Characterization of Walls Are Thin 1 Family in Cucumis sativus and Functional Identification of CsWAT1-20 in Response to Podosphaera xanthii
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Materials, Fungal Inoculation, and Growth Conditions
2.2. Identification and Characterization of WAT1s in Cucumber
2.3. Analysis of Structure and Motif and Phylogenetic of CsWAT1s
2.4. Chromosome Localization and Collinearity Analysis of CsWAT1s
2.5. Cis-Acting Elements Analysis of CsWAT1s Promoter
2.6. RT-qPCR Validation
2.7. Virus-Induced Gene Silence Test
2.8. Disease Index Investigation and P. xanthii Mycelia Staining
3. Results
3.1. Identification and Phylogeny Analysis of WAT1s in Cucumis sativus
3.2. Chromosome Localization and Collinearity Analysis of CsWAT1s
3.3. Gene Structure and Motif Distribution of CsWAT1s
3.4. Cis-Acting Elements Analysis of CsWAT1s Promoters
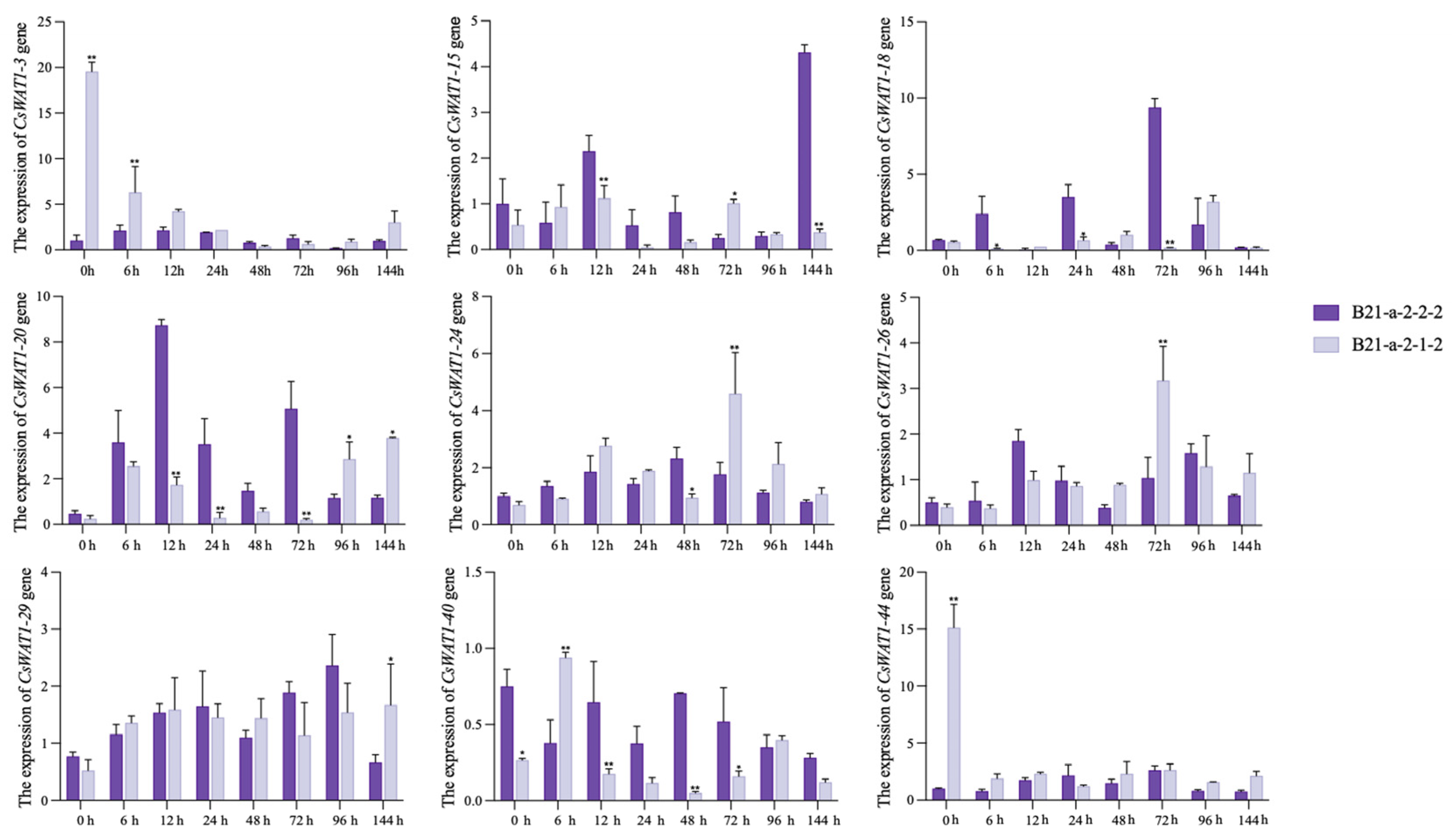
3.5. Expression Profiles of CsWAT1s in Response to P. xanthii Stress
3.6. Silencing of CsWAT1-20 in Cucumber Seedlings Contributes to P. xanthii Stress Tolerance
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Zhang, P.; Zhu, Y.; Zhou, S. Comparative analysis of powdery mildew resistant and susceptible. cultivated cucumber (Cucumis sativus L.) varieties to reveal the metabolic responses to Sphaerotheca fuliginea infection. BMC Plant Biol. 2021, 21, 24. [Google Scholar]
- Chikh-Rouhou, H.; Garcés-Claver, A.; Kienbaum, L.; Ben Belgacem, A.; Gómez-Guillamón, M.L. Resistance of Tunisian Melon Landraces to Podosphaera xanthii. Horticulturae 2022, 8, 1172. [Google Scholar] [CrossRef]
- Meng, X.; Yu, Y.; Song, T.; Yu, Y.; Cui, N.; Ma, Z.; Chen, L.; Fan, H. Transcriptome Sequence Analysis of the Defense Responses of Resistant and Susceptible Cucumber Strains to Podosphaera xanthii. Front. Plant Sci. 2022, 13, 872218. [Google Scholar] [CrossRef]
- Li, X.; Liu, Y.; He, Q.; Li, S.; Liu, W.; Lin, C.; Miao, W. A Candidate Secreted Effector Protein of Rubber Tree Powdery Mildew Fungus Contributes to Infection by Regulating Plant ABA Biosynthesis. Front. Microbiol. 2020, 11, 591387. [Google Scholar] [CrossRef]
- Zhang, D.; Wu, S.; Li, N.; Gao, J.; Liu, S.; Zhu, S.; Li, Z.; Ren, G.; Kuai, B. Chemical induction of leaf senescence and powdery mildew resistance involves ethylene-mediated chlorophyll degradation and ROS metabolism in cucumber. Hortic. Res. 2022, 9, uhac101. [Google Scholar] [CrossRef]
- Li, J.; Gu, C.; Yuan, Y.; Gao, Z.; Qin, Z.; Xin, M. Comparative transcriptome analysis revealed that auxin and cell wall biosynthesis play important roles in the formation of hollow hearts in cucumber. BMC Genom. 2024, 25, 36. [Google Scholar] [CrossRef]
- Dora, S.; Terrett, O.M.; Clara, S.R. Plant-microbe interactions in the apoplast: Communication at the plant cell wall. Plant Cell 2022, 34, 1532–1550. [Google Scholar] [CrossRef]
- Yu, Y.; Yu, Y.; Cui, N.; Ma, L.; Tao, R.; Ma, Z.; Meng, X.; Fan, H. Lignin biosynthesis regulated by CsCSE1 is required for Cucumis sativus defence to Podosphaera xanthii. Plant Physiol. Biochem. 2022, 186, 88–98. [Google Scholar] [CrossRef]
- Miedes, E.; Zarra, I.; Hoson, T.; Herbers, K.; Sonnewald, U.; Lorences, E.P. Xyloglucan endotransglucosylase and cell wall extensibility. J. Plant Physiol. 2011, 168, 196–203. [Google Scholar] [CrossRef]
- Fu, Y.; Win, P.; Zhang, H.; Li, C.; Shen, Y.; He, F.; Luo, K. PtrARF2.1 Is Involved in Regulation of Leaf Development and Lignin Biosynthesis in Poplar Trees. Int. J. Mol. Sci. 2019, 20, 4141. [Google Scholar] [CrossRef]
- Jiang, S.; Tian, X.; Huang, X.; Xin, J.; Yan, H. Physcomitrium patens CAD1 has distinct roles in growth and resistance to biotic stress. BMC Plant Biol. 2022, 22, 518. [Google Scholar] [CrossRef] [PubMed]
- Awwad, F.; Bertrand, G.; Grandbois, M.; Beaudoin, N. Auxin protects Arabidopsis thaliana cell suspension cultures from programmed cell death induced by the cellulose biosynthesis inhibitors thaxtomin A and isoxaben. BMC Plant Biol. 2019, 19, 512. [Google Scholar] [CrossRef] [PubMed]
- Lehman, T.A.; Sanguinet, K.A. Auxin and Cell Wall Crosstalk as Revealed by the Arabidopsis thaliana Cellulose Synthase Mutant Radially Swollen 1. Plant Cell Physiol. 2019, 60, 1487–1503. [Google Scholar] [CrossRef]
- Ranocha, P.; Dima, O.; Felten, J.; Freydier, A.; Hoffmann, L.; Ljung, K.; Lacombe, B.; Corratgé, C.; Thibaud, J.B.; Sundberg, B.; et al. WAT1 (WALLS ARE THIN1) defines a novel auxin transporter in plants and integrates auxin signaling in secondary wall formation in Arabidopsis fibers. BMC Proc. 2011, 5 (Suppl. S7), O24. [Google Scholar] [CrossRef]
- Gamas, P.; Niebel, F.d.C.; Lescure, N.; Cullimore, J. Use of a subtractive hybridization approach to identify new Medicago truncatula genes induced during root nodule development. Mol. Plant Microbe Interact. 1996, 9, 233–242. [Google Scholar] [CrossRef]
- Ranocha, P.; Denancé, N.; Vanholme, R.; Freydier, A.; Martinez, Y.; Hoffmann, L.; Köhler, L.; Pouzet, C.; Renou, J.P.; Sundberg, B.; et al. Walls are thin1 (WAT1), an Arabidopsis homolog of Medicago truncatula NODULIN21, is a tonoplast-localized protein required for secondary wall formation in fibers. Plant J. 2010, 63, 469–483. [Google Scholar] [CrossRef]
- Ranocha, P.; Dima, O.; Nagy, R.; Felten, J.; Corratgé-Faillie, C.; Novák, O.; Morreel, K.; Lacombe, B.; Martinez, Y.; Pfrunder, S.; et al. Arabidopsis WAT1 is a vacuolar auxin transport facilitator required for auxin homoeostasis. Nat. Commun. 2013, 4, 2625. [Google Scholar] [CrossRef]
- Denancé, N.; Sánchez-Vallet, A.; Goffner, D.; Molina, A. Disease resistance or growth: The role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 2013, 4, 155. [Google Scholar] [CrossRef]
- Tang, Y.; Zhang, Z.; Lei, Y.; Hu, G.; Liu, J.; Hao, M.; Chen, A.; Peng, Q.; Wu, J. Cotton WATs modulate SA biosynthesis and local lignin deposition participating in plant resistance against Verticillium dahlia. Front. Plant Sci. 2019, 10, 526. [Google Scholar] [CrossRef]
- Hanika, K.; Schipper, D.; Chinnappa, S.; Oortwijn, M.; Schouten, H.J.; Thomma, B.P.H.J.; Bai, Y. Impairment of Tomato WAT1 Enhances Resistance to Vascular Wilt Fungi Despite Severe Growth Defects. Front. Plant Sci. 2021, 12, 721674. [Google Scholar] [CrossRef]
- Koseoglou, E.; Hanika, K.; MohdNadzir, M.M.; Kohlen, W.; vanderWolf, J.M.; Visser, R.G.F.; Bai, Y. Inactivation of tomato WAT1 leads to reduced susceptibility to Clavibacter michiganensis through downregulation of bacterial virulence factors. Front. Plant Sci. 2023, 14, 1082094. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Ma, L.; Yu, Y.; Ma, Z.; Yin, Y.; Zhou, S.; Yu, Y.; Cui, N.; Meng, X.; Fan, H. Cucumis sativus CsbZIP90 suppresses Podosphaera xanthii resistance by modulating reactive oxygen species. Plant Sci. 2024, 339, 111945. [Google Scholar] [CrossRef] [PubMed]
- Ao, B.; Han, Y.; Wang, S.; Wu, F.; Zhang, J. Genome-Wide Analysis and Profile of UDP-Glycosyltransferases Family in Alfalfa (Medicago sativa L.) under Drought Stress. Int. J. Mol. Sci. 2022, 23, 7243. [Google Scholar] [CrossRef]
- Liao, G.; Li, Y.; Wang, H.; Liu, Q.; Zhong, M.; Jia, D.; Huang, C.; Xu, X. Genome-wide identification and expression profiling analysis of sucrose synthase (SUS) and sucrose phosphate synthase (SPS) genes family in Actinidia chinensis and A. eriantha. BMC Plant Biol. 2022, 22, 215. [Google Scholar] [CrossRef]
- Swarbreck, D.; Wilks, C.; Lamesch, P.; Berardini, T.Z.; Garcia-Hernandez, M.; Foerster, H.; Li, D.; Meyer, T.; Muller, R.; Ploetz, L.; et al. The Arabidopsis Information Resource (TAIR): Gene structure and function annotation. Nucleic Acids Res. 2008, 36, D1009–D1014. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, Y.; Altaf, M.A.; Hao, Y.; Zhou, G.; Li, X.; Zhu, J.; Ma, W.; Wang, Z.; Bao, W. Genome-wide identification of myeloblastosis gene family and its response to cadmium stress in Ipomoea aquatica. Front. Plant Sci. 2022, 13, 979988. [Google Scholar] [CrossRef]
- Baez, L.A.; Tichá, T.; Hamann, T. Cell wall integrity regulation across plant species. Plant Mol. Biol. 2022, 109, 483–504. [Google Scholar] [CrossRef]
- Bacete, L.; Mélida, H.; Miedes, E.; Molina, A. Plant cell wall-mediated immunity: Cell wall changes trigger disease resistance responses. Plant J. 2018, 93, 614–636. [Google Scholar] [CrossRef]
- Molina, A.; Jordá, L.; Torres, M.Á.; Martín-Dacal, M.; Berlanga, D.J.; Fernández-Calvo, P.; Gómez-Rubio, E.; Martín-Santamaría, S. Plant cell wall-mediated disease resistance: Current understanding and future perspectives. Mol. Plant 2024, 17, 699–724. [Google Scholar] [CrossRef]
- Del Corpo, D.; Fullone, M.R.; Miele, R.; Lafond, M.; Pontiggia, D.; Grisel, S.; Kieffer-Jaquinod, S.; Giardina, T.; Bellincampi, D.; Lionetti, V. AtPME17 is a functional Arabidopsis thaliana pectin methylesterase regulated by its PRO region that triggers PME activity in the resistance to Botrytis cinerea. Mol. Plant Pathol. 2020, 21, 1620–1633. [Google Scholar] [CrossRef]



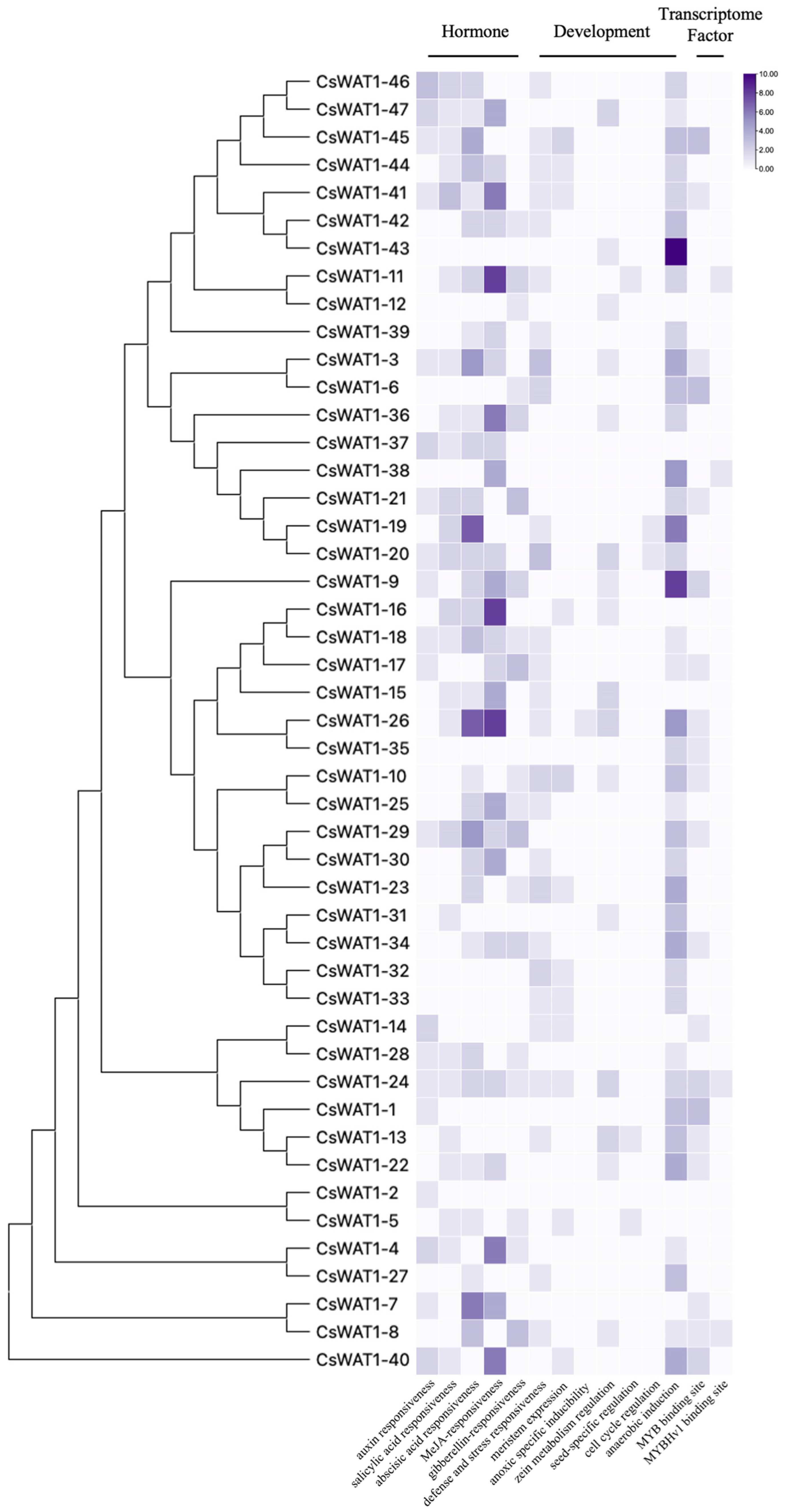

| Gene ID | Rename | Number of Amino Acids | Molecular Weight (kDa) | Theoretical pI | Gravy | Instability Index | Single Peptide |
|---|---|---|---|---|---|---|---|
| CsaV3_1G004090 | CsWAT1-1 | 362 | 40,240.51 | 7.58 | 0.559 | 36.93 | No |
| CsaV3_1G015750 | CsWAT1-2 | 367 | 40,023.02 | 9.34 | 0.402 | 33.84 | No |
| CsaV3_1G029400 | CsWAT1-3 | 369 | 40,737.24 | 8.86 | 0.615 | 42.95 | No |
| CsaV3_1G037450 | CsWAT1-4 | 354 | 39,043.52 | 9.32 | 0.670 | 32.07 | No |
| CsaV3_2G007260 | CsWAT1-5 | 584 | 65,291.84 | 8.84 | 0.451 | 31.17 | No |
| CsaV3_2G007860 | CsWAT1-6 | 369 | 40,123.75 | 8.21 | 0.791 | 42.45 | No |
| CsaV3_2G011920 | CsWAT1-7 | 394 | 43,412.44 | 9.20 | 0.561 | 42.31 | No |
| CsaV3_2G011970 | CsWAT1-8 | 386 | 43,122.36 | 9.40 | 0.314 | 33.06 | No |
| CsaV3_2G012900 | CsWAT1-9 | 360 | 40,729.15 | 9.16 | 0.544 | 39.5 | No |
| CsaV3_2G013970 | CsWAT1-10 | 386 | 42,020.27 | 9.58 | 0.742 | 33.34 | No |
| CsaV3_2G018030 | CsWAT1-11 | 374 | 41,491.28 | 8.90 | 0.367 | 33.12 | No |
| CsaV3_2G018040 | CsWAT1-12 | 314 | 34,780.27 | 9.07 | 0.596 | 36.94 | No |
| CsaV3_2G029170 | CsWAT1-13 | 396 | 42,845.51 | 8.95 | 0.591 | 35.73 | No |
| CsaV3_3G002650 | CsWAT1-14 | 342 | 37,246.73 | 9.03 | 0.842 | 39.66 | No |
| CsaV3_3G039950 | CsWAT1-15 | 397 | 43,687.78 | 9.13 | 0.534 | 29.42 | No |
| CsaV3_3G039960 | CsWAT1-16 | 305 | 33,692.49 | 6.93 | 0.372 | 32.50 | No |
| CsaV3_3G039970 | CsWAT1-17 | 387 | 42,729.21 | 9.37 | 0.669 | 36.12 | No |
| CsaV3_3G039980 | CsWAT1-18 | 381 | 42,023.38 | 9.36 | 0.728 | 33.98 | No |
| CsaV3_3G042150 | CsWAT1-19 | 349 | 37,839.84 | 8.89 | 0.724 | 31.54 | No |
| CsaV3_3G042160 | CsWAT1-20 | 356 | 38,697.67 | 8.35 | 0.650 | 33.29 | No |
| CsaV3_3G042170 | CsWAT1-21 | 361 | 39,111.1 | 8.34 | 0.672 | 37.53 | No |
| CsaV3_3G045560 | CsWAT1-22 | 394 | 42,528.02 | 9.23 | 0.640 | 33.71 | No |
| CsaV3_4G024710 | CsWAT1-23 | 375 | 41,352.42 | 9.23 | 0.547 | 32.18 | No |
| CsaV3_4G027990 | CsWAT1-24 | 374 | 40,087.27 | 8.50 | 0.699 | 30.19 | No |
| CsaV3_4G028340 | CsWAT1-25 | 374 | 40,756.59 | 9.30 | 0.670 | 27.13 | No |
| CsaV3_4G029290 | CsWAT1-26 | 441 | 49,378.12 | 8.94 | 0.350 | 35.64 | No |
| CsaV3_5G001460 | CsWAT1-27 | 362 | 39,127.52 | 8.77 | 0.702 | 30.82 | No |
| CsaV3_5G004250 | CsWAT1-28 | 360 | 38,969.85 | 9.02 | 0.851 | 37.27 | No |
| CsaV3_5G006960 | CsWAT1-29 | 308 | 34,086.16 | 9.66 | 0.308 | 34.56 | No |
| CsaV3_5G007970 | CsWAT1-30 | 377 | 41,435.33 | 8.99 | 0.627 | 36.52 | No |
| CsaV3_5G026330 | CsWAT1-31 | 376 | 40,929.42 | 9.38 | 0.522 | 26.25 | No |
| CsaV3_5G026340 | CsWAT1-32 | 355 | 39,086.41 | 9.43 | 0.622 | 24.44 | No |
| CsaV3_5G026350 | CsWAT1-33 | 357 | 39,279.63 | 9.51 | 0.65 | 30.04 | No |
| CsaV3_5G026360 | CsWAT1-34 | 374 | 40,691.15 | 9.56 | 0.471 | 30.09 | No |
| CsaV3_5G029570 | CsWAT1-35 | 406 | 45,391.56 | 9.35 | 0.365 | 32.63 | No |
| CsaV3_5G031760 | CsWAT1-36 | 374 | 41,277.68 | 8.62 | 0.601 | 34.41 | No |
| CsaV3_5G037340 | CsWAT1-37 | 348 | 38,209.47 | 9.16 | 0.778 | 33.58 | No |
| CsaV3_5G037350 | CsWAT1-38 | 377 | 41,006.99 | 8.25 | 0.492 | 40.00 | No |
| CsaV3_6G005240 | CsWAT1-39 | 365 | 41,024.8 | 9.57 | 0.502 | 25.22 | No |
| CsaV3_6G013640 | CsWAT1-40 | 332 | 37,085.6 | 8.98 | -0.714 | 56.11 | No |
| CsaV3_6G041910 | CsWAT1-41 | 417 | 46,071.84 | 9.48 | 0.463 | 31.34 | No |
| CsaV3_6G041920 | CsWAT1-42 | 277 | 31,060.88 | 9.75 | 0.442 | 42.34 | No |
| CsaV3_6G041930 | CsWAT1-43 | 288 | 31,413.17 | 9.46 | 0.639 | 30.95 | No |
| CsaV3_6G041950 | CsWAT1-44 | 360 | 39,766.27 | 9.45 | 0.606 | 31.37 | No |
| CsaV3_6G041960 | CsWAT1-45 | 425 | 47,743.04 | 9.59 | 0.505 | 30.60 | No |
| CsaV3_6G041970 | CsWAT1-46 | 273 | 30,516.62 | 9.26 | 0.606 | 30.32 | No |
| CsaV3_6G041980 | CsWAT1-47 | 362 | 39,794.29 | 8.96 | 0.581 | 31.84 | No |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hong, J.; Zhao, H.; Yuan, Y.; Wu, J.; Yu, Y.; Cui, N.; Meng, X.; Fan, H. Characterization of Walls Are Thin 1 Family in Cucumis sativus and Functional Identification of CsWAT1-20 in Response to Podosphaera xanthii. Horticulturae 2025, 11, 620. https://doi.org/10.3390/horticulturae11060620
Hong J, Zhao H, Yuan Y, Wu J, Yu Y, Cui N, Meng X, Fan H. Characterization of Walls Are Thin 1 Family in Cucumis sativus and Functional Identification of CsWAT1-20 in Response to Podosphaera xanthii. Horticulturae. 2025; 11(6):620. https://doi.org/10.3390/horticulturae11060620
Chicago/Turabian StyleHong, Jinghang, Hongyan Zhao, Youmei Yuan, Jinming Wu, Yang Yu, Na Cui, Xiangnan Meng, and Haiyan Fan. 2025. "Characterization of Walls Are Thin 1 Family in Cucumis sativus and Functional Identification of CsWAT1-20 in Response to Podosphaera xanthii" Horticulturae 11, no. 6: 620. https://doi.org/10.3390/horticulturae11060620
APA StyleHong, J., Zhao, H., Yuan, Y., Wu, J., Yu, Y., Cui, N., Meng, X., & Fan, H. (2025). Characterization of Walls Are Thin 1 Family in Cucumis sativus and Functional Identification of CsWAT1-20 in Response to Podosphaera xanthii. Horticulturae, 11(6), 620. https://doi.org/10.3390/horticulturae11060620




