Identification and Expression Profiles Reveal Key Myelocytomatosis (MYC) Involved in Drought, Chilling, and Salt Tolerance in Solanum lycopersicum
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide Identification of MYC Genes in Tomato
2.2. Prediction and Analysis of Physical and Chemical Properties
2.3. Phylogenetic Relationship, Gene Structure, and Conserved Motif Analysis
2.4. Promoter Analysis
2.5. Spatiotemporal Differential Expression, Interaction Network Analysis, and Signal Pathway Enrichment Analysis
2.6. Homology Modeling and Molecular Docking of SlMYC 3D Structure
2.7. qRT-PCR
2.8. LCI
2.9. Statistical Analysis
3. Results
3.1. Analysis of Whole Genome and Basic Physicochemical Properties of the SlMYC Family in Tomato
3.2. Phylogeny, Conserved Motif, and Structure Analysis of the SlMYC Family in Tomato
3.3. Promoter Analysis of the SlMYC Family
3.4. Expression and Pathway Enrichment of the SlMYC Family in Tomato Tissues
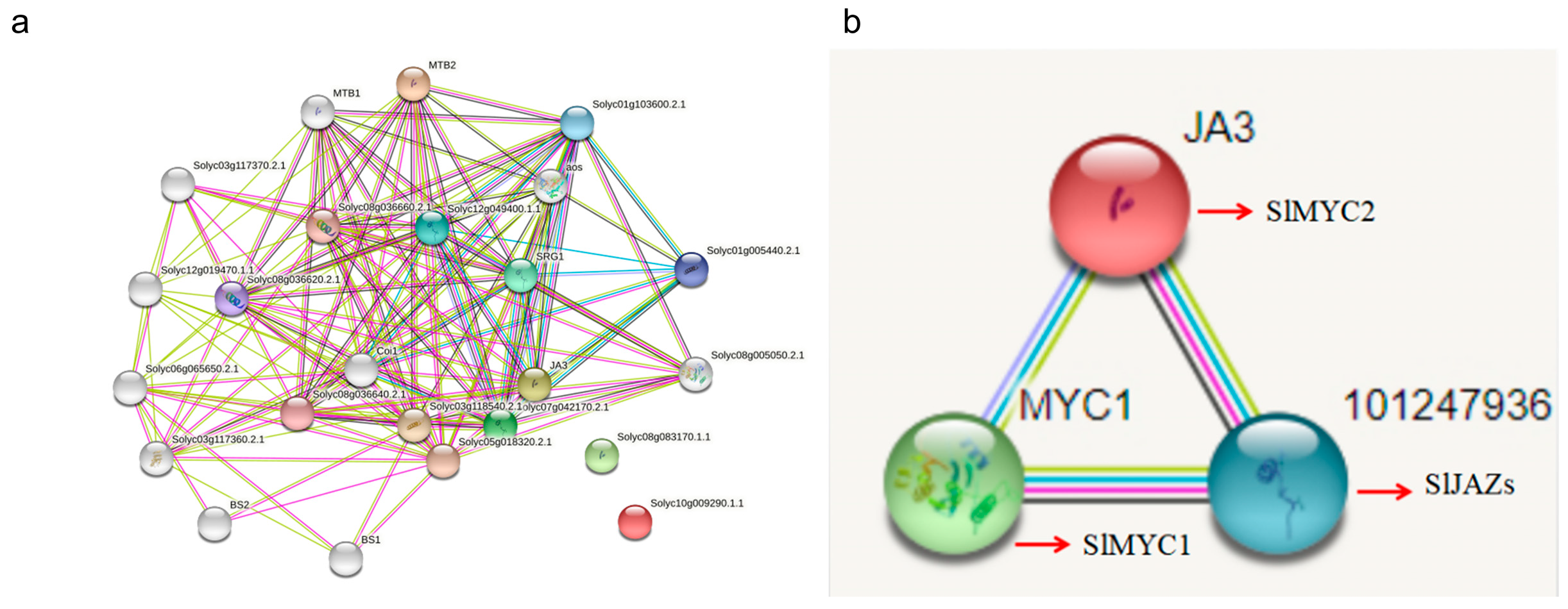
3.5. Interaction Network Analysis of the SlMYC Family
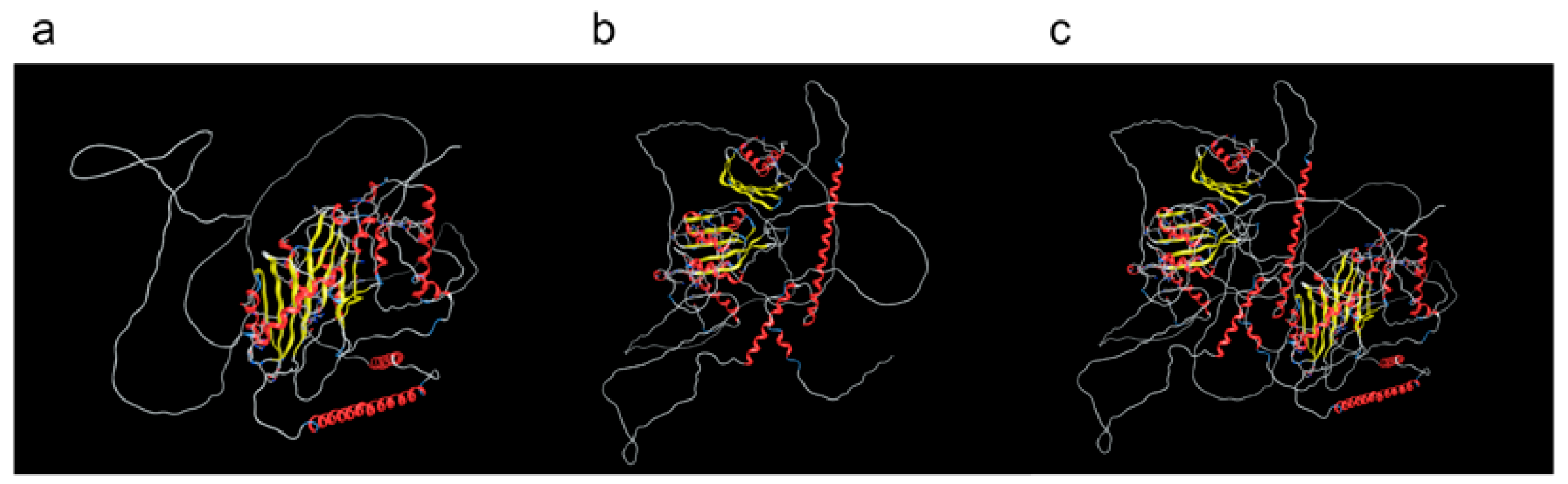
3.6. Protein 3D Models Predicted Docking with Protein Molecules
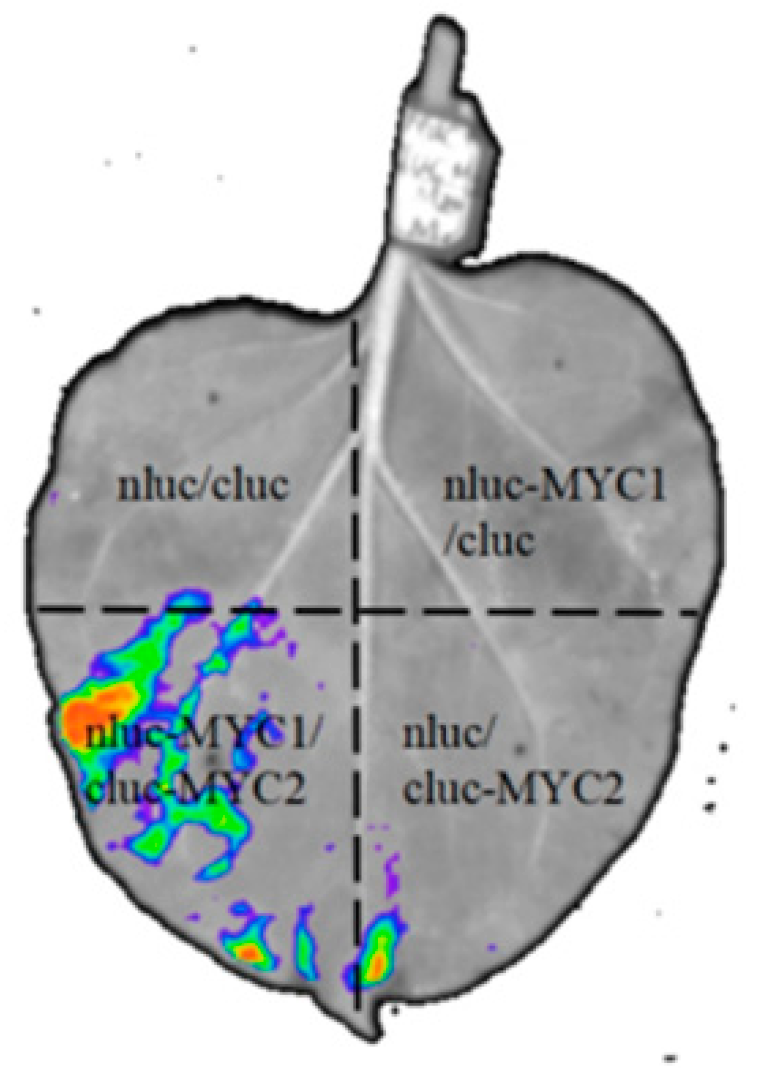
3.7. Interaction of SlMYC1 and SlMYC2
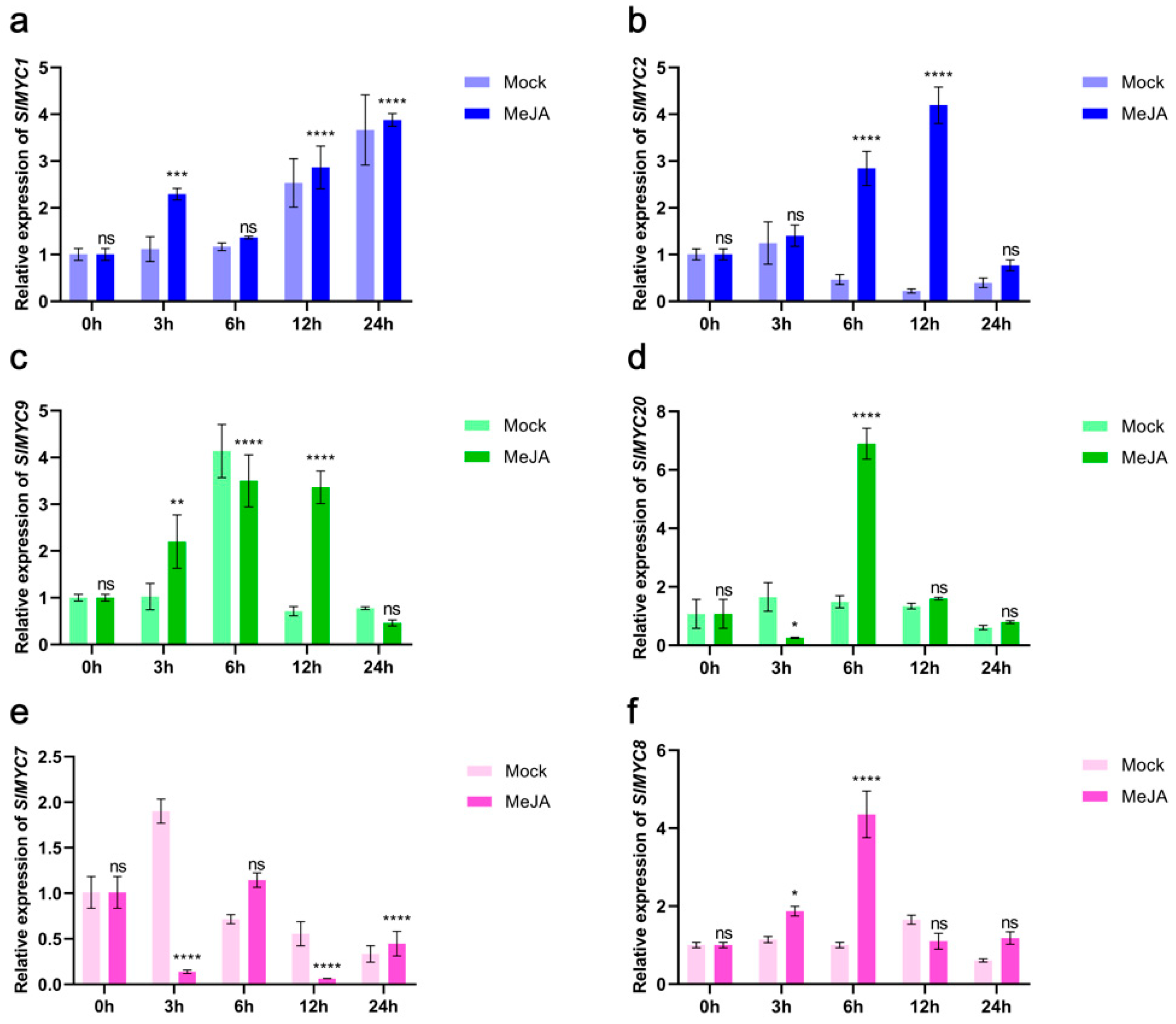
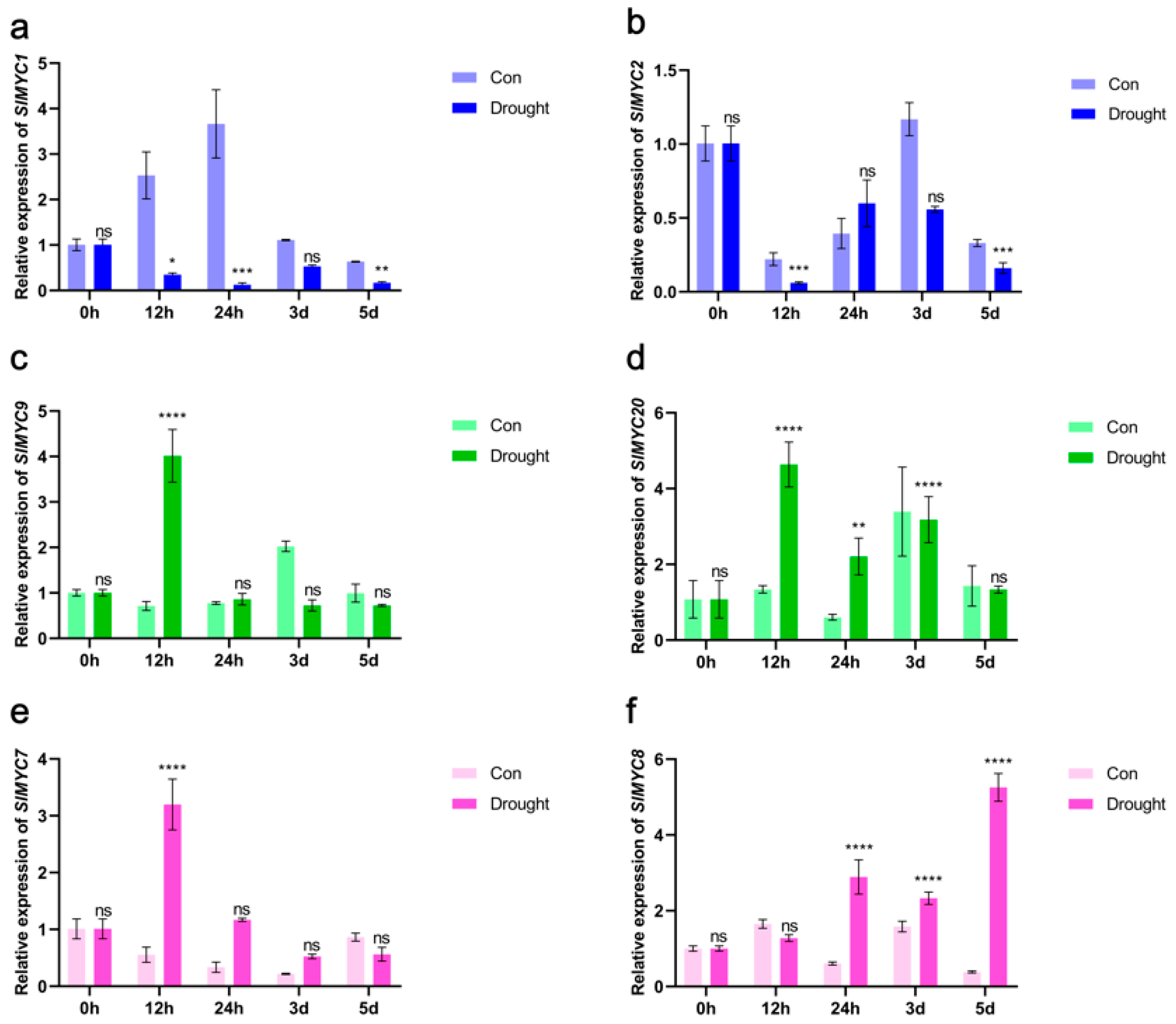
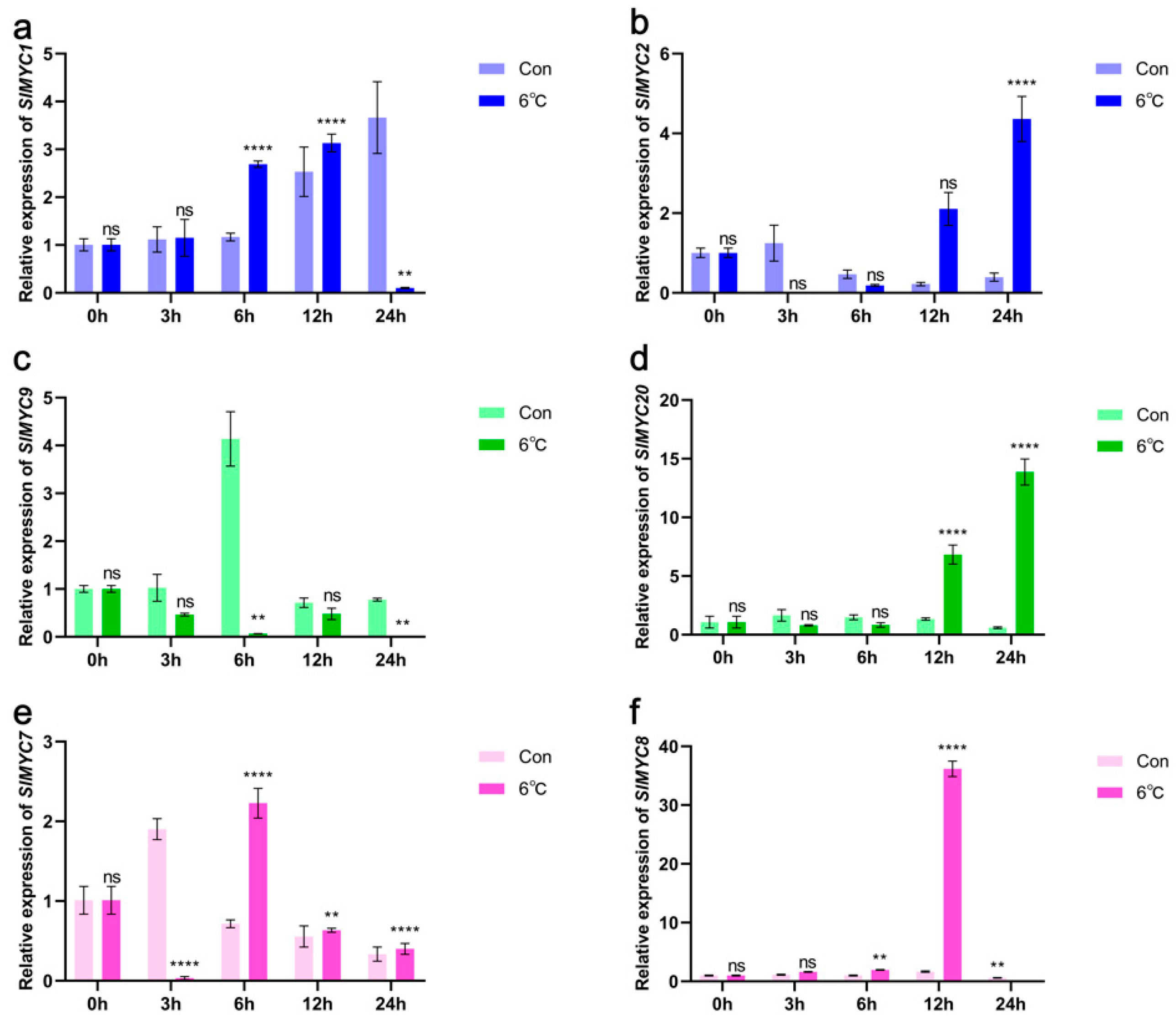
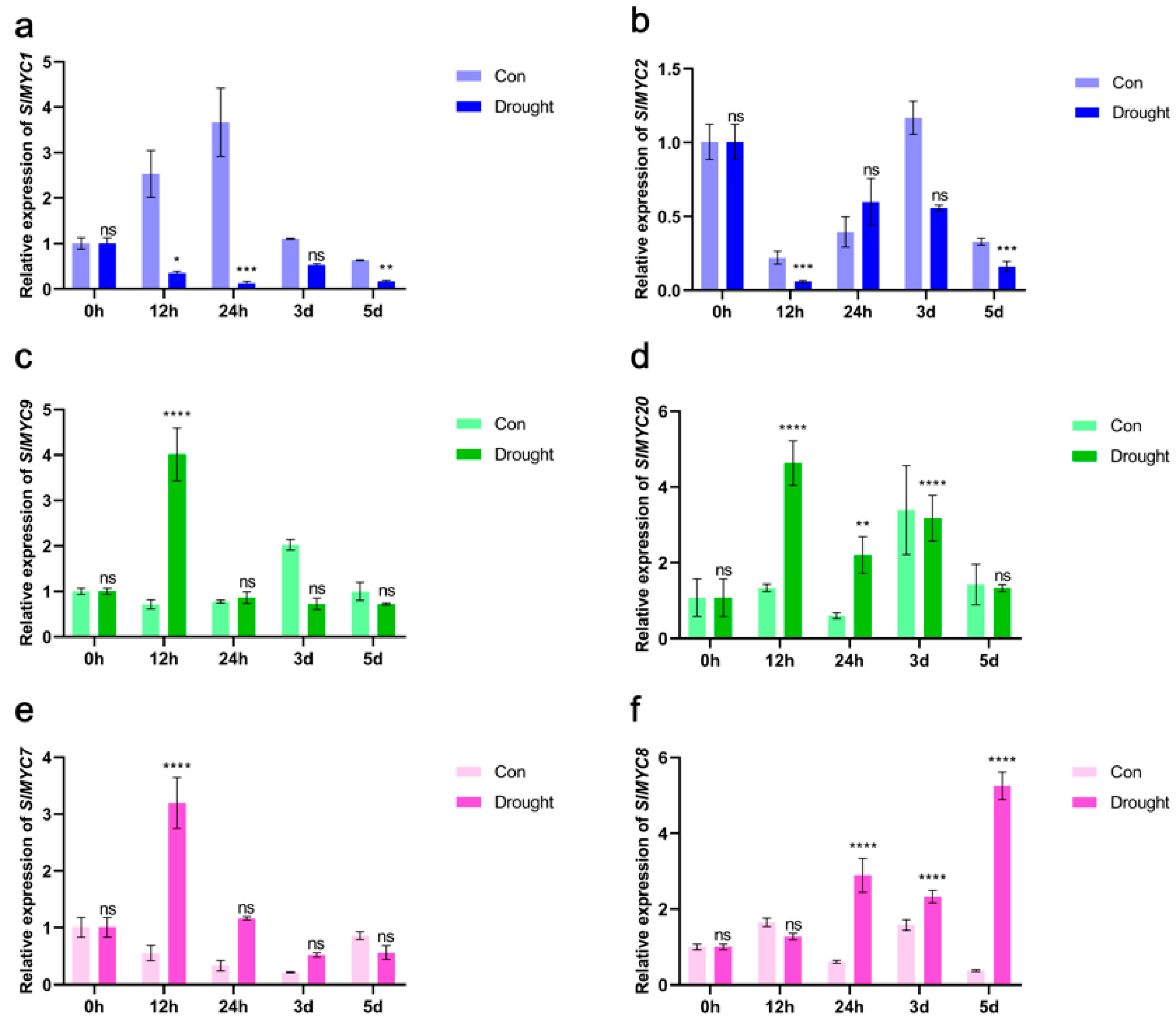
3.8. Expression of SlMYCs in Tomato
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Xiong, Y.; Sheen, J. The role of target of rapamycin signaling networks in plant growth and metabolism. Plant Physiol. 2014, 164, 499–512. [Google Scholar] [CrossRef] [PubMed]
- Harris, T.E.; Lawrence, J.C., Jr. TOR signaling. Sci. Signal. 2003, 2003, re15. [Google Scholar] [CrossRef] [PubMed]
- Ouziad, F.; Hildebrandt, U.; Schmelzer, E.; Bothe, H. Differential gene expressions in arbuscular mycorrhizal-colonized tomato grown under heavy metal stress. J. Plant Physiol. 2005, 162, 634–649. [Google Scholar] [CrossRef]
- Tanveer, K.; Gilani, S.; Hussain, Z.; Ishaq, R.; Adeel, M.; Ilyas, N. Effect of salt stress on tomato plant and the role of calcium. J. Plant Nutr. 2020, 43, 28–35. [Google Scholar] [CrossRef]
- Zhou, R.; Yu, X.; Ottosen, C.O.; Rosenqvist, E.; Zhao, L.; Wang, Y.; Yu, W.; Zhao, T.; Wu, Z. Drought stress had a predominant effect over heat stress on three tomato cultivars subjected to combined stress. BMC Plant Biol. 2017, 17, 24. [Google Scholar] [CrossRef]
- Wang, Y.; Mostafa, S.; Zeng, W.; Jin, B. Function and mechanism of jasmonic acid in plant responses to abiotic and biotic stresses. Int. J. Mol. Sci. 2021, 22, 8568. [Google Scholar] [CrossRef]
- Ruan, J.; Zhou, Y.; Zhou, M.; Yan, J.; Khurshid, M.; Weng, W.; Cheng, J.; Zhang, K. Jasmonic acid signaling pathway in plants. Int. J. Mol. Sci. 2019, 20, 2479. [Google Scholar] [CrossRef]
- Huang, H.; Liu, B.; Liu, L.; Song, S. Jasmonate action in plant growth and development. J. Exp. Bot. 2017, 68, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Chang, C.; Tucker, M.L. To grow old: Regulatory role of ethylene and jasmonic acid in senescence. Front. Plant Sci. 2015, 6, 20. [Google Scholar] [CrossRef]
- Song, S.; Qi, T.; Wasternack, C.; Xie, D. Jasmonate signaling and crosstalk with gibberellin and ethylene. Curr. Opin. Plant Biol. 2014, 21, 112–119. [Google Scholar] [CrossRef]
- Wasternack, C.; Hause, B. Jasmonates: Biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. Bot. 2013, 111, 1021–1058. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Xu, M.; Cai, X.; Han, Z.; Si, J.; Chen, D. Jasmonate signaling pathway modulates plant defense, growth, and their trade-offs. Int. J. Mol. Sci. 2022, 23, 3945. [Google Scholar] [CrossRef]
- Li, L.; Zhang, H.; Chai, X.; Lv, J.; Hu, L.; Wang, J.; Li, Z.; Yu, J.; Liu, Z. Genome-wide identification and expression analysis of the MYC transcription factor family and its response to sulfur stress in cabbage (Brassica oleracea L.). Gene 2022, 814, 146116. [Google Scholar] [CrossRef]
- Goossens, J.; Fernández-Calvo, P.; Schweizer, F.; Goossens, A. Jasmonates: Signal transduction components and their roles in environmental stress responses. Plant Mol. Biol. 2016, 91, 673–689. [Google Scholar] [CrossRef]
- Yan, Y.; Stolz, S.P.; Chételat, A.; Reymond, P.; Pagni, M.; Dubugnon, L.; Farmer, E.E. A downstream mediator in the growth repression limb of the jasmonate pathway. Plant Cell 2007, 19, 2470–2483. [Google Scholar] [CrossRef] [PubMed]
- TariqJaveed, M.; Farooq, T.; Al-Hazmi, A.S.; Hussain, M.D.; Rehman, A.U. Role of Trichoderma as a biocontrol agent (BCA) of phytoparasitic nematodes and plant growth inducer. J. Invertebr. Pathol. 2021, 183, 107626. [Google Scholar] [CrossRef]
- Yan, L.; Zhai, Q.; Wei, J.; Li, S.; Wang, B.; Huang, T.; Du, M.; Sun, J.; Kang, L.; Li, C.; et al. Role of tomato lipoxygenase D in wound-induced jasmonate biosynthesis and plant immunity to insect herbivores. PLoS Genet. 2013, 9, e1003964. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Song, L.; Gong, X.; Xu, J.; Li, M. Functions of jasmonic acid in plant regulation and response to abiotic stress. Int. J. Mol. Sci. 2020, 21, 1446. [Google Scholar] [CrossRef]
- Creelman, R.A.; Mullet, J.E. Jasmonic acid distribution and action in plants: Regulation during development and response to biotic and abiotic stress. Proc. Natl. Acad. Sci. USA 1995, 92, 4114–4119. [Google Scholar] [CrossRef]
- Delgado, C.; Mora-Poblete, F.; Ahmar, S.; Chen, J.-T.; Figueroa, C.R. Jasmonates and plant salt stress: Molecular players, physiological effects, and improving tolerance by using genome-associated tools. Int. J. Mol. Sci. 2021, 22, 3082. [Google Scholar] [CrossRef]
- Paes, B.; Carpinetti, P.d.A.; Fraga, O.T.; Rodrigues-Silva, P.L.; Fioresi, V.S.; de Camargos, L.F.; Ferreira, M.F.d.S. Abiotic stresses in plants and their markers: A practice view of plant stress responses and programmed cell death mechanisms. Plants 2022, 11, 1100. [Google Scholar] [CrossRef] [PubMed]
- Mostofa, M.G.; Rahman, M.M.; Ansary, M.M.U.; Keya, S.S.; Abdelrahman, M.; Miah, M.G.; Phan Tran, L.-S. Silicon in mitigation of abiotic stress-induced oxidative damage in plants. Crit. Rev. Biotechnol. 2021, 41, 918–934. [Google Scholar] [CrossRef]
- Ilyas, M.; Nisar, M.; Khan, N.; Hazrat, A.; Khan, A.H.; Hayat, K.; Fahad, S.; Khan, A.; Ullah, A. drought tolerance strategies in plants: A mechanistic approach. J. Plant Growth Regul. 2021, 40, 926–944. [Google Scholar] [CrossRef]
- Campos, M.L.; Kang, J.-H.; Howe, G.A. Jasmonate-triggered plant immunity. J. Chem. Ecol. 2014, 40, 657–675. [Google Scholar] [CrossRef] [PubMed]
- Llanes, A.S.; Andrade, A.M.; Alemano, S.G.; Luna, M.V. Alterations of endogenous hormonal levels in plants under drought and salinity. Am. J. Plant Sci. 2016, 7, 1357–1371. [Google Scholar] [CrossRef]
- Gupta, A.; Hisano, H.; Hojo, Y.; Matsuura, T.; Ikeda, Y.; Mori, I.C.; Senthil-Kumar, M. Global profiling of phytohormone dynamics during combined drought and pathogen stress in Arabidopsis thaliana reveals ABA and JA as major regulators. Sci. Rep. 2017, 7, 4017. [Google Scholar] [CrossRef] [PubMed]
- Aerts, N.; Pereira Mendes, M.; Van Wees, S.C. Multiple levels of crosstalk in hormone networks regulating plant defense. Plant J. 2021, 105, 489–504. [Google Scholar] [CrossRef]
- Kudla, J.; Becker, D.; Grill, E.; Hedrich, R.; Hippler, M.; Kummer, U.; Parniske, M.; Romeis, T.; Schumacher, K.J.N.P. Advances and current challenges in calcium signaling. New Phytol. 2018, 218, 414–431. [Google Scholar] [CrossRef]
- Heim, M.A.; Jakoby, M.; Werber, M.; Martin, C.; Weisshaar, B.; Bailey, P.C. The basic helix–loop–helix transcription factor family in plants: A genome-wide study of protein structure and functional diversity. Mol. Biol. Evol. 2003, 20, 735–747. [Google Scholar] [CrossRef]
- Hu, M.; Song, X.; Liu, Y.; Zhao, S. Analysis of expression patterns of MYC family members in Populus. For. Res. 2023, 36, 32–40. [Google Scholar]
- Song, C.; Cao, Y.; Dai, J.; Li, G.; Manzoor, M.A.; Chen, C.; Deng, H. The multifaceted roles of MYC2 in plants: Toward transcriptional reprogramming and stress tolerance by jasmonate signaling. Front. Plant Sci. 2022, 13, 868874. [Google Scholar] [CrossRef] [PubMed]
- Lorenzo, O.; Chico, J.M.; Saénchez-Serrano, J.J.; Solano, R. Jasmonate-insensitive1 encodes a MYC transcription factor essential to discriminate between different jasmonate-regulated defense responses in Arabidopsis. Plant Cell 2004, 16, 1938–1950. [Google Scholar] [CrossRef] [PubMed]
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar] [CrossRef]
- Li, R.; Weldegergis, B.T.; Li, J.; Jung, C.; Qu, J.; Sun, Y.; Qian, H.; Tee, C.; van Loon, J.J.; Dicke, M.; et al. Virulence factors of geminivirus interact with MYC2 to subvert plant resistance and promote vector performance. Plant Cell 2014, 26, 4991–5008. [Google Scholar] [CrossRef]
- Hua, B.; Chang, J.; Wu, M.; Xu, Z.; Zhang, F.; Yang, M.; Xu, H.; Wang, L.; Chen, X.; Wu, S. Mediation of JA signalling in glandular trichomes by the woolly/SlMYC1 regulatory module improves pest resistance in tomato. Plant Biol. J. 2021, 19, 375–393. [Google Scholar] [CrossRef]
- Zhai, Q.; Deng, L.; Li, C. Mediator subunit MED25: At the nexus of jasmonate signaling. Curr. Opin. Plant Biol. 2020, 57, 78–86. [Google Scholar] [CrossRef]
- Ming, R.; Zhang, Y.; Wang, Y.; Khan, M.; Dahro, B.; Liu, J. The JA-responsive MYC2-BADH-like transcriptional regulatory module in Poncirus trifoliata contributes to cold tolerance by modulation of glycine betaine biosynthesis. New Phytol. 2021, 229, 2730–2750. [Google Scholar] [CrossRef]
- Xu, J.; van Herwijnen, Z.O.; Dräger, D.B.; Sui, C.; Haring, M.A.; Schuurink, R.C. SlMYC1 Regulates Type VI Glandular Trichome Formation and Terpene Biosynthesis in Tomato Glandular Cells. Plant Cell 2018, 30, 2988–3005. [Google Scholar] [CrossRef] [PubMed]
- Reymond, P.; Bodenhausen, N.; Van Poecke, R.M.; Krishnamurthy, V.; Dicke, M.; Farmer, E.E. A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 2004, 16, 3132–3147. [Google Scholar] [CrossRef]
- Yadav, V.; Mallappa, C.; Gangappa, S.N.; Bhatia, S.; Chattopadhyay, S. A basic helix-loop-helix transcription factor in Arabidopsis, MYC2, acts as a repressor of blue light–mediated photomorphogenic growth. Plant Cell 2005, 17, 1953–1966. [Google Scholar] [CrossRef]
- Chen, Q.; Sun, J.; Zhai, Q.; Zhou, W.; Qi, L.; Xu, L.; Wang, B.; Chen, R.; Jiang, H.; Qi, J.; et al. The basic helix-loop-helix transcription factor MYC2 directly represses plethora expression during jasmonate-mediated modulation of the root stem cell niche in Arabidopsis. Plant Cell 2011, 23, 3335–3352. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Li, Y.; Pan, J.; Lou, D.; Hu, Y.; Yu, D. The bHLH transcription factors MYC2, MYC3, and MYC4 are required for jasmonate-mediated inhibition of flowering in Arabidopsis. Mol. Plant 2017, 10, 1461–1464. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Cheng, Y.; Wei, F.; Liu, Y.; Zhu, R.; Zhao, P.; Zhang, J.; Xiang, C.; Kang, E.; Shang, Z. Arabidopsis thaliana MYC2 and MYC3 are involved in ethylene-regulated hypocotyl growth as negative regulators. Int. J. Mol. Sci. 2024, 25, 8022. [Google Scholar] [CrossRef] [PubMed]
- Qi, T.; Song, S.; Ren, Q.; Wu, D.; Huang, H.; Chen, Y.; Fan, M.; Peng, W.; Ren, C.; Xie, D. The Jasmonate-ZIM-domain proteins interact with the WD-Repeat/bHLH/MYB complexes to regulate Jasmonate-mediated anthocyanin accumulation and trichome initiation in Arabidopsis thaliana. Plant Cell 2011, 23, 1795–1814. [Google Scholar] [CrossRef]
- Shan, X.; Zhang, Y.; Peng, W.; Wang, Z.; Xie, D. Molecular mechanism for jasmonate-induction of anthocyanin accumulation in Arabidopsis. J. Exp. Bot. 2009, 60, 3849–3860. [Google Scholar] [CrossRef]
- Qi, T.; Wang, J.; Huang, H.; Liu, B.; Gao, H.; Liu, Y.; Song, S.; Xie, D. Regulation of jasmonate-induced leaf senescence by antagonism between bHLH subgroup IIIe and IIId factors in Arabidopsis. Plant Cell 2015, 27, 1634–1649. [Google Scholar] [CrossRef]
- Zhu, X.; Chen, J.; Xie, Z.; Gao, J.; Ren, G.; Gao, S.; Zhou, X.; Kuai, B. Jasmonic acid promotes degreening via MYC2/3/4-and ANAC019/055/072-mediated regulation of major chlorophyll catabolic genes. Plant J. 2015, 84, 597–610. [Google Scholar] [CrossRef]
- Uji, Y.; Akimitsu, K.; Gomi, K. Identification of OsMYC2-regulated senescence-associated genes in rice. Planta 2017, 245, 1241–1246. [Google Scholar] [CrossRef]
- Uji, Y.; Taniguchi, S.; Tamaoki, D.; Shishido, H.; Akimitsu, K.; Gomi, K. Overexpression of OsMYC2 Results in the up-regulation of early JA-responsive genes and bacterial blight resistance in rice. Plant Cell Physiol. 2016, 57, 1814–1827. [Google Scholar] [CrossRef]
- Shoji, T.; Hashimoto, T.J.P.; Physiology, C. Tobacco MYC2 regulates jasmonate-inducible nicotine biosynthesis genes directly and by way of the NIC2-locus ERF genes. Plant Cell Physiol. 2011, 52, 1117–1130. [Google Scholar] [CrossRef]
- Zhang, H.; Bokowiec, M.T.; Rushton, P.J.; Han, S.; Timko, M.P. Tobacco Transcription Factors NtMYC2a and NtMYC2b Form Nuclear Complexes with the NtJAZ1 Repressor and Regulate Multiple Jasmonate-Inducible Steps in Nicotine Biosynthesis. Mol. Plant 2012, 5, 73–84. [Google Scholar] [CrossRef]
- An, J.; Wang, X.; Yao, J.; Ren, Y.; You, C.; Wang, X.; Hao, Y. Apple MdMYC2 reduces aluminum stress tolerance by directly regulating MdERF3 gene. Plant Soil 2017, 418, 255–266. [Google Scholar] [CrossRef]
- Li, T.; Xu, Y.; Zhang, L.; Ji, Y.; Tan, D.; Yuan, H.; Wang, A. The jasmonate-activated transcription factor MdMYC2 regulates ETHYLENE RESPONSE FACTOR and ethylene biosynthetic genes to promote ethylene biosynthesis during apple fruit ripening. Plant Cell 2017, 29, 1316–1334. [Google Scholar] [CrossRef]
- Lenka, S.K.; Nims, N.E.; Vongpaseuth, K.; Boshar, R.A.; Roberts, S.C.; Walker, E.L. Jasmonate-responsive expression of paclitaxel biosynthesis genes in Taxus cuspidata cultured cells is negatively regulated by the bHLH transcription factors TcJAMYC1, TcJAMYC2, and TcJAMYC4. Front. Plant Sci. 2015, 6, 115. [Google Scholar] [CrossRef] [PubMed]
- Zheng, L.; Gu, X.; Xiao, Y.; Wang, S.; Liu, L.; Pan, H.; Zhang, H. Antifungal activity of Bacillus mojavensis D50 against Botrytis cinerea causing postharvest gray mold of tomato. Sci. Hortic. 2023, 312, 111841. [Google Scholar] [CrossRef]
- Zheng, J.; Liao, Y.; Ye, J.; Xu, F.; Zhang, W.; Zhou, X.; Wang, L.; He, X.; Cao, Z.; Yi, Y.; et al. The transcription factor MYC2 positively regulates terpene trilactone biosynthesis through activating GbGGPPS expression in Ginkgo biloba. Hortic. Res. 2024, 11, uhae228. [Google Scholar] [CrossRef] [PubMed]
- Uimari, A.; Strommer, J. Myb26: A MYB-like protein of pea flowers with affinity for promoters of phenylpropanoid genes. Plant J. 1997, 12, 1273–1284. [Google Scholar] [CrossRef] [PubMed]
- Cao, L.; Liu, L.; Zhang, C.; Ren, W.; Zheng, J.; Tao, C.; Zhu, W.; Xiang, M.; Wang, L.; Liu, Y.; et al. The MYC2 and MYB43 transcription factors cooperate to repress HMA2 and HMA4 expression, altering cadmium tolerance in Arabidopsis thaliana. J. Hazard. Mater. 2024, 479, 135703. [Google Scholar] [CrossRef]
- Zuo, Z.; Kang, H.; Park, M.; Jeong, H.; Sun, H.; Song, P.; Lee, H. Zoysia japonica MYC type transcription factor ZjICE1 regulates cold tolerance in transgenic Arabidopsis. Plant Sci. 2019, 289, 110254. [Google Scholar] [CrossRef]
- Wu, X.; Xu, S.; Zhao, P.; Zhang, X.; Yao, X.; Sun, Y.; Fang, R.; Ye, J.J.P.P. The Orthotospovirus nonstructural protein NSs suppresses plant MYC-regulated jasmonate signaling leading to enhanced vector attraction and performance. PLoS Pathog. 2019, 15, e1007897. [Google Scholar] [CrossRef]
- Yu, C.; Lin, C.; Hwang, J. Predicting subcellular localization of proteins for gram-negative bacteria by support vector machines based on n-peptide compositions. Protein Sci. 2004, 13, 1402–1406. [Google Scholar] [CrossRef] [PubMed]
- Gillani, M.; Pollastri, G. Protein subcellular localization prediction tools. Comput. Struct. Biotechnol. J. 2024, 23, 1796–1807. [Google Scholar] [CrossRef] [PubMed]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef]
- Hu, B.; Jin, J.; Guo, A.-Y.; Zhang, H.; Luo, J.; Gao, G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics 2015, 31, 1296–1297. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Consortium, T.G. The tomato genome sequence provides insights into fleshy fruit evolution. Nature 2012, 485, 635. [Google Scholar] [CrossRef]
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589. [Google Scholar] [CrossRef]
- Chen, Q.; Zhou, S.; Qu, M.; Yang, Y.; Chen, Q.; Meng, X.; Fan, H. Cucumber (Cucumis sativus L.) translationally controlled tumor protein interacts with CsRab11A and promotes activation of target of rapamycin in response to Podosphaera xanthii. Plant J. 2024, 119, 332–347. [Google Scholar] [CrossRef]
- Chini, A.; Gimenez-Ibanez, S.; Goossens, A.; Solano, R. Redundancy and specificity in jasmonate signalling. Curr. Opin. Plant Biol. 2016, 33, 147–156. [Google Scholar] [CrossRef]
- Gasperini, D.; Chételat, A.; Acosta, I.F.; Goossens, J.; Pauwels, L.; Goossens, A.; Dreos, R.; Alfonso, E.; Farmer, E.E. Multilayered organization of jasmonate signalling in the regulation of root growth. PLoS Genet. 2015, 11, e1005300. [Google Scholar] [CrossRef]
- Havko, N.E.; Major, I.T.; Jewell, J.B.; Attaran, E.; Browse, J.; Howe, G.A. Control of carbon assimilation and partitioning by jasmonate: An accounting of growth–defense tradeoffs. Plants 2016, 5, 7. [Google Scholar] [CrossRef] [PubMed]
- Ramirez-Prado, J.S.; Latrasse, D.; Rodriguez-Granados, N.Y.; Huang, Y.; Manza-Mianza, D.; Brik-Chaouche, R.; Jaouannet, M.; Citerne, S.; Bendahmane, A.; Hirt, H.; et al. The Polycomb protein LHP 1 regulates Arabidopsis thaliana stress responses through the repression of the MYC 2-dependent branch of immunity. Plant J. 2019, 100, 1118–1131. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Ma, N.; Meng, X.; Zhang, S.; Wang, J.; Chai, S.; Meng, Q. A novel tomato MYC-type ICE1-like transcription factor, SlICE1a, confers cold, osmotic and salt tolerance in transgenic tobacco. Plant Physiol. Biol. 2013, 73, 309–320. [Google Scholar] [CrossRef]
- Shimomura, K.; Ohm, M.; San Thein, M. Collaborative exploration of Cucurbitaceae vegetable genetic resources in Myanmar in 2019. Annu. Rep. Explor. Introd. Plant Genet. Resour. 2020, 36, 148–158. [Google Scholar]
- Liu, Y.; Du, M.; Deng, L.; Shen, J.; Fang, M.; Chen, Q.; Lu, Y.; Wang, Q.; Li, C.; Zhai, Q. MYC2 regulates the termination of jasmonate signaling via an autoregulatory negative feedback loop. Plant Cell 2019, 31, 106–127. [Google Scholar] [CrossRef]
- Ali, M.S.; Baek, K.-H. Jasmonic acid signaling pathway in response to abiotic stresses in plants. Int. J. Mol. Sci. 2020, 21, 621. [Google Scholar] [CrossRef]
- Yang, Z.; Zhou, Y.; Wang, X.; Gu, S.; Yu, J.; Liang, G.; Yan, C.; Xu, C. Genomewide comparative phylogenetic and molecular evolutionary analysis of tubby-like protein family in Arabidopsis, rice, and poplar. Genomics 2008, 92, 246–253. [Google Scholar] [CrossRef]
- Han, Y.; Cheng, H.; Jiang, Y.; Wang, X.; Liu, X.; Zhang, D.; Wu, J.; Liu, L.; Yan, M.; Zhou, D. Identification and characterization of the BnFAR1/FHY3 gene family and expression analysis under shading and low-temperature responses in Brassica napus L. Agronomy 2024, 14, 202. Agronomy 2024, 14, 202. [Google Scholar] [CrossRef]
- Goossens, J.; Mertens, J.; Goossens, A. Role and functioning of bHLH transcription factors in jasmonate signalling. J. Exp. Bot. 2017, 68, 1333–1347. [Google Scholar] [CrossRef]
- Wasternack, C. Termination in jasmonate signaling by MYC2 and MTBs. Trends Plant Sci. 2019, 24, 667–669. [Google Scholar] [CrossRef]
- Meldau, S.; Ullman-Zeunert, L.; Govind, G.; Bartram, S.; Baldwin, I.T. MAPK-dependent JA and SA signalling in Nicotiana attenuata affects plant growth and fitness during competition with conspecifics. BMC Plant Biol. 2012, 12, 213. [Google Scholar] [CrossRef]
- Swinnen, G.; De Meyer, M.; Pollier, J.; Molina-Hidalgo, F.J.; Ceulemans, E.; De Clercq, R.; Bossche, R.V.; Fernández-Calvo, P.; Ron, M.; Pauwels, L.; et al. Constitutive steroidal glycoalkaloid biosynthesis in tomato is regulated by the clade IIIe basic helix-loop-helix transcription factors MYC1 and MYC2. BioRxiv 2020. [Google Scholar] [CrossRef]
- Yu, L.; Zhang, Y.; Ding, Q.; Wang, H.; Meng, X.; Fan, H.; Yu, Y.; Cui, N. The SlMYC1-TOR module regulates trichome formation and terpene biosynthesis in tomatoes (Solanum lycopersicum L.). J. Plant Growth Regul. 2024, 43, 3282–3294. [Google Scholar] [CrossRef]
- Zhang, Y.; Xing, H.; Wang, H.; Yu, L.; Yang, Z.; Meng, X.; Hu, P.; Fan, H.; Yu, Y.; Cui, N. SlMYC2 interacted with the SlTOR promoter and mediated JA signaling to regulate growth and fruit quality in tomato. Front Plant Sci. 2022, 13, 1013445. [Google Scholar] [CrossRef]
- Li, Y.; Yang, X.; Li, X. Role of jasmonate signaling pathway in resistance to dehydration stress in Arabidopsis. Acta Physiol. Plant. 2019, 41, 100. [Google Scholar] [CrossRef]
- Luo, L.; Wang, Y.; Qiu, L.; Han, X.; Zhu, Y.; Liu, L.; Man, M.; Li, F.; Ren, M.; Xing, Y. MYC2: A master switch for plant physiological processes and specialized metabolite synthesis. Int. J. Mol. Sci. 2023, 24, 3511. [Google Scholar] [CrossRef]
- Feng, Y.; Zeng, S.; Yan, J.; Li, K.; Xu, H. Genome-Wide analysis and expression of MYC family genes in tomato and the functional identification of SlMYC1 in response to salt and drought stress. Agronomy 2023, 13, 757. [Google Scholar] [CrossRef]
- Wang, R.; Yu, M.; Xia, J.; Xing, J.; Fan, X.; Xu, Q.; Cang, J.; Zhang, D. Overexpression of TaMYC2 confers freeze tolerance by ICE-CBF-COR module in Arabidopsis thaliana. Front Plant Sci. 2022, 13, 1042889. [Google Scholar] [CrossRef]
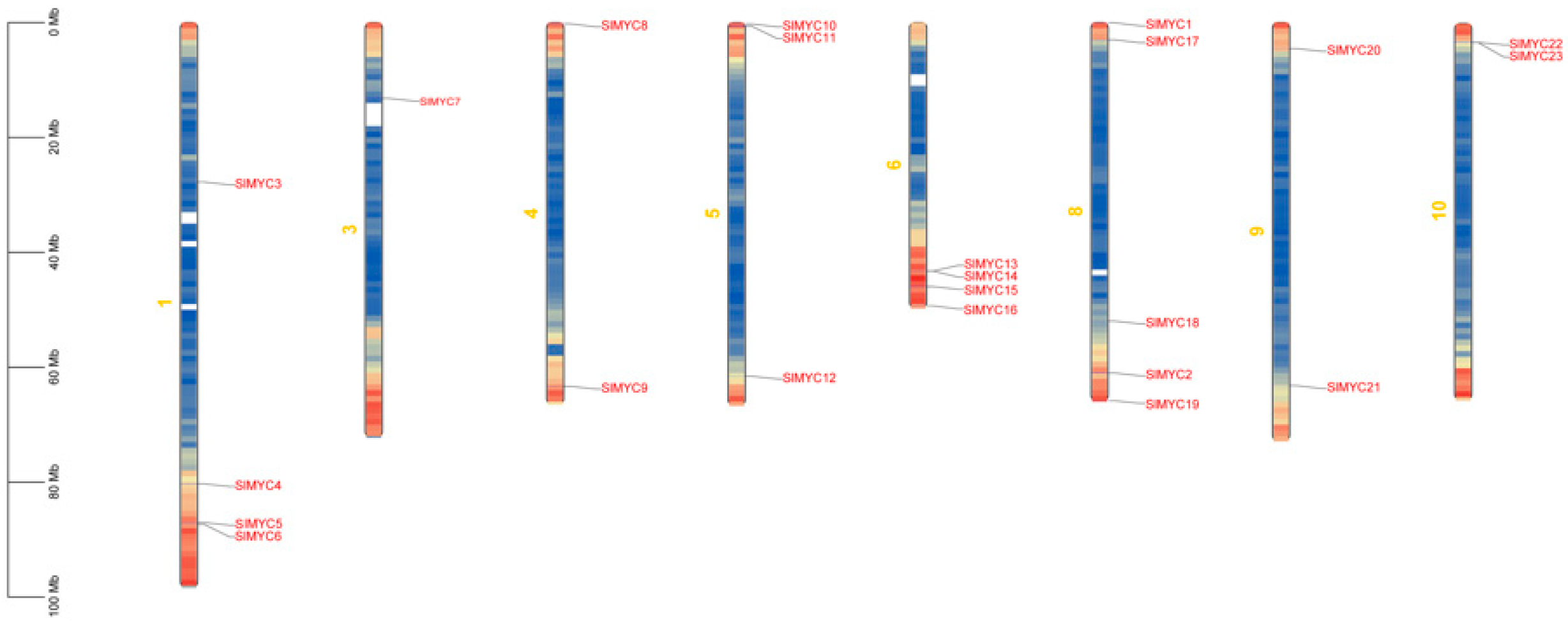
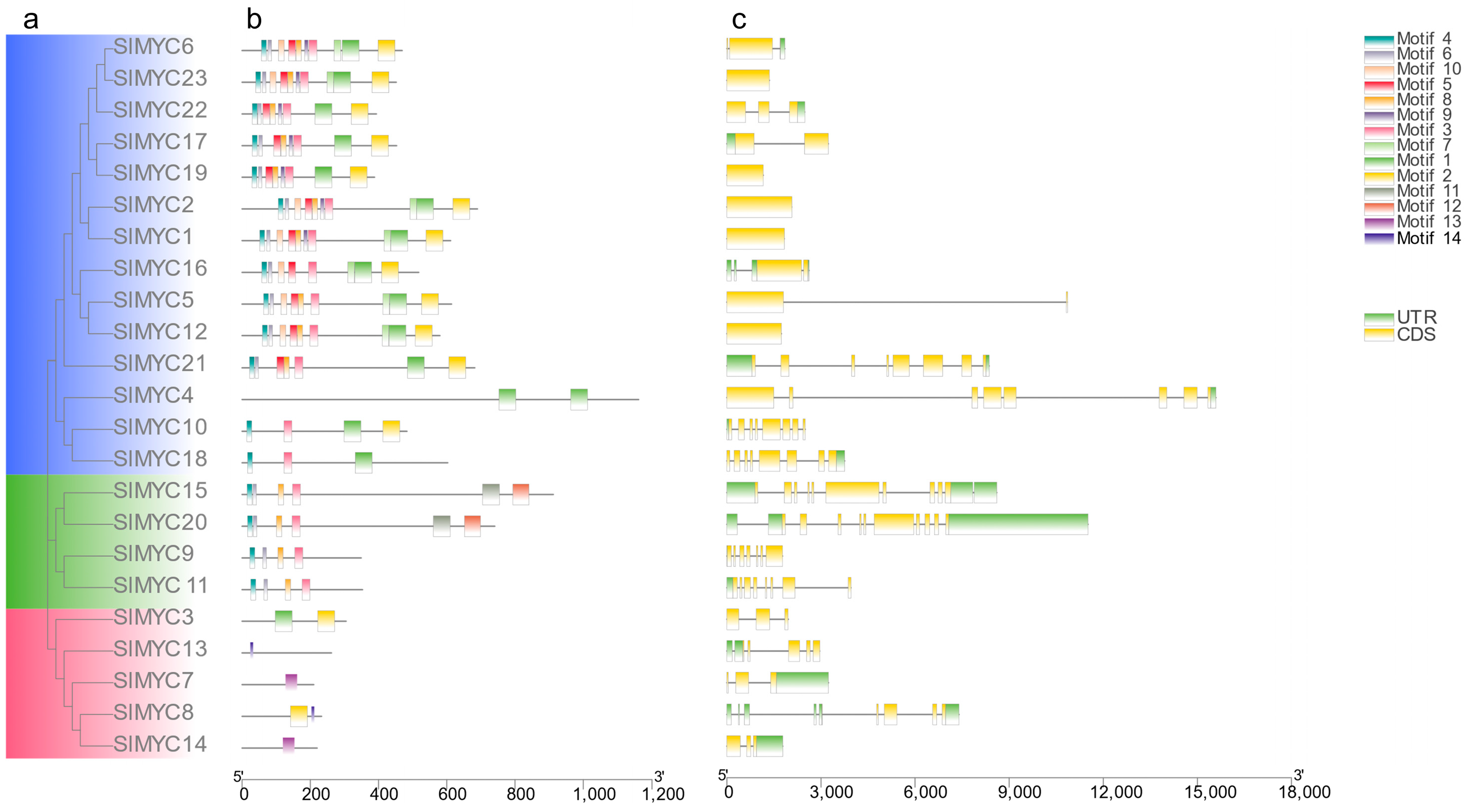
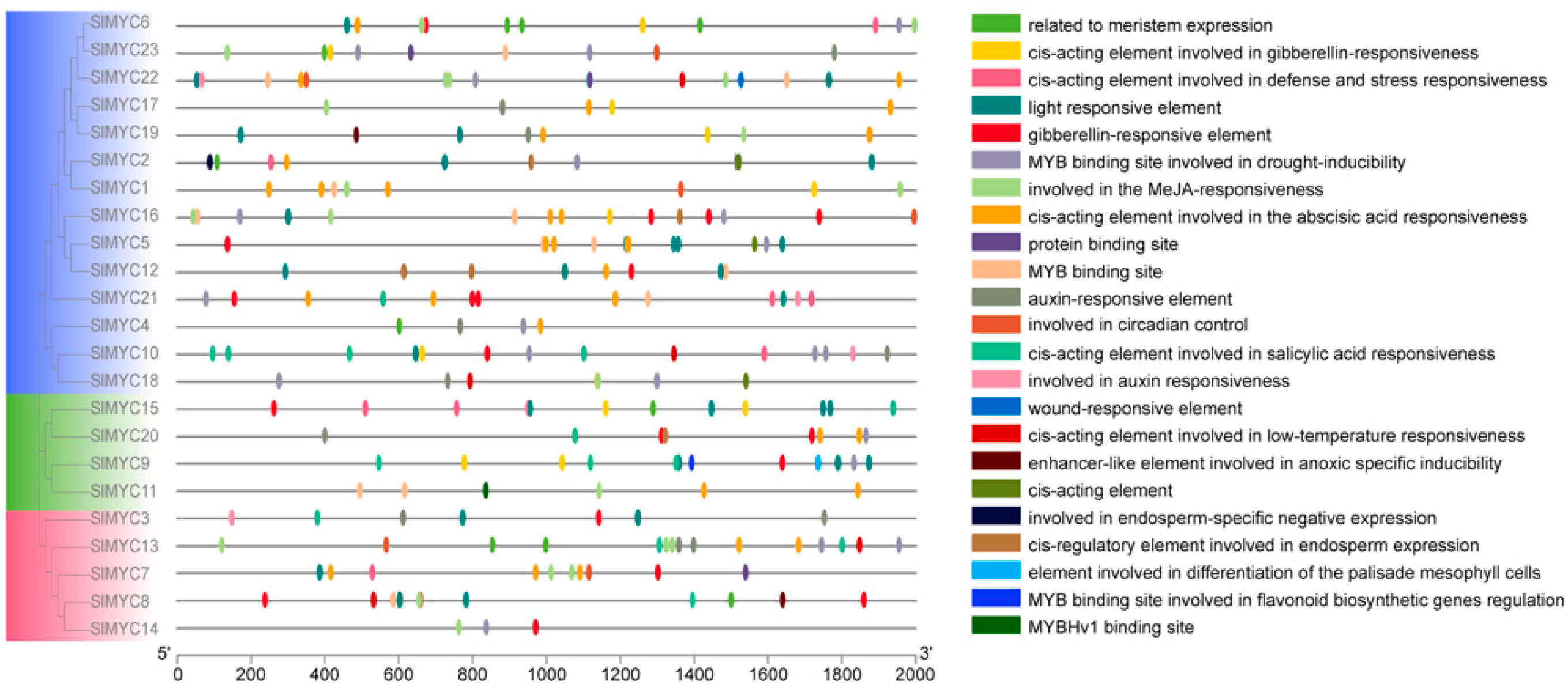

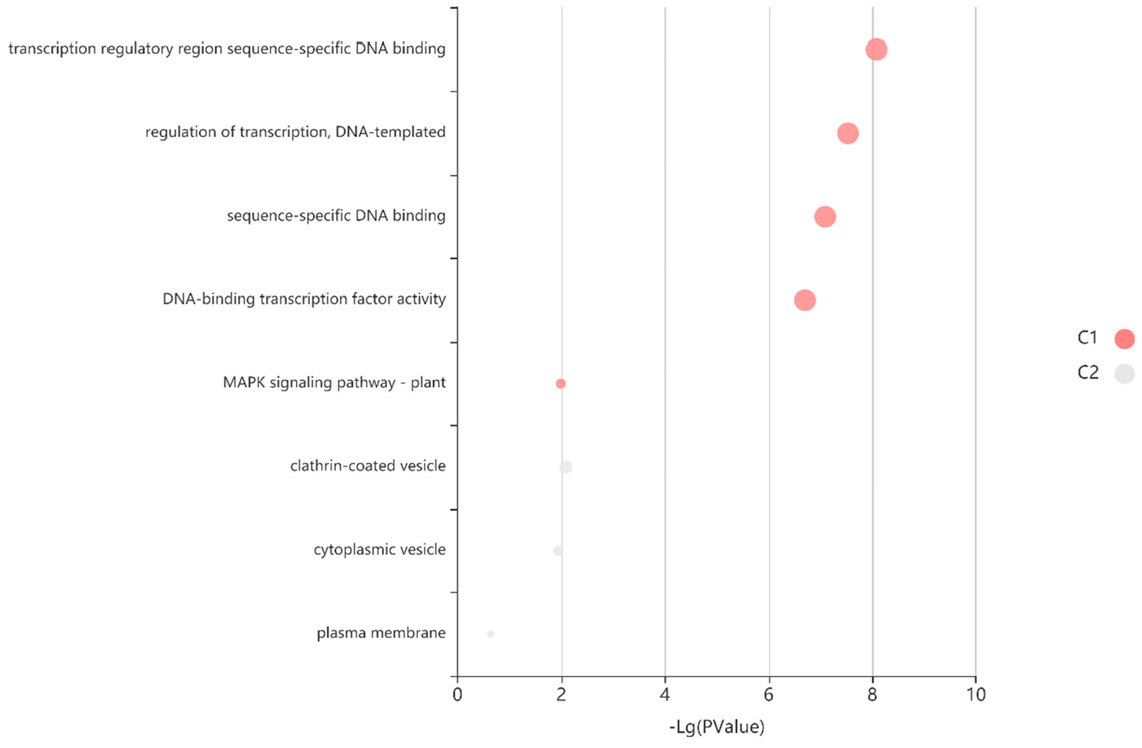
| Gene Name | EnsemblPlants ID | pI | Instability Index | Estimated Half-Life (h) | Deduced Protein Size (aa) | Molecular Weight (Da) | Gravy | Subcellular Location |
|---|---|---|---|---|---|---|---|---|
| SlMYC1 | Solyc08g005050.3.1 | 5.98 | 49.19 | 30 | 610 | 66,980.97 | −0.545 | Nucleus |
| SlMYC2 | Solyc08g076930.1.1 | 5.51 | 51.3 | 30 | 689 | 75,041.11 | −0.598 | Nucleus |
| SlMYC3 | Solyc01g020170.1.1 | 7.11 | 31. | 30 | 304 | 34,387.89 | −0.645 | Nucleus |
| SlMYC4 | Solyc01g081110.3.1 | 6.59 | 39.71 | 30 | 1160 | 130,949.9 | −0.366 | Nucleus |
| SlMYC5 | Solyc01g096050.3.1 | 7.60 | 42.61 | 30 | 613 | 68,254.22 | −0.506 | Nucleus |
| SlMYC6 | Solyc01g096370.3.1 | 5.29 | 37.55 | 30 | 468 | 51,573.11 | −0.348 | Nucleus |
| SlMYC7 | Solyc03g046570.3.1 | 5.00 | 53.29 | 30 | 209 | 22,589.12 | −0.425 | Nucleus |
| SlMYC8 | Solyc04g005280.3.1 | 5.59 | 48.42 | 30 | 232 | 25,362.96 | −0.750 | Nucleus |
| SlMYC9 | Solyc04g078520.3.1 | 6.27 | 45.27 | 30 | 348 | 39,536.46 | −0.484 | Chloroplast |
| SlMYC10 | Solyc05g005300.2.1 | 5.84 | 40.75 | 30 | 482 | 54,013.27 | −0.394 | Nucleus |
| SlMYC11 | Solyc05g005810.3.1 | 6.36 | 49.15 | 30 | 352 | 40,109.79 | −0.593 | Nucleus |
| SlMYC12 | Solyc05g050560.1.1 | 7.59 | 40.54 | 30 | 579 | 64,371.00 | −0.425 | Nucleus |
| SlMYC13 | Solyc06g069370.3.1 | 5.60 | 67.19 | 30 | 261 | 28,572.41 | −0.764 | Nucleus |
| SlMYC14 | Solyc06g069375.1.1 | 8.56 | 48.60 | 30 | 219 | 23,507.12 | −0.525 | Nucleus |
| SlMYC15 | Solyc06g074110.3.1 | 5.71 | 39.91 | 30 | 911 | 100,314.2 | −0.404 | Nucleus |
| SlMYC16 | Solyc06g083980.2.1 | 7.60 | 36.75 | 30 | 517 | 57,731.58 | −0.383 | Nucleus |
| SlMYC17 | Solyc08g008600.3.1 | 6.47 | 40.10 | 30 | 452 | 51,296.03 | −0.431 | Nucleus |
| SlMYC18 | Solyc08g062780.2.1 | 5.61 | 48.52 | 30 | 602 | 68,432.78 | −0.874 | Nucleus |
| SlMYC19 | Solyc08g083170.1.1 | 7.74 | 38.99 | 30 | 387 | 43,819.96 | −0.283 | Nucleus |
| SlMYC20 | Solyc09g011170.3.1 | 5.47 | 50.74 | 30 | 740 | 81,944.16 | −0.316 | Nucleus |
| SlMYC21 | Solyc09g065100.2.1 | 5.07 | 55.56 | 30 | 681 | 75,138.60 | −0.407 | Nucleus |
| SlMYC22 | Solyc10g009270.3.1 | 5.85 | 50.45 | 30 | 393 | 45,112.04 | −0.454 | Nucleus |
| SlMYC23 | Solyc10g009290.1.1 | 5.98 | 40.21 | 30 | 451 | 50,463.80 | −0.435 | Nucleus |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kang, C.; Cui, N.; Zhao, B.; Zou, Q.; Zhang, Y.; Bi, S.; Wu, Z.; Shao, M.; Qu, B. Identification and Expression Profiles Reveal Key Myelocytomatosis (MYC) Involved in Drought, Chilling, and Salt Tolerance in Solanum lycopersicum. Horticulturae 2025, 11, 693. https://doi.org/10.3390/horticulturae11060693
Kang C, Cui N, Zhao B, Zou Q, Zhang Y, Bi S, Wu Z, Shao M, Qu B. Identification and Expression Profiles Reveal Key Myelocytomatosis (MYC) Involved in Drought, Chilling, and Salt Tolerance in Solanum lycopersicum. Horticulturae. 2025; 11(6):693. https://doi.org/10.3390/horticulturae11060693
Chicago/Turabian StyleKang, Chenchen, Na Cui, Baozhen Zhao, Qingdao Zou, Yiming Zhang, Shiquan Bi, Zhongfen Wu, Meini Shao, and Bo Qu. 2025. "Identification and Expression Profiles Reveal Key Myelocytomatosis (MYC) Involved in Drought, Chilling, and Salt Tolerance in Solanum lycopersicum" Horticulturae 11, no. 6: 693. https://doi.org/10.3390/horticulturae11060693
APA StyleKang, C., Cui, N., Zhao, B., Zou, Q., Zhang, Y., Bi, S., Wu, Z., Shao, M., & Qu, B. (2025). Identification and Expression Profiles Reveal Key Myelocytomatosis (MYC) Involved in Drought, Chilling, and Salt Tolerance in Solanum lycopersicum. Horticulturae, 11(6), 693. https://doi.org/10.3390/horticulturae11060693






