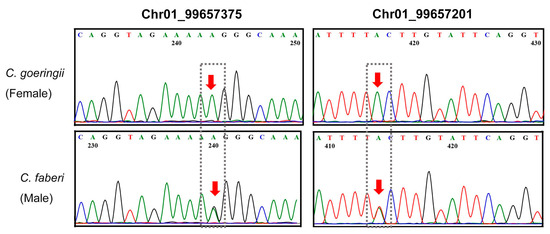
Figure 5.
Sanger sequencing results of SNP Chr01_99657375 (left) and SNP Chr01_99657201 (right) in C. goeringii and C. faberi. C. goeringii and C. faberi were the female parent and male parent for the hybrids, respectively. The red arrows indicate the SNP position. Different colors represent nucleotide bases (A: green, T: red, G: black, C: blue). The dashed boxes highlight the SNP loci showing clear base differences between the two species.
3.5. Applications of SSR-Base Hybrid Identification
Two polymorphic SSR markers—P1 (Chr01:78857480-78860461) and P8 (Chr01:93382182-93384869)—were selected to establish a robust hybrid identification system. A diagnostic panel of 30 progeny from interspecific crosses between C. goeringii and C. faberi was genotyped using these markers.
Capillary electrophoresis revealed distinct allele combinations in the progeny (Table 1). For marker P1, 12 individuals exhibited non-parental alleles (“129,141” or “129,144”), while marker P8 identified 10 individuals with novel alleles (“138,154” or “132,154”). These alleles were absent in both parental lines, confirming their status as false hybrids. Additionally, seven individuals showed homozygous “132” genotypes at marker P8, likely resulting from self-pollination of C. goeringii. By integrating data from both markers, we identified 12 true hybrids (40% of the tested population) that displayed heterozygous alleles inherited from both parental species.

Table 1.
Identification of hybrids from C. goeringii and C. faberi crosses using SSR markers.
This dual-SSR system provides a cost-effective, high-throughput solution for hybrid purity assessment in Cymbidium breeding programs, with clear diagnostic power to distinguish true hybrids from self-pollinated individuals or genetic contaminants.
3.6. Applications of SNP-Assisted Trait Prediction in Cymbidium Hybrids
Quantitative analysis of F1 hybrids from C. goeringii × C. faberi crosses revealed significant segregation of petal and lip length traits (Figure 6A). To develop a marker–trait association model, we randomly genotyped 15 individuals using Sanger sequencing for the Chr01_99657375 SNP locus. Genotyping results identified 11 homozygous (TT/AA) and 4 heterozygous (TC/AG) individuals.
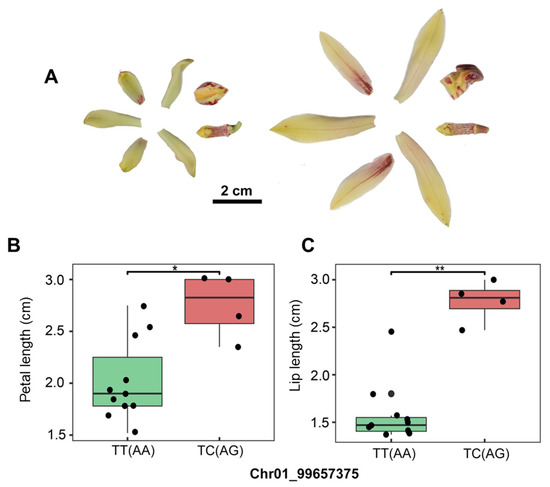
Figure 6.
Validation of SNP markers associated with petal and lip length using F1 individuals from C. goeringii × C. faberi crosses. (A) Anatomy of floral organs in large-flowered and small-flowered individuals. (B) The allele C genotype of Chr01_99657375 SNP was significantly associated with the petal length. The TT genotype showed shorter petal and lip length than the TC genotype. (C) The allele C genotype of Chr01_99657375 SNP was highly significantly associated with the lip length. The TT genotype showed shorter petal and lip length than the TC genotype. “*” indicates p < 0.05; “**” indicates p < 0.01.
Morphometric measurements of petal length and lip length using ImageJ software demonstrated distinct phenotypic differentiation between genotypes (Table S6). Boxplot analysis revealed that heterozygous (TC) individuals exhibited significantly longer petals and lips compared to homozygous (TT) counterparts (Figure 6B). Applying a binary classification system (Group A: ≤2 cm; Group B: >2.3 cm), the Chr01_99657375 SNP achieved 73% predictive accuracy for identifying long-petal/lip phenotypes.
These findings establish Chr01_99657375 as a diagnostic marker for marker-assisted selection (MAS) of ornamental traits in Cymbidium. By integrating this SNP into breeding pipelines, growers can significantly accelerate the development of cultivars with desired floral morphologies.
The boxplot analysis revealed that the lengths of petals and lips were significantly different between TT and TC genotypes (Figure 6B,C). Individuals with a heterozygous (TC) genotype presented a greater length of petals and lips. We defined individuals with petal and lip lengths less than or equal to 2 cm as Group A, and those with petals and lips greater than 2.3 cm as Group B. Chr01_99657375 SNP could achieve approximately 73% accuracy in identifying long or short petal and lip individuals. Thus, SNP Chr01_99657375 shows potential as a predictive marker associated with petal and lip length in Cymbidium, although further validation across diverse germplasm is needed to confirm its broader applicability.
4. Discussion
The genus Cymbidium represents one of the most economically important groups of ornamental orchids in global floriculture markets. Despite their commercial significance, breeding programs for C. goeringii and C. faberi have been constrained by insufficient genomic resources and limited understanding of the genetic architecture underlying key floral traits. This study employed RADseq to establish the first comprehensive molecular marker resource for these species, identifying 49,640 SSR loci and 405,416 SNPs. Two polymorphic SSR markers demonstrated exceptional diagnostic power for hybrid verification, while a validated SNP marker showed significant association with petal and lip length.
Our comprehensive genome-wide marker development in C. goeringii and C. faberi represents a transformative advance in orchid genomics, building upon yet substantially extending previous work in this field. While earlier studies in Cymbidium species were largely limited to small-scale SSR marker development [28,29], our identification of 49,640 SSR loci and 405,416 SNPs provides unprecedented resolution for genetic studies in these commercially important orchids. This achievement places our work on par with recent breakthroughs in other orchid genera such as Phalaenopsis [30] and Dendrobium [31], while specifically addressing the previously neglected but economically vital C. goeringii and C. faberi complex that dominates Asian orchid markets.
The transition/transversion ratios we observed (1.79–1.84) show remarkable consistency with values reported for other Cymbidium species, including C. ensifolium [32] and C. sinense [16], confirming evolutionary conservation of mutation patterns across this genus. However, our RADseq-based approach offers several significant advantages over previous methodologies. Compared to the array-based genotyping platform [10], our method achieves an eight-fold increase in marker density while substantially reducing costs, making it more accessible for breeding programs. Furthermore, compared to transcriptome-derived SNP markers that mainly target coding regions [33], our genome-wide markers cover both coding and non-coding regions, thereby increasing the potential to capture variants linked to complex traits, although functional validation remains necessary.
The practical applications of our markers demonstrate substantial improvements over existing methods. Our SSR-based hybrid identification system achieves high accuracy, a marked improvement over traditional morphological screening methods with typical reliability in Cymbidium [34]. This advancement is particularly significant given the growing importance of hybrid verification in commercial orchid breeding. Similarly, our trait-associated SNP marker (Chr01_99657375) shows superior predictive power (73% accuracy) for petal and lip length compared to similar markers developed in roses [35]. This enhanced performance likely stems from our innovative integration of RADseq with prior QTL mapping data combinatorial approach not previously employed in orchid marker development studies [36,37].
Importantly, the molecular basis underlying this SNP-trait association merits further exploration. The Chr01_99657375 and neighboring Chr01_99657201 SNPs identified in our study were located within the Qzhbc1-1 QTL interval previously associated with lip and petal length [18]. The observed polymorphisms between C. goeringii (female) and C. faberi (male) suggest a potential regulatory role in floral organ development. Previous studies have demonstrated that lip and petal morphology in orchids is predominantly controlled by MADS-box transcription factors, particularly members of the DEFICIENS and GLOBOSA-like subfamilies, which regulate lip and petal identity and growth. In Phalaenopsis equestris, for instance, specific DEF-like genes such as PeMADS4 have been shown to play critical roles in determining lip development, with expression patterns correlating with morphological changes in floral organs [38]. It is plausible that the SNPs identified in our study reside in or near regulatory regions modulating the expression of such floral development genes. Supporting this, genome-wide association studies in C. sinense have identified SNPs within MADS-box genes that are significantly associated with variations in floral traits, including petal shape and size [39]. These findings suggest that Chr01_99657375 and related variants could serve as functional markers for dissecting the genetic mechanisms underlying floral trait differentiation in Cymbidium.
However, several limitations of our study should be acknowledged in the context of recent methodological advances. Like all RADseq-based approaches, our study is subject to restriction enzyme bias, a constraint well-documented in recent vanilla orchid research [40]. Additionally, while our marker validation in controlled crosses provides robust proof-of-concept, broader field testing across diverse germplasm will be essential for assessing general applicability, as emphasized in contemporary ornamental crop breeding reviews [41]. Our sample size, though adequate for initial validation, falls short of the standards established by recent large-scale GWAS in C. sinense [16] that employed 195 accessions. Future studies could benefit from incorporating long-read sequencing technologies to complement our RADseq data and provide more complete genome coverage.
The significance of our findings is further underscored when considering recent advances in orchid genomics and breeding. Our work supports the ongoing recognition of Cymbidium as a valuable model for floral evolution studies [42], while offering preliminary tools that may assist commercial breeders, pending further validation in larger populations. The marker resources we developed overcome key limitations of previous array-based systems and transcriptome-derived markers [43], while offering superior cost-effectiveness compared to whole-genome sequencing approaches [44]. Importantly, our integrated marker–trait association strategy provides a replicable framework that could be adapted to other ornamental species, as suggested by recent reviews on molecular breeding in floriculture crops [45].
5. Conclusions
This study presents a comprehensive genome-wide identification and validation of SSR and SNP markers in C. goeringii and C. faberi using RADseq, significantly advancing the molecular breeding resources for these economically important orchids. The identification of 49,640 SSR loci and 405,416 high-quality SNPs provides a valuable genomic toolkit for future genetic studies and breeding applications in Cymbidium. Notably, the discovery of 17,637 SSR loci conserved between both species offers promising targets for cross-species marker development, while the species-specific SSR and SNP variants enable precise genetic discrimination. The successful validation of two diagnostic SSR markers (Chr01:78857480-78860461 and Chr01:93382182-93384869) establishes an efficient system for hybrid identification in interspecific crosses, addressing a critical need in Cymbidium breeding programs. Furthermore, the SNP marker Chr01_99657375 demonstrates significant predictive value for petal and lip length traits, achieving 73% accuracy in phenotype classification.
These findings not only corroborate previous QTL mapping studies in orchids [46,47], but also provide practical molecular tools for marker-assisted selection in Cymbidium. The integration of these markers into breeding pipelines will facilitate the development of novel cultivars with improved ornamental characteristics. Future research should focus on functional validation of candidate genes linked to these markers and the expansion of marker applications to additional Cymbidium species and traits. This work lays a solid foundation for advancing genomic-assisted breeding strategies in orchids and contributes to shortening the breeding cycle in Cymbidium.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae11060622/s1, Supplementary Figure S1: The electrophoretogram of eight SSR markers amplified among hybrids from Cymbidium goeringii and C. faberi crosses; Supplementary Table S1: Primer information of SSR markers; Supplementary Table S2: Primer information of SNP markers; Supplementary Table S3: Genome-wide identification of SSR markers from the C. goeringii ‘Huanghemei’ and C. faberi ‘Laojiping’; Supplementary Table S4: Distribution of different SSR motif types in each chromosome; Supplementary Table S5: Genome-wide identification of SSR markers from the C. goeringii ‘Huanghemei’ and C. faberi ‘Laojiping’; Supplementary Table S6: The data of petal and lip length for 15 F1 hybrids. Supplementary File S1: Sequences used for SSR primer design and their genomic coordinates; Supplementary File S2: Sequences used for SNP primer design and their genomic coordinates.
Author Contributions
Conceptualization, X.Z. and D.P.; methodology, M.C., Y.L., and C.J.; software, X.Z., M.C., C.J., and X.J.; data curation, M.C., Y.L., C.J., X.C., F.W., and Y.D.; writing—original draft preparation, M.C. and Y.L.; writing—reviewing and editing, X.Z., D.P., M.C., Y.L., F.W., and X.J.; validation, X.Z., D.P., and M.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the National Key Research and Development Program of China (grant No. 2023YFD1600504) and the Natural Science Foundation of Fujian Province, China (grant No. 2024J01394).
Data Availability Statement
The genome of Cymbidium goeringii used as the reference in this study was reported by Sun et al. (2021) [15]. The RADseq data generated in this study have been deposited in the National Genomics Data Center (NGDC) under the accession number [PRJCA040609].
Acknowledgments
We acknowledge the technical support of laboratory staff during the laboratory experiments.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Bowman, J.L.; Smyth, D.R.; Meyerowitz, E.M. Genetic interactions among floral homeotic genes of Arabidopsis. Development 1991, 112, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Mondragón-Palomino, M.; Theißen, G. Conserved differential expression of paralogous DEFICIENS-and GLOBOSA-like MADS-box genes in the flowers of Orchidaceae: Refining the ‘orchid code’. Plant J. 2011, 66, 1008. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.-Y.; Kao, N.-H.; Li, J.-Y.; Hsu, W.-H.; Liang, Y.-L.; Wu, J.-W.; Yang, C.-H. Characterization of the possible roles for B class MADS box genes in regulation of perianth formation in orchid. Plant Physiol. 2010, 152, 837–853. [Google Scholar] [CrossRef]
- Yang, F.; Guo, Y.; Li, J.; Lu, C.; Wei, Y.; Gao, J.; Xie, Q.; Jin, J.; Zhu, G. Genome-wide association analysis identified molecular markers and candidate genes for flower traits in Chinese orchid (Cymbidium sinense). Hortic. Res. 2023, 10, uhad206. [Google Scholar] [CrossRef]
- Shi, Y.; Zhu, H.; Zhang, J.; Bao, M.; Zhang, J. Development and validation of molecular markers for double flower of Prunus mume. Sci. Hortic. 2023, 310, 111761. [Google Scholar] [CrossRef]
- Tan, C.; Zhang, H.; Chen, H.; Guan, M.; Zhu, Z.; Cao, X.; Ge, X.; Zhu, B.; Chen, D. First report on development of genome-wide microsatellite markers for stock (Matthiola incana L.). Plants 2023, 12, 748. [Google Scholar] [CrossRef]
- Flint-Garcia, S.A.; Thornsberry, J.M.; Buckler, E.S. Structure of linkage disequilibrium in plants. Annu. Rev. Plant Biol. 2003, 54, 357–374. [Google Scholar] [CrossRef] [PubMed]
- García, C.; Guichoux, E.; Hampe, A. A comparative analysis between SNPs and SSRs to investigate genetic variation in a juniper species (Juniperus phoenicea ssp. turbinata). Tree Genet. Genomes 2018, 14, 87. [Google Scholar] [CrossRef]
- Nam, D.; Cha, M.J.; Kim, Y.D.; Awasthi, M.; Do, Y.; Kong, S.G.; Chung, K.W. Microsatellite Dataset for Cultivar Discrimination in Spring Orchid (Cymbidium goeringii). Genes 2023, 14, 1610. [Google Scholar] [CrossRef] [PubMed]
- Shen, B.; Shen, A.; Tan, Y.; Liu, L.; Li, S.; Tan, Z. Development of KASP markers, SNP fingerprinting and population genetic analysis of Cymbidium ensifolium (L.) Sw. germplasm resources in China. Front. Plant Sci. 2025, 15, 1460603. [Google Scholar] [CrossRef] [PubMed]
- Reis, J.M.; Pereira, R.J.; Coelho, P.S.; Leitao, J.M. Assessment of Wild Rocket (Diplotaxis tenuifolia (L.) DC.) Germplasm Accessions by NGS Identified SSR and SNP Markers. Plants 2022, 11, 3482. [Google Scholar] [CrossRef] [PubMed]
- Moreau, E.L.P.; Medberry, A.N.; Honig, J.A.; Molnar, T.J. Genetic diversity analysis of big-bracted dogwood (Cornus florida and C. kousa) cultivars, interspecific hybrids, and wild-collected accessions using RADseq. PLoS ONE 2024, 19, e0307326. [Google Scholar] [CrossRef]
- Shen, B.; Shen, A.; Liu, L.; Tan, Y.; Li, S.; Tan, Z. Assembly and comparative analysis of the complete multichromosomal mitochondrial genome of Cymbidium ensifolium, an orchid of high economic and ornamental value. BMC Plant Biol. 2024, 24, 255. [Google Scholar] [CrossRef]
- Ai, Y.; Li, Z.; Sun, W.H.; Chen, J.; Zhang, D.Y.; Ma, L.; Zhang, Q.H.; Chen, M.K.; Zheng, Q.D.; Liu, J.F.; et al. The Cymbidium genome reveals the evolution of unique morphological traits. Hortic. Res. 2021, 8, 255. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Chen, G.Z.; Huang, J.; Liu, D.K.; Xue, F.; Chen, X.L.; Chen, S.Q.; Liu, C.G.; Liu, H.; Ma, H.; et al. The Cymbidium goeringii genome provides insight into organ development and adaptive evolution in orchids. Ornament. Plant Res. 2021, 1, 1–13. [Google Scholar] [CrossRef]
- Zhang, Y.; Li, C.; An, Y.; Zhang, X.; Zhou, Y. Integrated transcriptome and metabolome analysis reveals the anthocyanin biosynthesis mechanisms in blueberry (Vaccinium corymbosum L.) leaves under different light qualities. Front. Plant Sci. 2022, 13, 1073332. [Google Scholar] [CrossRef] [PubMed]
- Elshire, R.J.; Glaubitz, J.C.; Sun, Q.; Poland, J.A.; Kawamoto, K.; Buckler, E.S.; Mitchell, S.E. A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 2011, 6, e19379. [Google Scholar] [CrossRef]
- Jiang, X. Construction of Bin Map and QTL Mapping of Flower Traits in F1 Population of Cymbidium goeringii ‘Huanghemei’ × C. faberi ‘Laojipin’; Fujian Agriculture and Forestry University: Fuzhou, China, 2024. [Google Scholar]
- Baird, N.A.; Etter, P.D.; Atwood, T.S.; Currey, M.C.; Shiver, A.L.; Lewis, Z.A.; Selker, E.U.; Cresko, W.A.; Johnson, E.A. Rapid SNP discovery and genetic mapping using sequenced RAD markers. PLoS ONE 2008, 3, e3376. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Zerbino, D.R.; Birney, E. Velvet: Algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008, 18, 821–829. [Google Scholar] [CrossRef]
- Kohany, O.; Gentles, A.J.; Hankus, L.; Jurka, J. Annotation, submission and screening of repetitive elements in Repbase: RepbaseSubmitter and Censor. BMC Bioinform. 2006, 7, 474. [Google Scholar] [CrossRef] [PubMed]
- Beier, S.; Thiel, T.; Münch, T.; Scholz, U.; Mascher, M. MISA-web: A web server for microsatellite prediction. Bioinformatics 2017, 33, 2583–2585. [Google Scholar] [CrossRef]
- Ouellette, L.A.; Reid, R.W.; Blanchard, S.G.; Brouwer, C.R. LinkageMapView—Rendering high-resolution linkage and QTL maps. Bioinformatics 2018, 34, 306–307. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed]
- McKenna, A.; Hanna, M.; Banks, E.; Sivachenko, A.; Cibulskis, K.; Kernytsky, A.; Garimella, K.; Altshuler, D.; Gabriel, S.; Daly, M.; et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010, 20, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Danecek, P.; Auton, A.; Abecasis, G.; Albers, C.A.; Banks, E.; DePristo, M.A.; Handsaker, R.E.; Lunter, G.; Marth, G.T.; Sherry, S.T.; et al. The variant call format and VCFtools. Bioinformatics 2011, 27, 2156–2158. [Google Scholar] [CrossRef]
- Moe, K.T.; Zhao, W.; Song, H.-S.; Kim, Y.-H.; Chung, J.-W.; Cho, Y.-I.; Park, P.H.; Park, H.-S.; Chae, S.-C.; Park, Y.-J. Development of SSR markers to study diversity in the genus Cymbidium. Biochem. Syst. Ecol. 2010, 38, 585–594. [Google Scholar] [CrossRef]
- Li, X.; Jin, F.; Jin, L.; Jackson, A.; Huang, C.; Li, K.; Shu, X. Development of Cymbidium ensifolium genic-SSR markers and their utility in genetic diversity and population structure analysis in Cymbidiums. BMC Genet. 2014, 15, 124. [Google Scholar] [CrossRef]
- Tsai, C.-C.; Shih, H.-C.; Wang, H.-V.; Lin, Y.-S.; Chang, C.-H.; Chiang, Y.-C.; Chou, C.-H. RNA-Seq SSRs of Moth Orchid and Screening for Molecular Markers across Genus Phalaenopsis (Orchidaceae). PLoS ONE 2015, 10, e0141761. [Google Scholar] [CrossRef]
- Yi, S.S.; Huang, M.Z.; Yang, G.S.; Niu, J.H.; Lu, S.J.; Yin, J.M.; Zhang, Z.Q. Development and characterization of expressed sequence-tagged simple sequence repeat markers in Anoectochilus roxburghii. J. Am. Soc. Hortic. Sci. 2022, 147, 349–357. [Google Scholar] [CrossRef]
- Li, X.B.; Luo, J.; Yan, T.L.; Xiang, L.; Jin, F.; Qin, D.H.; Sun, C.B.; Xie, M. Deep sequencing-based analysis of the Cymbidium ensifolium floral transcriptome. PLoS ONE 2013, 8, e85480. [Google Scholar] [CrossRef] [PubMed]
- Azodi, C.B.; Pardo, J.; VanBuren, R.; de los Campos, G.; Shiu, S.H. Transcriptome-based prediction of complex traits in maize. Plant Cell 2020, 32, 139–151. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Park, H.R.; Lee, A.J.; Nam, D.; Lee, D.G.; Do, Y.; Chung, K.W. Genetic authentication of cultivars with flower-variant types using SSR markers in spring orchid, Cymbidium goeringii. Hortic. Environ. Biotechnol. 2020, 61, 577–590. [Google Scholar] [CrossRef]
- Schulz, D.; Linde, M.; Debener, T. Detection of reproducible major effect QTL for petal traits in garden roses. Plants 2021, 10, 897. [Google Scholar] [CrossRef]
- Chutimanitsakun, Y.; Nipper, R.W.; Cuesta-Marcos, A.; Cistué, L.; Corey, A.; Filichkina, T.; Johnson, E.A.; Hayes, P.M. Construction and application for QTL analysis of a Restriction Site Associated DNA (RAD) linkage map in barley. BMC Genom. 2011, 12, 4. [Google Scholar] [CrossRef]
- Fuentes-Utrilla, P.; Goswami, C.; Cottrell, J.E.; Pong-Wong, R.; Law, A.; A’Hara, S.W.; Lee, S.J.; Woolliams, J.A. QTL analysis and genomic selection using RADseq derived markers in Sitka spruce: The potential utility of within family data. Tree Genet. Genomes 2017, 13, 33. [Google Scholar] [CrossRef]
- Teo, X.; Toh, J.R.H.; Yeoh, H.H.; Su, M.M.J.; Yu, Y.; Chua, H.W.; Lim, C.K.; Kwan, K.M.; Yu, Y.; Wang, M.C. Dissecting the function of MADS-box transcription factors in orchid reproductive development. Front. Plant Sci. 2019, 10, 1474. [Google Scholar] [CrossRef]
- Wang, S.L.; Viswanath, K.K.; Tong, C.G.; An, H.R.; Chen, F.C. Floral induction and flower development of orchids. Front. Plant Sci. 2019, 10, 1258. [Google Scholar] [CrossRef]
- Rasoamanalina, R.O.L.; Mirzaei, K.; El Jaziri, M.; Rasamiravaka, T.; Ramarosandratana, A.V.; Bertin, P. Bertin, Diversity and structure assessment of the genetic resources in a germplasm collection from a vanilla breeding programme in Madagascar. Genet. Resour. Crop Evol. 2023, 70, 548–557. [Google Scholar]
- Baloch, F.S.; Altaf, M.T.; Liaqat, W.; Bedir, M.; Nadeem, M.A.; Cömertpay, G.; Çoban, N.; Habyarimana, E.; Barutçular, C.; Cerit, I.; et al. Recent advancements in the breeding of sorghum crop: Current status and future strategies for marker-assisted breeding. Front. Genet. 2023, 14, 1150616. [Google Scholar] [CrossRef]
- Yang, F.X.; Gao, J.; Wei, Y.L.; Ren, R.; Zhang, G.Q.; Lu, C.Q.; Jin, J.P.; Ai, Y.; Wang, Y.Q.; Chen, L.J.; et al. The genome of Cymbidium sinense revealed the evolution of orchid traits. Plant Biotechnol. J. 2021, 19, 2501–2516. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Zong, J.; Luo, Z.J.; Chen, M.J.; Zhao, X.X.; Zhang, D.B.; Qi, Y.P.; Yuan, Z. Development of a RAD-seq based DNA polymorphism identification software, AgroMarker Finder, and its application in rice marker-assisted breeding. PLoS ONE 2016, 11, e0147187. [Google Scholar] [CrossRef] [PubMed]
- Li, C.R.; Dong, N.; Zhao, Y.; Wu, S.S.; Liu, Z.J.; Zhai, J. A review for the breeding of orchids: Current achievements and prospects. Hortic. Plant J. 2021, 7, 380–392. [Google Scholar] [CrossRef]
- Bhattarai, K.; Van Huylenbroeck, J. Breeding, genetics, and genomics of ornamental plants. Horticulturae 2022, 8, 148. [Google Scholar] [CrossRef]
- Hsu, C.-C.; Chen, S.-Y.; Chiu, S.-Y.; Lai, C.-Y.; Lai, P.-H.; Shehzad, T.; Wu, W.-L.; Chen, W.-H.; Paterson, A.H.; Chen, H.-H. High-density genetic map and genome-wide association studies of aesthetic traits in Phalaenopsis orchids. Sci. Rep. 2022, 12, 3346. [Google Scholar] [CrossRef]
- Li, D.-M.; Zhu, G.-F. High-density genetic linkage map construction and QTLs Identification Associated with four leaf-related traits in lady’s slipper orchids (Paphiopedilum concolor × Paphiopedilum hirsutissimum). Horticulturae 2022, 8, 842. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).