Genome-Wide Analysis of the ATGs Family in Watermelon and the Involvement of ATG8s in Graft Union Formation
Abstract
1. Introduction
2. Materials and Methods
2.1. The Plant Materials and Stress Treatments
2.2. Identification of ClaATGs Gene Family Members
2.3. Analysis of Protein Physicochemical Properties of ClaATGs Gene Family Members
2.4. Amino Acid Sequence Comparison and Phylogenetic Tree Construction of ClaATGs Family Members
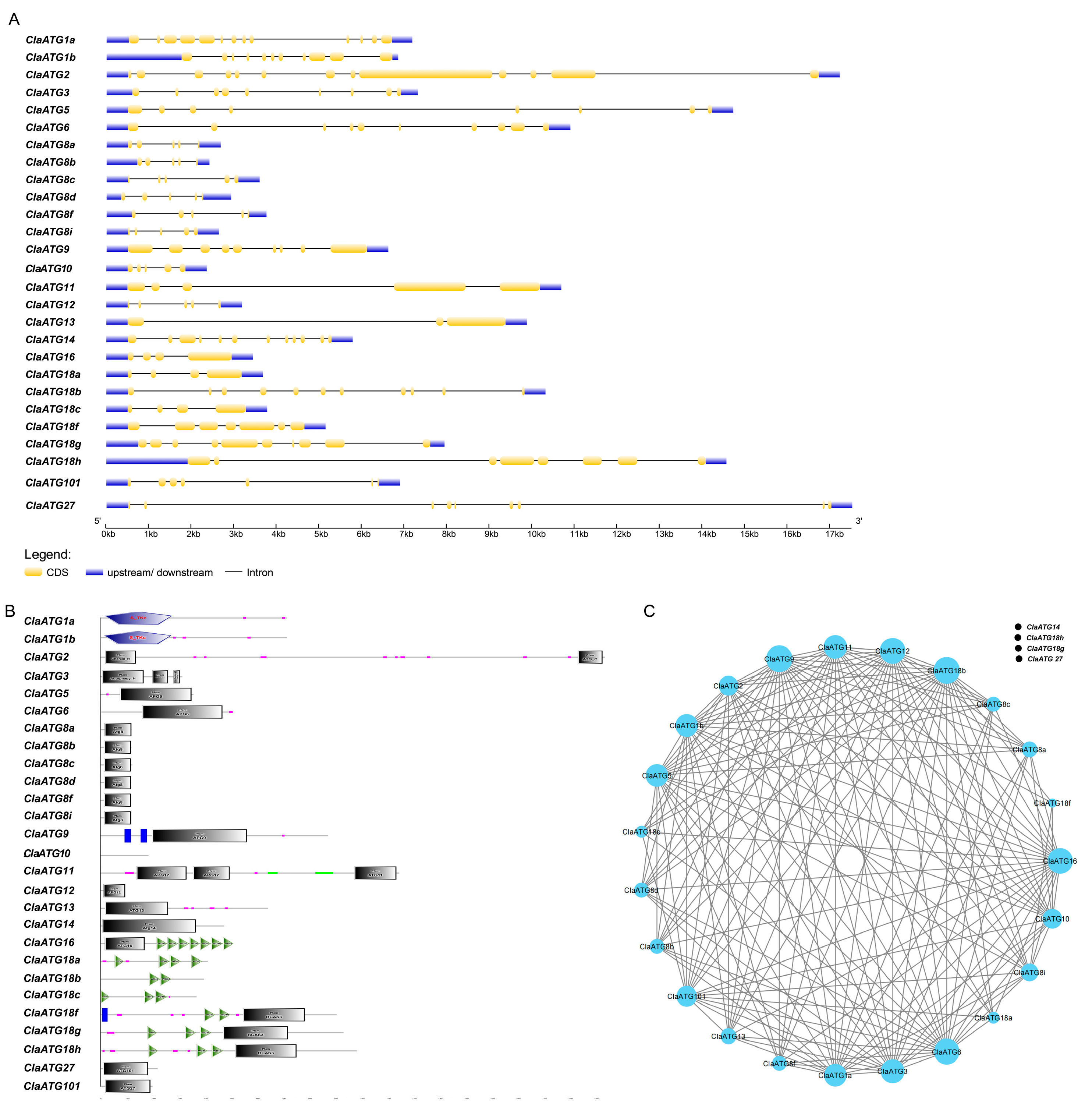
2.5. Gene Structure, Protein Structural Domain Distribution, and Protein Interaction Network Construction of ClaATGs Family Members
2.6. Cis-Acting Element Analysis of ClaATGs Family Members
2.7. Total RNA Extraction, Reverse Transcription and qRT-PCR Analysis
2.8. Measurement of Plant Physiological Parameters
2.9. MDC Staining
2.10. Observation of Grafting Interface Healing
2.11. Data Analysis
3. Results
3.1. Bioinformatics Analysis of 27 ClaATGs in the Watermelon Genome
3.2. Genes Structure and Protein Motif Analysis of Genes Encoding ClaATGs
3.3. Promoter Elements Analysis of ClaATGs and Respond to Exogenous Sugars
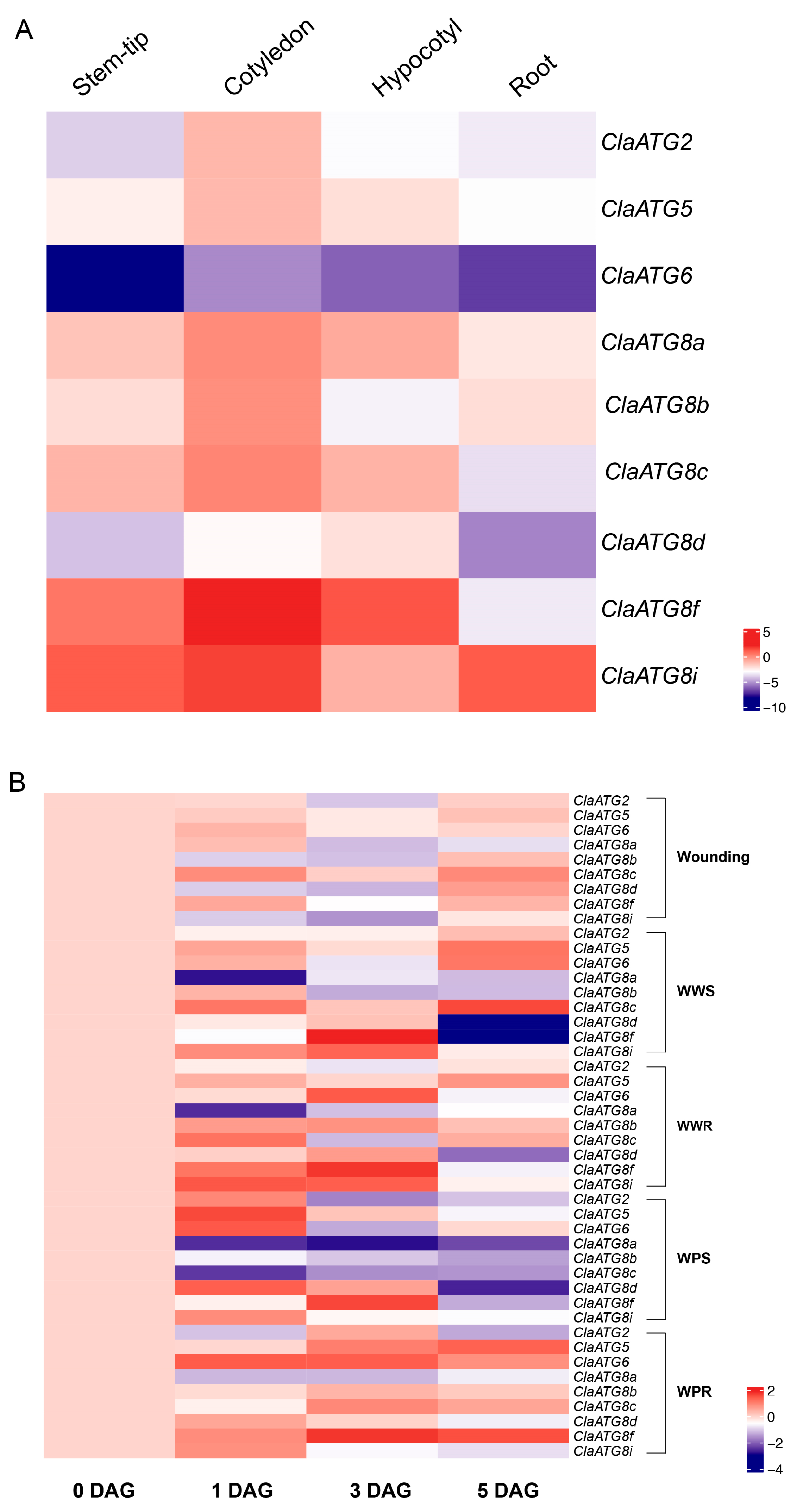
3.4. Watermelon ClaATGs Expression Profiles at Various Tissues and Wounding Responses
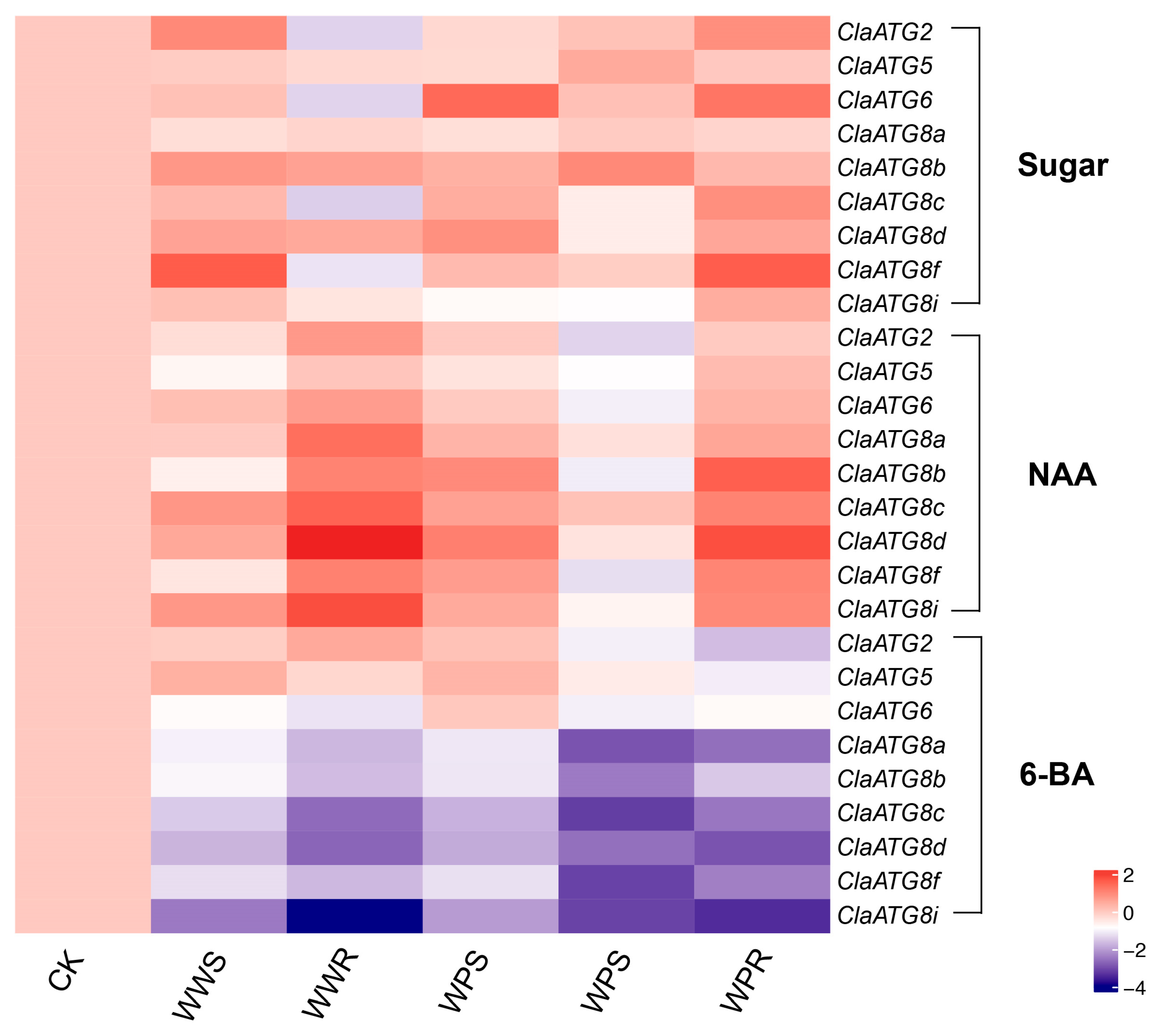
3.5. Watermelon ClaATG Expression Profiles in Response to Various Treatments at the Grafting Union
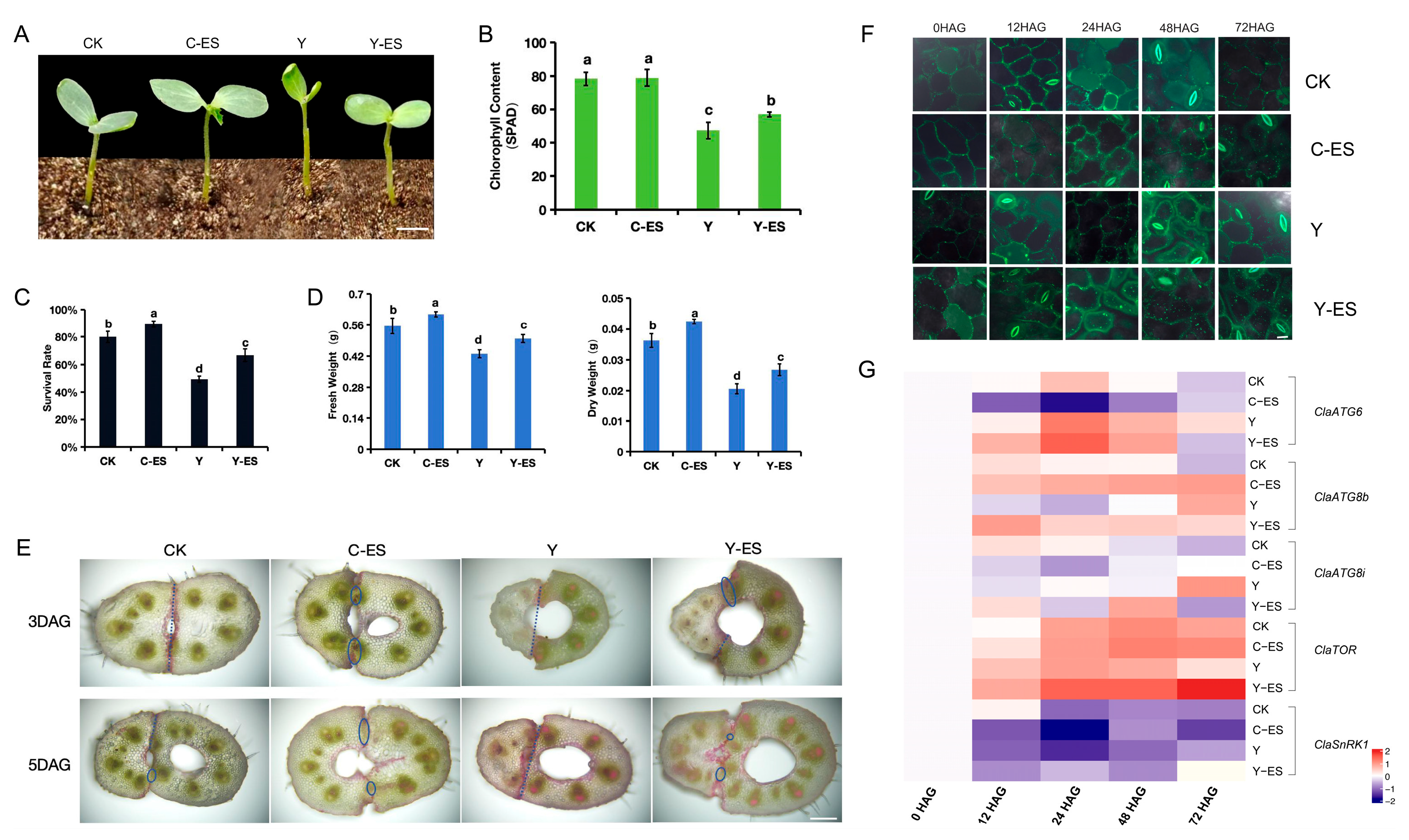
3.6. Autophagy Monitoring During Heterograft Graft Union Formation After an Exogenous Sugar Treatment
4. Discussion
4.1. ClaATGs Involved in the Growth and Development of Watermelon
4.2. Expression of ClaATGs Under Various Treatments
4.3. ClaATGs Involved in Graft Union Formation Under Sugar Starvation in Watermelon Plants
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Kanne, J.V.; Ishikawa, M.; Bressendorff, S.; Ansbol, J.; Hasebe, M.; Rodriguez, E.; Petersen, M. Overexpression of ATG8/LC3 enhances wound-induced somatic reprogramming in Physcomitrium patens. Autophagy 2022, 18, 1463–1466. [Google Scholar] [CrossRef] [PubMed]
- Bassham, D.C. Plant autophagy--more than a starvation response. Curr. Opin. Plant Biol. 2007, 10, 587–593. [Google Scholar] [CrossRef] [PubMed]
- Katz, E.; Chamovitz, D.A. Wounding of Arabidopsis leaves induces indole-3-carbinol-dependent autophagy in roots of Arabidopsis thaliana. Plant J. 2017, 91, 779–787. [Google Scholar] [CrossRef]
- Slavikova, S.; Ufaz, S.; Avin-Wittenberg, T.; Levanony, H.; Galili, G. An autophagy-associated Atg8 protein is involved in the responses of Arabidopsis seedlings to hormonal controls and abiotic stresses. J. Exp. Bot. 2008, 59, 4029–4043. [Google Scholar] [CrossRef]
- Sláviková, S.; Shy, G.; Yao, Y.; Glozman, R.; Levanony, H.; Pietrokovski, S.; Elazar, Z.; Galili, G. The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J. Exp. Bot. 2005, 56, 2839–2849. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Yang, W.; Yue, J.; Liu, Y.; Pei, D.; Wang, H. The Responses of Wheat Autophagy and ATG8 Family Genes to Biotic and Abiotic Stresses. J. Plant Growth Regul. 2020, 39, 867–876. [Google Scholar] [CrossRef]
- Xia, T.; Xiao, D.; Liu, D.; Chai, W.; Gong, Q.; Wang, N.N. Heterologous Expression of ATG8c from Soybean Confers Tolerance to Nitrogen Deficiency and Increases Yield in Arabidopsis. PLoS ONE 2012, 7, e37217. [Google Scholar] [CrossRef]
- Minina, E.A.; Moschou, P.N.; Vetukuri, R.R.; Sanchez-Vera, V.; Cardoso, C.; Liu, Q.; Elander, P.H.; Dalman, K.; Beganovic, M.; Yilmaz, J.L.; et al. Transcriptional stimulation of rate-limiting components of the autophagic pathway improves plant fitness. J. Exp. Bot. 2018, 69, 1415–1432. [Google Scholar] [CrossRef]
- Yu, X.-Q.; Su, W.; Zhang, H.; Niu, M.; Liu, X.; Li, Z.; Liu, C.; Wang, H.-L.; Yin, W.; Xia, X. Genome-wide analysis of autophagy-related gene family and PagATG18a enhances salt tolerance by regulating ROS homeostasis in poplar. Int. J. Biol. Macromol. 2023, 224, 1524–1540. [Google Scholar] [CrossRef]
- Wei, Y.; Liu, W.; Hu, W.; Liu, G.; Wu, C.; Liu, W.; Zeng, H.; He, C.; Shi, H. Genome-wide analysis of autophagy-related genes in banana highlights MaATG8s in cell death and autophagy in immune response to Fusarium wilt. Plant Cell Rep. 2017, 36, 1237–1250. [Google Scholar] [CrossRef]
- Kakanj, P.; Bhide, S.; Moussian, B.; Leptin, M. Autophagy-mediated plasma membrane removal promotes the formation of epithelial syncytia. Embo J. 2022, 41, e109992. [Google Scholar] [CrossRef] [PubMed]
- Mijaljica, D.; Spada, F.; Klionsky, D.J.J.; Harrison, I.P.P. Autophagy is the key to making chronic wounds acute in skin wound healing. Autophagy 2023, 19, 2578–2584. [Google Scholar] [CrossRef] [PubMed]
- Miao, L.; Li, S.; Bai, L.; Anwar, A.; Li, Y.; He, C.; Yu, X. Effect of grafting methods on physiological change of graft union formation in cucumber grafted onto bottle gourd rootstock. Sci. Hortic. 2019, 244, 249–256. [Google Scholar] [CrossRef]
- Kurotani, K.I.; Tabata, R.; Kawakatsu, Y.; Sugita, R.; Notaguchi, M. Autophagy is Induced During Plant Grafting for Wound Healing; Cold Spring Harbor Laboratory: New York, NY, USA, 2020. [Google Scholar]
- Pu, Y.; Luo, X.; Bassham, D.C. TOR-Dependent and -Independent Pathways Regulate Autophagy in Arabidopsis thaliana. Front. Plant Sci. 2017, 8, 1204. [Google Scholar] [CrossRef] [PubMed]
- Janse van Rensburg, H.C.; Van den Ende, W.; Signorelli, S. Autophagy in Plants: Both a Puppet and a Puppet Master of Sugars. Front. Plant Sci. 2019, 10, 1664. [Google Scholar] [CrossRef]
- Miao, L.; Li, Q.; Sun, T.-S.; Chai, S.; Wang, C.; Bai, L.; Sun, M.; Li, Y.; Qin, X.; Zhang, Z.; et al. Sugars promote graft union development in the heterograft of cucumber onto pumpkin. Hortic. Res. 2021, 8, 146. [Google Scholar] [CrossRef]
- Li, Y.; Wang, N.; Feng, J.; Liu, Y.; Wang, H.; Deng, S.; Dong, W.; Liu, X.; Lv, B.; Sun, J.; et al. Enhancing genetic transformation efficiency in cucurbit crops through AtGRF5 overexpression: Mechanistic insights and applications. J. Integr. Plant Biol. 2025. [Google Scholar] [CrossRef]
- Kong, Q.; Yuan, J.; Gao, L.; Zhao, S.; Jiang, W.; Huang, Y.; Bie, Z. Identification of suitable reference genes for gene expression normalization in qRT-PCR analysis in watermelon. PLoS ONE 2014, 9, e90612. [Google Scholar] [CrossRef]
- Wang, P.; Nolan, T.M.; Yin, Y.; Bassham, D.C. Identification of transcription factors that regulate ATG8 expression and autophagy in Arabidopsis. Autophagy 2020, 16, 123–139. [Google Scholar] [CrossRef]
- Yang, C.; Shen, W.; Yang, L.; Sun, Y.; Li, X.; Lai, M.; Wei, J.; Wang, C.; Xu, Y.; Li, F.; et al. HY5-HDA9 module transcriptionally regulates plant autophagy in response to light-to-dark conversion and nitrogen starvation. Mol. Plant 2020, 13, 515–531. [Google Scholar] [CrossRef]
- Xia, K.; Liu, T.; Ouyang, J.; Wang, R.; Fan, T.; Zhang, M. Genome-Wide Identification, Classification, and Expression Analysis of Autophagy-Associated Gene Homologues in Rice (Oryza sativa L.). DNA Res. 2011, 18, 363–377. [Google Scholar] [CrossRef] [PubMed]
- Zhai, Y.; Guo, M.; Wang, H.; Lu, J.; Liu, J.; Zhang, C.; Gong, Z.; Lu, M. Autophagy, a Conserved Mechanism for Protein Degradation, Responds to Heat, and Other Abiotic Stresses in Capsicum annuum L. Front. Plant Sci. 2016, 7, 131. [Google Scholar] [CrossRef]
- Han, Y.; Yang, Y.; Wang, Y.; Elsheery, N.I.; Ding, G. Genome-wide identification and expression analysis of autophagy genes in cucumber. Res. Sq. 2021. [Google Scholar] [CrossRef]
- Wang, H.; Ding, Z.; Gou, M.; Hu, J.; Wang, Y.; Wang, L.; Wang, Y.; Di, T.; Zhang, X.; Hao, X.; et al. Genome-wide identification, characterization, and expression analysis of tea plant autophagy-related genes (CsARGs) demonstrates that they play diverse roles during development and under abiotic stress. BMC Genom. 2021, 22, 121. [Google Scholar] [CrossRef]
- Qin, G.; Ma, Z.; Zhang, L.; Xing, S.; Hou, X.; Deng, J.; Liu, J.; Chen, Z.; Qu, L.-J.; Gu, H. Arabidopsis AtBECLIN 1/AtAtg6/AtVps30 is essential for pollen germination and plant development. Cell Res. 2007, 17, 249–263. [Google Scholar] [CrossRef]
- Di Berardino, J.; Marmagne, A.; Berger, A.; Yoshimoto, K.; Cueff, G.; Chardon, F.; Masclaux-Daubresse, C.; Reisdorf-Cren, M. Autophagy controls resource allocation and protein storage accumulation in Arabidopsis seeds. J. Exp. Bot. 2018, 69, 1403–1414. [Google Scholar] [CrossRef] [PubMed]
- Nair, U.; Yen, W.-L.; Mari, M.; Cao, Y.; Xie, Z.; Baba, M.; Reggiori, F.; Klionsky, D.J. A role for Atg8-PE deconjugation in autophagosome biogenesis. Autophagy 2012, 8, 780–793. [Google Scholar] [CrossRef]
- Nieto-Torres, J.L.; Leidal, A.M.; Debnath, J.; Hansen, M. Beyond Autophagy: The Expanding Roles of ATG8 Proteins. Trends Biochem. Sci. 2021, 46, 673–686. [Google Scholar] [CrossRef] [PubMed]
- Xie, Z.; Nair, U.; Klionsky, D.J. Atg8 controls phagophore expansion during autophagosome formation. Mol. Biol. Cell 2008, 19, 3290–3298. [Google Scholar] [CrossRef]
- Wang, Y.; Yu, B.; Zhao, J.; Guo, J.; Li, Y.; Han, S.; Huang, L.; Du, Y.; Hong, Y.; Tang, D.; et al. Autophagy Contributes to Leaf Starch Degradation. Plant Cell 2013, 25, 1383–1399. [Google Scholar] [CrossRef]
- Yan, H.; Liu, Y.; Zhang, K.; Song, J.; Xu, W.; Su, Z. Chromatin State-Based Analysis of Epigenetic H3K4me3 Marks of Arabidopsis in Response to Dark Stress. Front. Genet. 2019, 10, 306. [Google Scholar] [CrossRef]
- Huang, X.; Zheng, C.; Liu, F.; Yang, C.; Zheng, P.; Lu, X.; Tian, J.; Chung, T.; Otegui, M.S.; Xiao, S.; et al. Genetic Analyses of the Arabidopsis ATG1 Kinase Complex Reveal Both Kinase-Dependent and Independent Autophagic Routes during Fixed-Carbon Starvation. Plant Cell 2019, 31, 2973–2995. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, E.; Chevalier, J.; Olsen, J.; Ansbol, J.; Kapousidou, V.; Zuo, Z.; Svenning, S.; Loefke, C.; Koemeda, S.; Drozdowskyj, P.S.; et al. Autophagy mediates temporary reprogramming and dedifferentiation in plant somatic cells. Embo J. 2020, 39, e103315. [Google Scholar] [CrossRef]
- Saera-Vila, A.; Kish, P.E.; Louie, K.W.; Grzegorski, S.J.; Klionsky, D.J.; Kahana, A. Autophagy regulates cytoplasmic remodeling during cell reprogramming in a zebrafish model of muscle regeneration. Autophagy 2016, 12, 1864–1875. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Zhao, Y.; Wang, B.; Han, Y.; Li, Y.; Prusky, D.; Bi, Y. Soluble sugars, organic acids and energy metabolism involved in the wound healing of muskmelons elicited by benzothiadiazole. Postharvest Biol. Technol. 2023, 199, 112277. [Google Scholar] [CrossRef]
- Li, K.; Liang, Y.; Xing, L.; Mao, J.; Liu, Z.; Dong, F.; Meng, Y.; Han, M.; Zhao, C.; Bao, L.; et al. Transcriptome Analysis Reveals Multiple Hormones, Wounding and Sugar Signaling Pathways Mediate Adventitious Root Formation in Apple Rootstock. Int. J. Mol. Sci. 2018, 19, 2201. [Google Scholar] [CrossRef]
- Jarvis, P.; Lopez-Juez, E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 2013, 14, 787–802. [Google Scholar] [CrossRef]
- Hirota, T.; Izumi, M.; Wada, S.; Makino, A.; Ishida, H. Vacuolar Protein Degradation via Autophagy Provides Substrates to Amino Acid Catabolic Pathways as an Adaptive Response to Sugar Starvation in Arabidopsis thaliana. Plant Cell Physiol. 2018, 59, 1363–1376. [Google Scholar] [CrossRef]
- Izumi, M.; Nakamura, S.; Otomo, K.; Ishida, H.; Hidema, J.; Nemoto, T.; Hagihara, S. Autophagosome development and chloroplast segmentation occur synchronously for piecemeal degradation of chloroplasts. Elife 2024, 12, RP93232. [Google Scholar] [CrossRef]
- Dobrenel, T.; Caldana, C.; Hanson, J.; Robaglia, C.; Vincentz, M.; Veit, B.; Meyer, C. TOR Signaling and Nutrient Sensing. Annu. Rev. Plant Biol. 2016, 67, 261–285. [Google Scholar] [CrossRef]
- Ananieva, E.A.; Gillaspy, G.E.; Ely, A.; Burnette, R.N.; Erickson, F.L. Interaction of the WD40 domain of a myoinositol polyphosphate 5-phosphatase with SnRK1 links inositol, sugar, and stress signaling. Plant Physiol. 2008, 148, 1868–1882. [Google Scholar] [CrossRef] [PubMed]
- Baena-González, E.; Rolland, F.; Thevelein, J.M.; Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 2007, 448, 938–943. [Google Scholar] [CrossRef] [PubMed]
- Ramon, M.; Dang, T.V.T.; Broeckx, T.; Hulsmans, S.; Crepin, N.; Sheen, J.; Rolland, F. Default Activation and Nuclear Translocation of the Plant Cellular Energy Sensor SnRK1 Regulate Metabolic Stress Responses and Development. Plant Cell 2019, 31, 1614–1632. [Google Scholar] [CrossRef] [PubMed]
- Versluys, M.; Tarkowski, L.P.; Van den Ende, W. Fructans As DAMPs or MAMPs: Evolutionary Prospects, Cross-Tolerance, and Multistress Resistance Potential. Front. Plant Sci. 2017, 7, 2061. [Google Scholar] [CrossRef]


| Gene Name | Gene ID | ORF (bp) | AA | MW (KDa) | pI | Instability Index | Aliphatic Index | Loc | SignalP | TMHs |
|---|---|---|---|---|---|---|---|---|---|---|
| ClaATG1a | Cla97C06G111650 | 2154 | 717 | 79.78 | 6.06 | unstable | 91.28 | Nucleus | NO | 0 |
| ClaATG1b | Cla97C10G204720 | 2154 | 717 | 79.75 | 6.78 | unstable | 87.46 | Chloroplast | NO | 0 |
| ClaATG2 | Cla97C05G092440 | 5823 | 1940 | 214.86 | 5.51 | unstable | 86.57 | Nucleus | NO | 0 |
| ClaATG3 | Cla97C06G111850 | 948 | 315 | 35.54 | 4.80 | unstable | 83.49 | Cytoplasm | NO | 0 |
| ClaATG5 | Cla97C05G092850 | 1077 | 358 | 40.58 | 5.00 | unstable | 92.51 | Cytoplasm | NO | 0 |
| ClaATG6 | Cla97C06G111550 | 1530 | 509 | 58.23 | 6.23 | stable | 70.35 | Nucleus | NO | 0 |
| ClaATG8a | Cla97C03G055830 | 357 | 118 | 13.65 | 8.78 | unstable | 84.32 | Cytoplasm | NO | 0 |
| ClaATG8b | Cla97C01G021580 | 372 | 123 | 13.76 | 9.12 | unstable | 88.05 | Cytoplasm | NO | 0 |
| ClaATG8c | Cla97C02G035300 | 360 | 119 | 13.73 | 8.62 | unstable | 84.37 | Cytoplasm | NO | 0 |
| ClaATG8d | Cla97C01G004150 | 357 | 118 | 13.69 | 8.71 | stable | 85.93 | Cytoplasm | NO | 0 |
| ClaATG8f | Cla97C06G112490 | 354 | 117 | 13.51 | 7.89 | unstable | 82.48 | Cytoplasm | NO | 0 |
| ClaATG8i | Cla97C09G167010 | 357 | 118 | 13.85 | 6.73 | stable | 68.47 | Nucleus | NO | 0 |
| ClaATG9 | Cla97C07G129250 | 2628 | 875 | 100.98 | 6.14 | unstable | 82.00 | Plasmid | NO | 5 |
| ClaATG10 | Cla97C09G171110 | 558 | 185 | 21.53 | 5.88 | unstable | 78.00 | Nucleus | NO | 0 |
| ClaATG11 | Cla97C02G040060 | 3450 | 1149 | 129.83 | 5.57 | unstable | 82.66 | Nucleus | NO | 0 |
| ClaATG12 | Cla97C07G140800 | 288 | 95 | 10.51 | 9.21 | unstable | 88.21 | Cytoplasm | NO | 0 |
| ClaATG13 | Cla97C01G014550 | 1935 | 644 | 71.20 | 9.07 | unstable | 69.21 | Nucleus | NO | 0 |
| ClaATG14 | Cla97C11G215480 | 1431 | 476 | 53.61 | 8.88 | unstable | 76.43 | Chloroplast | NO | 0 |
| ClaATG16 | Cla97C06G127290 | 1536 | 511 | 56.19 | 6.17 | stable | 91.84 | Cytoplasm | NO | 0 |
| ClaATG18a | Cla97C09G168700 | 1242 | 413 | 45.61 | 6.60 | unstable | 80.48 | Cytoplasm | NO | 0 |
| ClaATG18b | Cla97C11G218570 | 1113 | 370 | 40.39 | 6.75 | unstable | 98.08 | Chloroplast | NO | 0 |
| ClaATG18c | Cla97C11G212430 | 1200 | 399 | 44.27 | 6.18 | stable | 88.17 | Nucleus | NO | 0 |
| ClaATG18f | Cla97C05G096000 | 2730 | 909 | 98.78 | 8.34 | unstable | 86.42 | Chloroplast | NO | 1 |
| ClaATG18g | Cla97C09G169290 | 2808 | 935 | 102.22 | 5.81 | unstable | 79.60 | Chloroplast | NO | 0 |
| ClaATG18h | Cla97C03G066990 | 2964 | 987 | 107.01 | 5.83 | unstable | 75.73 | Chloroplast | NO | 0 |
| ClaATG101 | Cla97C01G015650 | 663 | 220 | 25.82 | 6.15 | unstable | 92.50 | Nucleus | NO | 0 |
| ClaATG27 | Cla97C07G133080 | 603 | 200 | 22.03 | 8.77 | unstable | 97.35 | Cytoplasm | NO | 1 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ding, F.; Cheng, S.; Fan, S.; Fan, X.; Chen, X.; Zhang, J.; Zhang, Y.; Li, Y.; Miao, L. Genome-Wide Analysis of the ATGs Family in Watermelon and the Involvement of ATG8s in Graft Union Formation. Horticulturae 2025, 11, 619. https://doi.org/10.3390/horticulturae11060619
Ding F, Cheng S, Fan S, Fan X, Chen X, Zhang J, Zhang Y, Li Y, Miao L. Genome-Wide Analysis of the ATGs Family in Watermelon and the Involvement of ATG8s in Graft Union Formation. Horticulturae. 2025; 11(6):619. https://doi.org/10.3390/horticulturae11060619
Chicago/Turabian StyleDing, Fei, Siqi Cheng, Shaoshuai Fan, Xiulan Fan, Xiaonuan Chen, Jianan Zhang, Yixin Zhang, Yansu Li, and Li Miao. 2025. "Genome-Wide Analysis of the ATGs Family in Watermelon and the Involvement of ATG8s in Graft Union Formation" Horticulturae 11, no. 6: 619. https://doi.org/10.3390/horticulturae11060619
APA StyleDing, F., Cheng, S., Fan, S., Fan, X., Chen, X., Zhang, J., Zhang, Y., Li, Y., & Miao, L. (2025). Genome-Wide Analysis of the ATGs Family in Watermelon and the Involvement of ATG8s in Graft Union Formation. Horticulturae, 11(6), 619. https://doi.org/10.3390/horticulturae11060619






