Abstract
The U-box E3 ubiquitin ligase (PUB) gene family plays an important role in regulating plant responses to abiotic stress. Poncirus trifoliata (trifoliate orange), a citrus rootstock with notable cold, drought, and salt tolerance, serves as an excellent model for studying stress-responsive genes. In this study, a total of 47 PUB genes (PtrPUBs) were identified in the trifoliate orange genome. Chromosomal distribution analysis indicated that PtrPUB genes were unevenly distributed across nine trifoliate orange chromosomes. Phylogenetic tree analysis indicated that 170 PUB proteins from trifoliate orange, Arabidopsis thaliana, and tomato were clustered into five subfamilies. Gene structure, conserved domain, and motif analyses revealed diverse exon–intron and motif organizations of PtrPUB genes, suggesting potential functional differentiation among PtrPUBs. Cis-acting analysis indicated that the promoters of PtrPUB genes harbor elements related to hormone signaling (ABA, MeJA), drought stress, and low-temperature responses. Transcriptomic data and qRT-PCR results suggested that PtrPUB genes are responsive to ABA and dehydration treatments. This study provides a foundation for understanding the functional roles of PUB genes in trifoliate orange and offers insights for improving stress tolerance in citrus breeding programs.
1. Introduction
Poncirus trifoliata (2n = 2x = 18), commonly known as trifoliate orange, is a deciduous citrus relative native to China [1]. It is widely used as a rootstock in citrus production due to its notable resistance to various abiotic stresses, particularly cold, drought, and salinity [2,3,4]. This hardiness provides a significant advantage in temperate climates where traditional citrus species often struggle to survive. Despite its agricultural importance, the molecular mechanisms underlying stress tolerance in trifoliate orange remain poorly understood.
Plants are often exposed to various abiotic stresses such as salinity, drought, and low temperatures, which can lead to oxidative stress, ultimately hindering their growth and development. To survive under these adverse environmental conditions, plants have evolved intricate and highly efficient adaptive mechanisms [5]. Previous studies have identified four major signal transduction pathways involved in plant responses to abiotic stress: transcriptional regulation, post-transcriptional modifications, epigenetic regulation, and post-translational modifications [6]. Among these, ubiquitination is a key post-translational modification, playing a central role in regulating stress-related responses in plants.
The U-box (PUB) protein family, which is part of the E3 ubiquitin ligase group, plays a crucial role in the regulation of plant stress responses. For instance, Cho et al. reported that Arabidopsis PUB22 and PUB23 can ubiquitinate RPN12a under drought stress, enhancing the plant’s drought resistance [7]. Wang et al. revealed that PUB25 and PUB26 ubiquitinate MYB15 under cold induction, playing a precise regulatory role in the low-temperature stress response of Arabidopsis [8]. Bergler et al. suggested that PUB18/PUB19 may function as regulatory components in cooperation during ABA induction or salt stress [9]. Li et al. indicated that PUB12/PUB13 negatively regulate SA accumulation and positively regulate JA/ET signaling pathways, thereby modulating plant resistance [10]. AtPUB46 and AtPUB48, highly homologous genes encoding U-box E3 ligases, play essential roles in the plant’s response to water stress [10]. In Malus domestica (apple), MdPUB23 positively regulates the cold stress response in apple by promoting the ubiquitination and degradation of the positive regulator MdICE1 through the 26S proteasome pathway [11].
In Arabidopsis, approximately 61 PUB genes have been identified [12], while 77 PUB genes are present in Oryza sativa (rice) [13], 62 in Solanum lycopersicum (tomato) [14], 93 in Gossypium raimondii (cotton) [15], 91 in Musa acuminata (banana) [16], 61 in Medicago truncatula [17], 101 in Brassica rapa (cabbage) [18], and 125 in Glycine max (soybean) [19]. In apple, 69 PUB genes have been classified into seven subgroups based on phylogenetic analysis of PUB proteins from both apple and Arabidopsis [20]. In Pyrus bretschneideri (pear), 62 PUB genes have been identified and grouped into five categories [21]. However, information on PUB genes in trifoliate orange remains limited, hindering the functional characterization of these genes in this species.
In this study, we used bioinformatics tools to identify and characterize the PUB genes in the trifoliate orange genome. A phylogenetic analysis was conducted, and gene structures were examined. Additionally, conserved motifs within these genes were identified. We further investigated the expression patterns of selected PUB genes under dehydration and ABA stress treatments to explore their potential roles in stress responses. The findings will contribute to a deeper understanding of the molecular mechanisms underlying stress tolerance in trifoliate orange and provide valuable insights for enhancing stress resistance in citrus breeding programs.
2. Materials and Methods
2.1. Genome-Wide Identification of PUB Gene Family in Trifoliate Orange
We downloaded the trifoliate orange genome (Poncirus trifoliata v1.0) from the Citrus Pan-genome to Breeding Database (http://citrus.hzau.edu.cn/, accessed on 8 March 2025). To identify the PUB gene family in trifoliate orange, we employed BLAST and HMMsearch methods. We obtained the Hidden Markov Model (HMM) of the U-box domain (PF04564) from the Pfam database (https://pfam.xfam.org/, accessed on 8 March 2025). Then, the seed file of the U-box domain (PF04564) was used to search the candidate PUB genes in the whole-genome protein database of trifoliate orange using HMMsearch (v3.4) software (E-value < 1 × 10−5). In addition, we downloaded the protein sequences of PUB genes of tomato and Arabidopsis from previous studies [12,14]. The PUB protein sequences from Arabidopsis and tomato were used as queries to search the poplar protein database using BLASTP (E-value < 1 × 10−5, identity > 30%). Redundant hits identified by both BLASTP and HMMsearch were removed.
To verify the presence and integrity of the U-box domain, all putative PUB genes were analyzed using the SMART database (http://smart.embl-heidelberg.de/, accessed on 8 March 2025) and the Conserved Domain Database (CDD) (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 8 March 2025). After filtering, a total of 47 PUB genes were confirmed in the trifoliate orange genome.
Furthermore, key physicochemical characteristics of the corresponding PUB proteins—including amino acid composition, molecular mass, theoretical isoelectric point (pI), instability index, aliphatic index, and GRAVY (grand average of hydropathicity)—were computed using the ExPASy ProtParam tool (https://web.expasy.org/protparam/, accessed on 8 March 2025). The subcellular localization of these PUB proteins was predicted using WoLF PSORT (https://wolfpsort.hgc.jp/, accessed on 8 March 2025). The chromosomal distribution of PUB genes in the trifoliate orange genome was visualized using TBtools (v2.210) [22].
2.2. Multiple Sequence Alignment and Phylogenetic Analysis
We collected PUB protein sequences of tomato, Arabidopsis, and trifoliate orange [12,14]. A total of 170 PUB protein sequences were downloaded. Multiple sequence alignment was performed using ClustalW (v2.1) software with the default parameters. Subsequently, a Neighbor-Joining (NJ) phylogenetic tree was constructed using MEGA (v10.2.2) software with 1000 bootstrap replications [23]. Finally, the Evolview tool (https://evolgenius.info//evolview-v2/#login, accessed on 8 March 2025) was used to visualize the phylogenetic tree of PUB proteins [24].
2.3. Gene Structure, Conserved Domain, and Motif Analysis
To explore the structural composition (including introns, exons, and UTRs) of the trifoliate orange PUB gene family, gene structure information was extracted from the GFF file of the trifoliate orange genome using in-house scripts. The conserved motifs in the PtrPUB genes were identified using the MEME suite (http://meme-suite.org/tools/meme, accessed on 8 March 2025). A total of 20 motifs were identified. We used the CDD website (https://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi, accessed on 8 March 2025) to predict the conserved domain of the trifoliate orange PUB gene family. Gene structure, conserved domain, and motif results were visualized with TBtools software.
2.4. Cis-Acting Element Analysis
The 2000 bp upstream region of PtrPUB genes was defined as the putative promoter sequence. The promoter sequences of PtrPUBs were extracted using the getfasta function in Bedtools [25]. Cis-acting elements within the promoter sequence were predicted using the PlantCare tool [26]. Based on the functional annotations of these cis-acting elements, key elements were selected for further analysis, and elements with similar functional annotations were grouped together. Cis-acting elements were visualized with TBtools software.
2.5. Collinearity and Ka/Ks Analysis
Homologous gene pairs of PtrPUBs were detected using BLAST (v2.13.0+) software with an all-vs-all approach. Synteny regions were then identified with the Multiple Collinearity Scan toolkit (MCScanX) (v1.0.0) [27], based on the results from the all-vs-all BLAST analysis. The classification of gene duplication types was performed using the duplicate_gene_classifier program, which categorized the PUB genes into five distinct types: whole-genome duplication (WGD) or segmental duplication, tandem duplication (TD), proximal duplication (PD), dispersed duplication (DD), and singleton duplication (SD). A Circos plot was created to illustrate the distribution of syntenic gene pairs [28]. The non-synonymous (Ka) and synonymous (Ks) substitution rates for homologous gene pairs were calculated with KaKs_Calculator 2.0 (parameter: -m YN; -c).
2.6. Plant Materials, Growth Conditions, ABA and Dehydration Treatment
To investigate the expression patterns of PtrPUB genes, 3-month-old plants of trifoliate orange (Poncirus trifoliata) were exposed to different stress treatments. For ABA treatment, plants were treated with 100 mL of solutions containing 100 mM abscisic acid (ABA). For dehydration treatment, leaves from the trifoliate orange plants were detached and placed on filter papers at room temperature to simulate drought treatment. Leaf samples were collected at six time points (0, 3, 6, 9, 12, 24 h) [29], immediately frozen in liquid nitrogen, and stored at −80 °C.
2.7. RNA-Seq Analysis
The collected leaf samples were used for RNA-seq according to the method described in a previous study [30]. FastQC (v0.12.1) software was used to perform quality control of raw data [31]. Adapters and low-quality reads were trimmed using trimmomatic (v0.39) software [32]. Bowtie2 (v2.5.4) software was used to construct the genome index file for trifoliate orange [33]. Tophat2 (v2.0.0) software was used to perform reads mapping using trifoliate orange genome [1]. FPKM values were used to evaluate the expression level of genes. FPKM values were measured by FeatureCount (v2.0.6) software [34]. Then, we used the pheatmap (v1.0.12) package (R language) to display the expression pattern of the PtrPUB gene family.
2.8. RNA Extraction and Quantitative Reverse Transcription PCR Analysis
Total RNA was extracted from leaf samples under stress conditions using the Plant Total RNA Isolation Kit Plus (FOREGENE, Chengdu, China). cDNA was synthesized from RNA using the PrimeScript™ RT Reagent Kit (Takara Bio, Dalian, China). Quantitative RT-PCR (qRT-PCR) was performed on a Roche LightCycler® 480 II (Roche, Mannheim, Germany) with the LightCycler® SYBR GREEN I Master Mix kit (Roche, Shanghai, China). Fifteen pairs of specific primers were designed using Primer5.0 software (Table S2). The qRT-PCR reaction system and protocol were based on our previous study [35,36]. The relative expression levels of PtrPUBs were calculated using the 2−∆∆CT method [37].
3. Results
3.1. Whole-Genome Identification of PUB Genes in Trifoliate Orange
In this study, the PUB gene family in trifoliate orange was identified using BLASTP and HMMER (v3.4) software (see Section 2). The accuracy of the candidate PUB genes was further evaluated using the CDD and SMART tools. Finally, a total of 47 PUB genes were identified in the trifoliate orange genome. The number of PUB genes in trifoliate orange (47 genes) is smaller than in tomato (62 genes) and Arabidopsis (61 genes) [12,14]. These 47 PUB genes were named according to their order of location: PtrPUB1 to PtrPUB47 (Table 1).

Table 1.
The members of the PUB gene family in trifoliate orange.
As shown in Table 1, the physicochemical properties of the PtrPUB protein sequences were analyzed, including the amino acid count, molecular weight (MW), isoelectric point (pI), instability index, aliphatic index, and grand average of hydropathicity. The lengths of the PUB proteins ranged from 281 amino acids (PtrPUB17) to 3181 amino acids (PtrPUB15), with an average length of 645 amino acids. The molecular weights of the PUB proteins varied from 32 kDa (PtrPUB17) to 358 kDa (PtrPUB15). The pI values ranged from 5.16 (PtrPUB8) to 9.07 (PtrPUB24). Thirteen PtrPUB proteins had an instability index of less than 40, indicating their stability, while the remaining 34 were predicted to be unstable (instability index > 40). Subcellular localization predictions suggested that 11 PtrPUB proteins are predicted to be localized in the nucleus, 21 in the cytoplasm, 9 in the chloroplast, 2 in the endoplasmic reticulum, two in the plasma membrane, 1 in the Golgi apparatus, and 1 in the mitochondria. These findings suggest that PtrPUB proteins may function in various subcellular compartments, potentially participating in diverse regulatory processes.
3.2. The Chromosomal Distribution of PUB Genes in Trifoliate Orange Genome
The genomic positions for PUB genes were extracted using in-house scripts.The chromosomal distribution of PtrPUB genes across the trifoliate orange genome was visualized using TBtools (Figure 1). A total of 41 PtrPUB genes were unevenly distributed across nine trifoliate orange chromosomes. Six genes were located on scaffold/contigs (chrUn) and are not displayed in Figure 1. Chromosome 15 had the highest number of PtrPUB genes, containing seven genes, followed by chromosome 3 with six genes. Chromosomes 2, 5, 6, and 7 each had five PtrPUB genes. Chromosome 8 contained four PtrPUB genes. Chromosome 9 contained three PtrPUB genes, and chromosome 1 had only one gene.
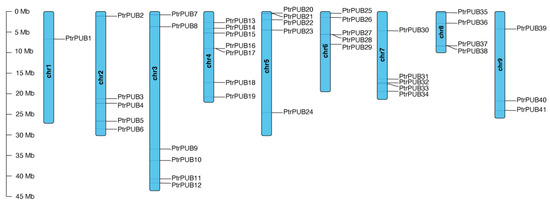
Figure 1.
Chromosomal distribution of PUB genes in the trifoliate orange genome.
3.3. Phylogenetic Analysis of PUB Genes in Trifoliate Orange, Arabidopsis, and Tomato
To explore the evolutionary relationships of PUB genes in trifoliate orange, a phylogenetic tree (Neighbor-Joining, NJ) was constructed using the MEGA software, based on PUB proteins from trifoliate orange (47 genes), tomato (62 genes), and Arabidopsis (61 genes) (Figure 2a). The PUB protein sequences for tomato and Arabidopsis were obtained from previous studies [12,14]. According to the phylogenetic tree, 170 PUB genes from the three species were grouped into five subfamilies: Group I, Group II, Group III, Group IV, and Group V. Group IV contained the largest number of PUB genes, with 55 members, while Group V had the smallest, with only 8 PUB genes. Overall, PUB genes from trifoliate orange and tomato were more closely related, as they clustered together in a single subclade (e.g., PtrPUB10 and SlU-box48), suggesting a closer evolutionary relationship between trifoliate orange and tomato compared to Arabidopsis.
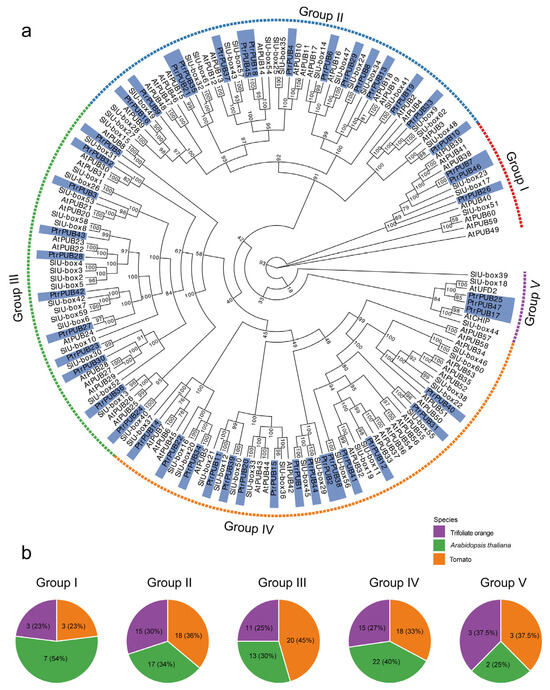
Figure 2.
Phylogenetic analysis of the PUB gene family. (a) A Neighbor-Joining (NJ) tree using 170 PUB proteins from three species, including trifoliate orange, tomato, and Arabidopsis. The phylogenetic tree was generated by MEGA software. Based on their phylogenetic relationships, the 170 PUB proteins were divided into five groups, named Group I, Group II, Group III, Group IV, and Group V. (b) Five pie charts represent the percentage of PUB proteins from the three species within each of the five groups.
It is noteworthy that the number of PUB genes in tomato is quite similar to that in Arabidopsis, while the number of PUB genes in trifoliate orange is smaller than that in tomato and Arabidopsis. To investigate whether any groups of PUB genes in trifoliate orange have undergone expansion or loss during evolution, we compared the number of PUB genes in each group for each species. In trifoliate orange, Groups I, II, III, IV, and V contain 3, 15, 11, 15, and 3 PtrPUB genes, respectively. In tomato, Groups I, II, III, IV, and V contain 3, 18, 20, 18, and 3 SIU-box genes, respectively. In Arabidopsis, Groups I, II, III, IV, and V contain 7, 17, 13, 22, and 2 AtPUB genes, respectively (Figure 2b). Compared to tomato and Arabidopsis, trifoliate orange exhibits significant gene loss in Groups II, III, and IV.
3.4. Gene Structure, Conserved Domain, and Motif Analysis of PUB Gene Family
To gain a deeper understanding of the characteristics of PUB genes in trifoliate orange, we analyzed the gene structures, conserved domains, and motif compositions of the PUB gene family. A phylogenetic tree of 47 PtrPUB genes was constructed as the basis for classification (Figure 3a). Using the MEME Suite, we identified 20 conserved motifs in the PUB gene family (Figure 3b), which were named Motif 1 through Motif 20. The motif analysis indicated that members within the same subgroup shared similar motif compositions, while distinct subgroups exhibited unique motif patterns. We identified six types of conserved domains in the PUB family members: RING-Ubox_PUB, RING_Ubox superfamily, Ubox, RING-Ubox_CHIP, RING-Ubox_UBE4A, and RING-Ubox_WDSUB1-like. These findings support the accuracy of PUB family identification (Figure 3c). Gene structure plays a crucial role in regulating gene expression and function. Thus, we examined the exon–intron structures of the 47 PUB genes (Figure 3d). The number of exons (coding sequences, CDS) varied from 1 to 50 (PtrPUB15). Fourteen PUB genes are single-exon genes, including PtrPUB3, PtrPUB7, PtrPUB13, PtrPUB19, PtrPUB23, PtrPUB26, PtrPUB27, PtrPUB28, PtrPUB30, PtrPUB32, PtrPUB36, PtrPUB42, PtrPUB43, and PtrPUB46. Thirty-one PUB genes contain UTR regions. These structural variations highlight the functional diversity of PUB genes in trifoliate orange. Notably, PtrPUB genes within the same subclades exhibited similar gene structure characteristics.
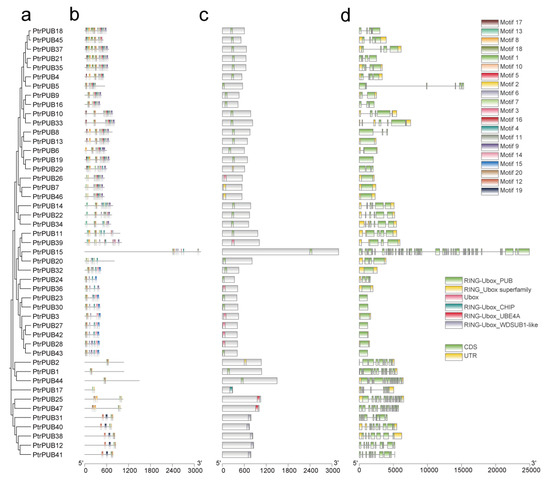
Figure 3.
The phylogenetic tree, conserved motifs, conserved domains, and gene structures of the PUB gene family in trifoliate orange. (a) A Neighbor-Joining (NJ) phylogenetic tree was constructed using the 47 PUB protein sequences from trifoliate orange, with a bootstrap value of 1000. (b) Conserved motif analysis of PUB proteins, where 20 unique motifs were identified via the MEME online tool, named motif1–motif20. (c) Conserved domain analysis of PUB proteins. The conserved domains were predicted using the CDD tool from the NCBI website. (d) Exon–intron structure of PUB genes, visualized based on genome annotations. Untranslated regions (UTRs) are shown as yellow boxes, coding sequences (CDSs) as green boxes, and introns as black lines.
3.5. Cis-Acting Element Analysis of PtrPUB Genes
Cis-acting regulatory elements in gene promoters provide important clues about transcriptional regulation and potential functional roles [14]. Transcription factors can regulate the expression levels of target genes by binding to the cis-acting elements in their promoters, playing a crucial role in specific biological processes. To further explore the function of PtrPUB genes, we predicted the cis-acting elements within their putative promoter regions using the PlantCARE database. As a result, a total of 42 cis-acting elements were identified, and we selected 20 noteworthy elements for further analysis. These 20 cis-acting elements are linked to stress responses, hormone regulation, and plant growth and development. As shown in Figure 4a, various distribution patterns of cis-acting elements were observed in the promoter regions of PtrPUB genes, suggesting that the PUB gene family in trifoliate orange may be involved in diverse biological processes.

Figure 4.
Analysis of cis-acting elements in the putative promoter regions of 47 PtrPUB genes. (a) The distribution of 20 cis-acting elements within the putative promoters of the PUB gene family in trifoliate orange is shown. (b) The frequency of the 20 cis-acting elements within the putative promoters of PtrPUB genes is shown.
Notably, we found all PtrPUB genes contained cis-acting elements related to hormone regulation, such as methyl jasmonate (MeJA), salicylic acid (SA), auxin, and abscisic acid (ABA). In our study, 39 PtrPUB genes were identified as ABA-responsive, suggesting that the PUB gene family may play a significant role in ABA-mediated stress resistance (Figure 4b). Thirty-nine PtrPUB genes were identified as MeJA-responsive, suggesting that the PUB gene family may play a significant role in resistance under MeJA treatment. Notably, a MYB binding site involved in drought response was predicted in 22 PtrPUB genes, indicating that these genes may be regulated by MYB transcription factors in response to drought stress.
3.6. Synteny Analysis of PUB Gene Family in Trifoliate Orange
To investigate the expansion and evolutionary history of the PUB gene family in trifoliate orange, we conducted a collinearity analysis using MCScanX (v1.0.0) software. In the trifoliate orange genome, 21,155 genes were derived from tandem duplication, 8349 genes were derived from dispersed duplication, 4228 genes were derived from whole-genome duplication (WGD) or segmental duplication, and 2769 genes were derived from proximal duplication (Figure 5a). Regarding the PUB gene family, 21 PUB genes were produced through dispersed duplication, 13 PUB genes were produced through WGD or segmental duplication, 12 PUB genes were produced through tandem duplication, and 1 PUB gene was produced through singleton duplication (Figure 5b). These findings suggest that dispersed duplications were the primary contributors to the expansion of the PUB gene family in trifoliate orange. Additionally, we identified homologous genes using MCScanX software (Figure 5c). A total of 2307 homologous gene pairs were identified in the trifoliate orange genome, and six homologous gene pairs were identified in the trifoliate orange PUB gene family, comprising 12 homologous genes. To determine the selection pressures influencing the evolution of PtrLBD genes, we calculated the ka/ks ratios for six PtrLBD paralogs using the KaKs_Calculator tool. A ka/ks ratio of 1 suggests neutral selection, a ratio less than 1 suggests negative (purifying) selection, and a ratio greater than 1 suggests positive (Darwinian) selection. The results showed that all ka/ks ratios for the PtrLBD paralogs were below 1 (Table 2), indicating that negative selection has been the dominant force driving the evolution of the PtrLBD gene family.
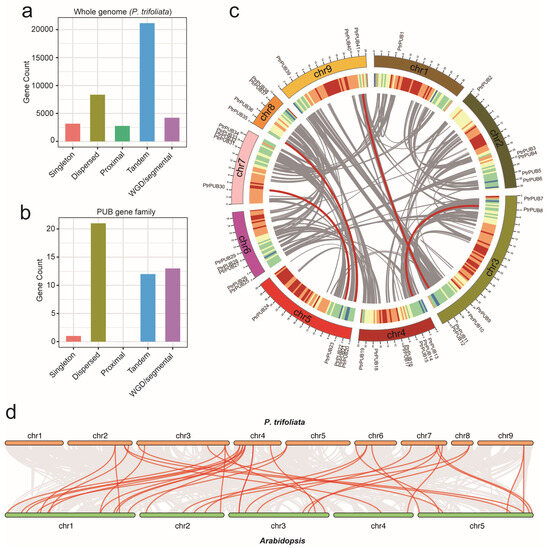
Figure 5.
The evolution analysis of the PUB gene family in trifoliate orange. The number of whole-genome genes (a) and PUB genes (b) across five types of gene duplication events in the trifoliate orange genome, including WGD (whole-genome duplication) or segmental duplication, singleton, dispersed duplication, proximal duplication, and tandem duplication. (c) The distribution pattern synteny analysis of the PUB gene family. From the outermost circle inward: chromosomal distribution; gene density (window size = 500k); the circles were generated using Circos (version 0.69.2) software. The red lines indicate collinear gene pairs of PUB genes, while the grey lines represent collinearity within the trifoliate orange genome. (d) Collinear gene pairs of PUB genes between trifoliate orange and Arabidopsis.

Table 2.
Ka, Ks, and Ka/Ks ratio of the PUB syntenic gene pairs in trifoliate orange.
To gain a deeper understanding of the evolutionary features of the PUB gene family, we created comparative synteny maps for trifoliate orange and Arabidopsis. As shown in Figure 5d, 27 orthologous gene pairs were identified between trifoliate orange and Arabidopsis. All ka/ks ratios for the PUB paralogs between trifoliate orange and Arabidopsis were below 1. This finding indicates that, while these gene pairs are syntenic, they may have experienced substantial structural and functional modifications during the evolutionary divergence of trifoliate orange and Arabidopsis (Table S1).
3.7. Expression Pattern Analysis of PtrPUB Genes Under Abiotic Stresses
Previous studies have extensively documented the involvement of the PUB gene family in various abiotic stresses [38]. To investigate the functions of the PUB gene family in trifoliate orange, we performed ABA and dehydration treatments for seedling samples of trifoliate orange. We measured the expression levels of PtrPUB genes at 0 h, 3 h, 6 h, 9 h, and 24 h after ABA and dehydration treatments using RNA-seq (Table S3). In ABA treatment (Figure 6a), multiple PUB genes showed a marked increase in expression at various time points. PtrPUB14 was upregulated at 3 h after ABA treatment; PtrPUB37 was upregulated at 6 h after ABA treatment; PtrPUB15 was upregulated at 9 h after ABA treatment; PtrPUB1, PtrPUB32, PtrPUB33, PtrPUB34, and PtrPUB38 were upregulated at 12 h after ABA treatment; and PtrPUB6, PtrPUB18, PtrPUB29, and PtrPUB45 were upregulated at 12 h after ABA treatment. Therefore, the PUB gene family in trifoliate orange plays a vital role in the process of ABA response.
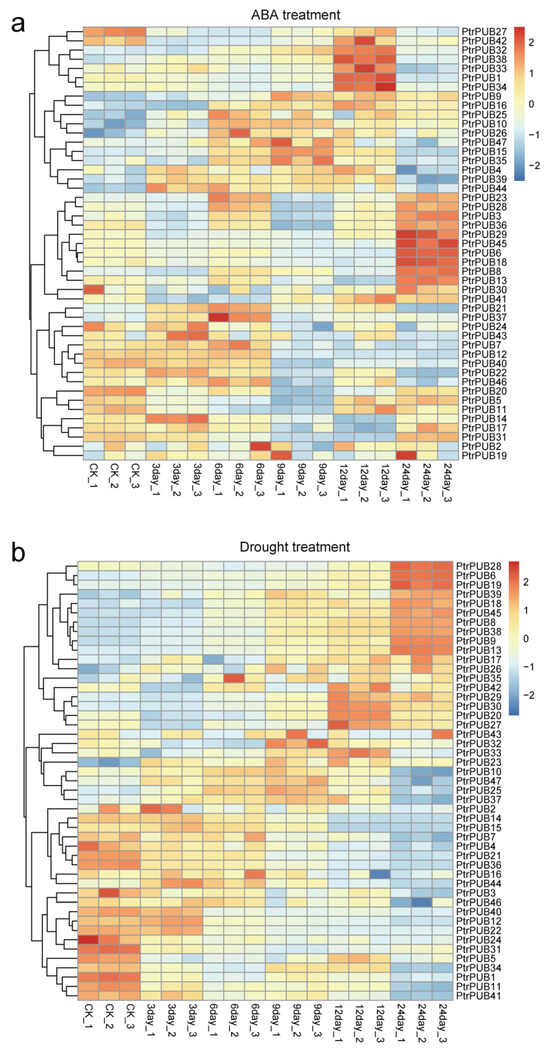
Figure 6.
Expression patterns of PUB genes in trifoliate orange under abiotic stress. (a) Heatmap of PUB gene expression in trifoliate orange under ABA treatment at different time points (3 h, 6 h, 9 h, 12 h, and 24 h). (b) Heatmap of PUB gene expression in trifoliate orange under dehydration treatment (3 h, 6 h, 9 h, 12 h, and 24 h). The color scale represents relative expression levels, with red indicating high expression and blue indicating low expression.
In the dehydration treatment (Figure 6b), 20 PUB genes showed higher expression levels under control conditions (CK) and exhibited downregulated expression following dehydration treatment, suggesting that these PUB genes may have a negative response to dehydration stress. Eight PUB genes (PtrPUB43, PtrPUB32, PtrPUB33, PtrPUB23, PtrPUB10, PtrPUB47, PtrPUB25, PtrPUB37) were upregulated at 3 h, 6 h, and 9 h after dehydration treatment. Five PUB genes (PtrPUB42, PtrPUB29, PtrPUB30, PtrPUB20, PtrPUB27) were upregulated at 12 h after dehydration treatment. Ten PUB genes (PtrPUB28, PtrPUB6, PtrPUB19, PtrPUB39, PtrPUB18, PtrPUB45, PtrPUB8, PtrPUB38, PtrPUB9, PtrPUB13) were upregulated at 24 h after dehydration treatment. The PUB gene family in trifoliate orange is crucial for the response to dehydration stress. Moreover, different PUB genes exhibit functional diversification, with distinct expression profiles at various time points.
3.8. Quantitative Real-Time PCR (qRT-PCR) Analysis of PtrPUB Genes Under Abiotic Stresses
To investigate the role of PtrPUB genes in response to ABA and dehydration stress, we selected 13 PtrPUB genes for qRT-PCR analysis to assess their expression patterns under ABA and dehydration treatments. In response to ABA treatment (Figure 7a), we observed significant upregulation of PtrPUB8, PtrPUB10, PtrPUB15, PtrPUB23, PtrPUB30, PtrPUB36, and PtrPUB45, suggesting their involvement in ABA signaling. These genes showed the highest expression levels at 9 h of ABA treatment. For dehydration stress (Figure 7b), ten PtrPUB genes were upregulated, while three genes (PtrPUB1, PtrPUB15, and PtrPUB36) were downregulated. Among the upregulated genes, PtrPUB6, PtrPUB8, PtrPUB13, PtrPUB28, PtrPUB31, and PtrPUB35 exhibited the highest expression levels at 24 h, while PtrPUB10, PtrPUB18, PtrPUB23, and PtrPUB30 returned to baseline expression by 24 h. PtrPUB8, PtrPUB10, PtrPUB23, PtrPUB30, and PtrPUB45 were upregulated in both ABA and dehydration treatments, whereas PtrPUB15 and PtrPUB36 exhibited opposite expression patterns under ABA and dehydration stress. These results suggested the functional diversity of PUB genes under different stress conditions. Overall, these findings indicated that PtrPUB genes play a crucial role in mediating the plant’s response to ABA and dehydration stress.
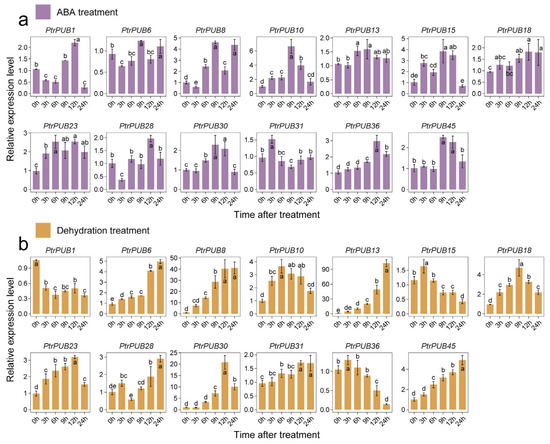
Figure 7.
The expression level of 13 PtrPUB genes in trifoliate orange under ABA and dehydration treatments. (a) For ABA treatment, the seedlings were placed in 100 µM ABA solution for 0, 3, 6, 9, 12, and 24 h. (b) For dehydration treatment, the seedlings of trifoliate orange were placed on dry filter paper for 0, 3, 6, 9, 12, and 24 h, simulating drought treatment. Different letters above bars indicate significant differences at p < 0.05 according to Duncan’s multiple range test.
4. Discussion
With the advancement of whole-genome sequencing technologies, an increasing number of plant genomes have been assembled and released, leading to the identification of U-box gene families in a wide range of species. In Arabidopsis thaliana, 61 PUB genes have been identified [12], while 77 have been found in Oryza sativa (rice) [13], 62 in Solanum lycopersicum (tomato) [14], 93 in Gossypium raimondii (cotton) [15], 91 in Musa acuminata (banana) [16], 61 in Medicago truncatula [17], 101 in Brassica rapa (cabbage) [18], 125 in Glycine max (soybean) [19], 69 in Malus domestica (apple) [20], and 62 in Pyrus bretschneideri (pear) [21]. In this study, we identified 47 PUB genes (PtrPUBs) in the Poncirus trifoliata genome and performed a comprehensive structural analysis. Our findings expand the diversity of the PUB gene family among plants and provide a theoretical basis for future functional studies of PUB genes in trifoliate orange. Compared with other fruit tree species such as apple and pear, which contain 69 and 62 PUB genes, respectively, trifoliate orange contains a relatively smaller number of PUB genes. This variation may result from species-specific genomic characteristics or differences in the frequency of whole-genome duplication (WGD) events. Previous studies have reported that both apple and pear have undergone two WGD events [39,40], whereas trifoliate orange has experienced only a single WGD event [1], which may partly explain the reduced number of PUB genes in its genome.
Phylogenetic analysis revealed that the 170 PUB proteins identified from trifoliate orange (47), tomato (62) [14], and Arabidopsis (61) [12] were clustered into five subfamilies: Group I, Group II, Group III, Group IV, and Group V. This classification is consistent with phylogenetic patterns reported in other plant species. For example, 69 PUB genes in apple were grouped into seven clades [20], 62 in pear were clustered into five clades [21], and 125 in soybean were divided into six subfamilies [19]. Based on the phylogenetic relationships, PUB proteins from trifoliate orange (PtrPUBs) showed a closer evolutionary relationship with those from tomato (SIU-box proteins) than those from Arabidopsis, which aligns with the closer taxonomic relationship between pear and tomato. Among the five subfamilies, the number of PUB genes in Groups I and V was relatively comparable across the three species. However, Groups II, III, and IV contained fewer members in trifoliate orange, suggesting that gene loss may have occurred in these groups during trifoliate orange evolution. These results provide further insight into the lineage-specific contraction of the PUB gene family in trifoliate orange.
In this study, cis-element analysis of the putative promoter regions indicated that PtrPUB genes may be involved in a wide range of biological processes, including defense and stress responses (e.g., drought and low temperature), light responsiveness, regulation of flavonoid biosynthesis, hormonal signaling (e.g., abscisic acid, methyl jasmonate, and salicylic acid), as well as plant growth and development. In Arabidopsis, AtPUB41 is essential for Arabidopsis drought stress resilience [41]. In our study, qRT-PCR confirmed that ten PtrPUB genes were induced under dehydration, implying a potential role in enhancing drought resilience in P. trifoliata.
Notably, 39 PtrPUB genes were predicted to be responsive to ABA, suggesting an important role in abscisic acid-mediated stress signaling. This is in line with previous reports showing that PUB genes participate in ABA-related pathways. For instance, DSG1, which encodes a U-box domain protein, was shown to regulate cell division and elongation by responding to multiple hormones, including auxin, salicylic acid, and ethylene [42]. ABA is well known for its role in reducing grain yield under water stress during the reproductive stage by increasing sterility and reducing grain filling in cereal crops [43,44,45,46]. RNA-seq and qRT-PCR analysis provided evidence on PUB genes (PtrPUB14, PtrPUB37, PtrPUB1, PtrPUB32, PtrPUB33, PtrPUB34, PtrPUB38, PtrPUB6, PtrPUB18, PtrPUB29, PtrPUB45) related to ABA response. MeJA is a key regulator in plant responses to both biotic and abiotic stresses. It enhances plant defense, modulates hormonal balance, and alleviates oxidative and osmotic stress through the activation of antioxidant systems and osmoprotectant signaling pathways [47]. Furthermore, tissue-specific expression patterns revealed that most of the PtrPUB genes exhibited distinct spatial expression profiles in root, stem, and leaf tissues (Figure S1). Together, these findings highlight the potential roles of PtrPUB genes in hormone-mediated signaling pathways and stress adaptation mechanisms in trifoliate orange.
Transcription factors play a critical role in controlling gene expression regulation [48]. In our study, several MYB binding sites were identified in the promoter regions of PtrPUB genes, including an MYB binding site involved in drought inducibility, an MYB binding site involved in light responsiveness, and an MYB binding site involved in flavonoid biosynthetic gene regulation. For example, 10 PtrPUB genes contained the MYB binding site involved in light responsiveness, 6 PtrPUB genes contained the MYB binding site involved in flavonoid biosynthetic gene regulation, and 22 PtrPUB genes contained the MYB binding site involved in drought inducibility. These results suggest that PtrPUB genes may be transcriptionally regulated by MYB transcription factors and potentially participate in MYB-mediated signaling pathways.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/horticulturae11060604/s1: Figure S1. Expression patterns of 13 PtrPUB genes in different tissues (leaf, stem, root) of trifoliate orange. Different letters above bars indicate significant differences at p < 0.05 according to Duncan’s multiple range test; Table S1: Ka, Ks, and Ka/Ks ratio of the PUB syntenic gene pairs between trifoliate orange and Arabidopsis; Table S2: Primer sequences used in this study; Table S3: The expression levels of 47 PtrPUB genes under ABA and dehydration treatment.
Author Contributions
Conceptualization, B.S., S.J., and J.S.; methodology, B.S., S.J., and X.G.; validation, S.J. and X.G.; formal analysis, B.S., S.J., and X.G.; investigation, Y.L. and D.L.; writing—original draft preparation, B.S.; writing—review and editing, J.S., D.L., and Y.L.; visualization, L.Y., W.H., and L.K.; project administration, J.S.; funding acquisition, J.S. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (32302481), the Major Discipline Academic and Technical Leaders Training Program of Jiangxi Province (20243BCE51157), the Natural Science Foundation of Jiangxi Province (20242BAB20292), and the Open Funds of The National Key Laboratory for Germplasm Innovation & Utilization of Horticultural Crops (Horti-KF-2023-15).
Data Availability Statement
Data are contained within the article and Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
Abbreviations
The following abbreviations are used in this manuscript:
| PUB | Plant U-box gene |
| ABA | Abscisic acid |
| MeJA | Methyl jasmonate |
| qRT-PCR | Quantitative real-time PCR |
References
- Peng, Z.; Bredeson, J.V.; Wu, G.A.; Shu, S.; Rawat, N.; Du, D.; Parajuli, S.; Yu, Q.; You, Q.; Rokhsar, D.S.; et al. A chromosome-scale reference genome of trifoliate orange (Poncirus trifoliata) provides insights into disease resistance, cold tolerance and genome evolution in Citrus. Plant J. 2020, 104, 1215–1232. [Google Scholar] [CrossRef] [PubMed]
- Boava, L.P.; Cristofani-Yaly, M.; Mafra, V.S.; Kubo, K.; Kishi, L.T.; Takita, M.A.; Ribeiro-Alves, M.; Machado, M.A. Global gene expression of Poncirus trifoliata, Citrus sunki and their hybrids under infection of Phytophthora parasitica. BMC Genom. 2011, 12, 39. [Google Scholar] [CrossRef] [PubMed]
- Gong, X.-Q.; Liu, J.-H. Genetic transformation and genes for resistance to abiotic and biotic stresses in Citrus and its related genera. Plant Cell Tissue Organ Cult. (PCTOC) 2013, 113, 137–147. [Google Scholar] [CrossRef]
- Dahro, B.; Li, C.; Liu, J.-H. Overlapping responses to multiple abiotic stresses in citrus: From mechanism understanding to genetic improvement. Hortic. Adv. 2023, 1, 4. [Google Scholar] [CrossRef]
- Gong, Z.; Xiong, L.; Shi, H.; Yang, S.; Herrera-Estrella, L.R.; Xu, G.; Chao, D.-Y.; Li, J.; Wang, P.-Y.; Qin, F.; et al. Plant abiotic stress response and nutrient use efficiency. Sci. China Life Sci. 2020, 63, 635–674. [Google Scholar] [CrossRef]
- Hirayama, T.; Shinozaki, K. Research on plant abiotic stress responses in the post-genome era: Past, present and future. Plant J. 2010, 61, 1041–1052. [Google Scholar] [CrossRef]
- Cho, S.K.; Ryu, M.Y.; Song, C.; Kwak, J.M.; Kim, W.T. Arabidopsis PUB22 and PUB23 Are Homologous U-Box E3 Ubiquitin Ligases That Play Combinatory Roles in Response to Drought Stress. Plant Cell 2008, 20, 1899–1914. [Google Scholar] [CrossRef]
- Wang, X.; Ding, Y.; Li, Z.; Shi, Y.; Wang, J.; Hua, J.; Gong, Z.; Zhou, J.M.; Yang, S. PUB25 and PUB26 Promote Plant Freezing Tolerance by Degrading the Cold Signaling Negative Regulator MYB15. Dev. Cell 2019, 51, 222–235.e5. [Google Scholar] [CrossRef]
- Bergler, J.; Hoth, S. Plant U-box armadillo repeat proteins AtPUB18 and AtPUB19 are involved in salt inhibition of germination in Arabidopsis. Plant Biol. 2011, 13, 725–730. [Google Scholar] [CrossRef]
- Li, W.; Ahn, I.-P.; Ning, Y.; Park, C.-H.; Zeng, L.; Whitehill, J.G.A.; Lu, H.; Zhao, Q.; Ding, B.; Xie, Q.; et al. The U-Box/ARM E3 Ligase PUB13 Regulates Cell Death, Defense, and Flowering Time in Arabidopsis. Plant Physiol. 2012, 159, 239–250. [Google Scholar] [CrossRef]
- Wang, D.-R.; Zhang, X.-W.; Xu, R.-R.; Wang, G.-L.; You, C.-X.; An, J.-P. Apple U-box-type E3 ubiquitin ligase MdPUB23 reduces cold-stress tolerance by degrading the cold-stress regulatory protein MdICE1. Hortic. Res. 2022, 9, uhac171. [Google Scholar] [CrossRef] [PubMed]
- Wiborg, J.; O’Shea, C.; Skriver, K. Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem. J. 2008, 413, 447–457. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.-R.; Park, C.H.; Venu, R.C.; Gough, J.; Wang, G.-L. Classification, Expression Pattern, and E3 Ligase Activity Assay of Rice U-Box-Containing Proteins. Mol. Plant 2008, 1, 800–815. [Google Scholar] [CrossRef] [PubMed]
- Sharma, B.; Taganna, J. Genome-wide analysis of the U-box E3 ubiquitin ligase enzyme gene family in tomato. Sci. Rep. 2020, 10, 9581. [Google Scholar] [CrossRef]
- Lu, X.; Shu, N.; Wang, D.; Wang, J.; Chen, X.; Zhang, B.; Wang, S.; Guo, L.; Chen, C.; Ye, W. Genome-wide identification and expression analysis of PUB genes in cotton. BMC Genom. 2020, 21, 213. [Google Scholar] [CrossRef]
- Hu, H.; Dong, C.; Sun, D.; Hu, Y.; Xie, J. Genome-Wide Identification and Analysis of U-Box E3 Ubiquitin-Protein Ligase Gene Family in Banana. Int. J. Mol. Sci. 2018, 19, 3874. [Google Scholar] [CrossRef]
- Song, J.; Mo, X.; Yang, H.; Yue, L.; Song, J.; Mo, B. The U-box family genes in Medicago truncatula: Key elements in response to salt, cold, and drought stresses. PLoS ONE 2017, 12, e0182402. [Google Scholar] [CrossRef]
- Wang, C.; Duan, W.; Riquicho, A.R.; Jing, Z.; Liu, T.; Hou, X.; Li, Y. Genome-wide survey and expression analysis of the PUB family in Chinese cabbage (Brassica rapa ssp. pekinesis). Mol. Genet. Genom. 2015, 290, 2241–2260. [Google Scholar] [CrossRef]
- Wang, N.; Liu, Y.; Cong, Y.; Wang, T.; Zhong, X.; Yang, S.; Li, Y.; Gai, J. Genome-Wide Identification of Soybean U-Box E3 Ubiquitin Ligases and Roles of GmPUB8 in Negative Regulation of Drought Stress Response in Arabidopsis. Plant Cell Physiol. 2016, 57, 1189–1209. [Google Scholar] [CrossRef]
- Wang, K.; Yang, Q.; Lanhuang, B.; Lin, H.; Shi, Y.; Dhanasekaran, S.; Godana, E.A.; Zhang, H. Genome-wide investigation and analysis of U-box Ubiquitin–Protein ligase gene family in apple: Expression profiles during Penicillium expansum infection process. Physiol. Mol. Plant Pathol. 2020, 111, 101487. [Google Scholar] [CrossRef]
- Wang, C.; Song, B.; Dai, Y.; Zhang, S.; Huang, X. Genome-wide identification and functional analysis of U-box E3 ubiquitin ligases gene family related to drought stress response in Chinese white pear (Pyrus bretschneideri). BMC Plant Biol. 2021, 21, 235. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef]
- Zhang, H.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W.-H. EvolView, an online tool for visualizing, annotating and managing phylogenetic trees. Nucleic Acids Res. 2012, 40, W569–W572. [Google Scholar] [CrossRef]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef] [PubMed]
- Rombauts, S.; Déhais, P.; Van Montagu, M.; Rouzé, P. PlantCARE, a plant cis-acting regulatory element database. Nucleic Acids Res. 1999, 27, 295–296. [Google Scholar] [CrossRef]
- Wang, Y.; Tang, H.; DeBarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef]
- Song, J.; Sun, P.; Kong, W.; Xie, Z.; Li, C.; Liu, J.-H. SnRK2.4-mediated phosphorylation of ABF2 regulates expression and putrescine accumulation under drought stress. New Phytol. 2023, 238, 216–236. [Google Scholar] [CrossRef]
- Wei, T.; Wang, Y.; Liu, J.-H. Comparative transcriptome analysis reveals synergistic and disparate defense pathways in the leaves and roots of trifoliate orange (Poncirus trifoliata) autotetraploids with enhanced salt tolerance. Hortic. Res. 2020, 7, 88. [Google Scholar] [CrossRef]
- Brown, J.; Pirrung, M.; McCue, L.A. FQC Dashboard: Integrates FastQC results into a web-based, interactive, and extensible FASTQ quality control tool. Bioinformatics 2017, 33, 3137–3139. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Langdon, W.B. Performance of genetic programming optimised Bowtie2 on genome comparison and analytic testing (GCAT) benchmarks. BioData Min. 2015, 8, 1–7. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The Subread aligner: Fast, accurate and scalable read mapping by seed-and-vote. Nucleic Acids Res. 2013, 41, e108. [Google Scholar] [CrossRef]
- Song, B.; Tang, Z.; Li, X.; Li, J.; Zhang, M.; Zhao, K.; Liu, H.; Zhang, S.; Wu, J. Mining and evolution analysis of lateral organ boundaries domain (LBD) genes in Chinese white pear (Pyrus bretschneideri). BMC Genom. 2020, 21, 644. [Google Scholar] [CrossRef]
- Gong, X.; Zhao, L.; Song, X.; Lin, Z.; Gu, B.; Yan, J.; Zhang, S.; Tao, S.; Huang, X. Genome-wide analyses and expression patterns under abiotic stress of NAC transcription factors in white pear (Pyrus bretschneideri). BMC Plant Biol. 2019, 19, 161. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of Relative Gene Expression Data Using Real-Time Quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Stone, S.L. The role of ubiquitin and the 26S proteasome in plant abiotic stress signaling. Front. Plant Sci. 2014, 5, 135. [Google Scholar] [CrossRef]
- Wu, J.; Wang, Z.; Shi, Z.; Zhang, S.; Ming, R.; Zhu, S.; Khan, M.A.; Tao, S.; Korban, S.S.; Wang, H. The genome of the pear (Pyrus bretschneideri Rehd.). Genome Res. 2013, 23, 396–408. [Google Scholar] [CrossRef]
- Sun, X.; Jiao, C.; Schwaninger, H.; Chao, C.T.; Ma, Y.; Duan, N.; Khan, A.; Ban, S.; Xu, K.; Cheng, L.; et al. Phased diploid genome assemblies and pan-genomes provide insights into the genetic history of apple domestication. Nat. Genet. 2020, 52, 1423–1432. [Google Scholar] [CrossRef]
- Sharma, A.; Goldfarb, S.; Raveh, D.; Bar-Zvi, D. Arabidopsis ubiquitin ligase PUB41 positively regulates ABA-mediated seed dormancy and drought response. Physiol. Mol. Biol. Plants 2024, 30, 1819–1827. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Xing, Y.; Lou, Q.; Feng, P.; Liu, S.; Zhu, M.; Yin, W.; Fang, S.; Lin, Y.; Zhang, T.; et al. Dwarf and short grain 1, encoding a putative U-box protein regulates cell division and elongation in rice. J. Plant Physiol. 2017, 209, 84–94. [Google Scholar] [CrossRef] [PubMed]
- Morgan, J.M. Possible role of abscisic acid in reducing seed set in water-stressed wheat plants. Nature 1980, 285, 655–657. [Google Scholar] [CrossRef]
- Ober, E.S.; Setter, T.L.; Madison, J.T.; Thompson, J.F.; Shapiro, P.S. Influence of Water Deficit on Maize Endosperm Development 1: Enzyme Activities and RNA Transcripts of Starch and Zein Synthesis, Abscisic Acid, and Cell Division. Plant Physiol. 1991, 97, 154–164. [Google Scholar] [CrossRef]
- Yang, J.C.; Zhang, J.H.; Ye, Y.X.; Wang, Z.Q.; Zhu, Q.S.; Liu, L.J. Involvement of abscisic acid and ethylene in the responses of rice grains to water stress during filling. Plant Cell Environ. 2004, 27, 1055–1064. [Google Scholar] [CrossRef]
- Yang, J.; Zhang, J.; Liu, K.; Wang, Z.; Liu, L. Abscisic Acid and Ethylene Interact in Rice Spikelets in Response to Water Stress During Meiosis. J. Plant Growth Regul. 2007, 26, 318–328. [Google Scholar] [CrossRef]
- Yu, X.; Zhang, W.; Zhang, Y.; Zhang, X.; Lang, D.; Zhang, X. The roles of methyl jasmonate to stress in plants. Funct. Plant Biol. 2019, 46, 197–212. [Google Scholar] [CrossRef]
- Wang, G.; Wang, F.; Huang, Q.; Li, Y.; Liu, Y.; Wang, Y. Understanding Transcription Factor Regulation by Integrating Gene Expression and DNase I Hypersensitive Sites. Biomed. Res. Int. 2015, 2015, 757530. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).