Abstract
Heat shock transcription factor (Hsf) plays a crucial role in the signal transduction pathways of plants in response to drought stress. However, studies exploring the specific functions and mechanisms of action of the Hsf family in tea plants (Camellia sinensis L.) remain limited. In this study, we identified 31 members of the CsHsf family from the C. sinensis genome. CsHsf10 was determined to be a potential drought-resistant candidate gene by screening 10 highly expressed genes in mature leaves and confirming results through RT-qPCR. Correlation analysis indicates that CsHsf10 may enhance the drought resistance of tea plants by participating in the tea polyphenol synthesis pathway and regulating the expression of antioxidant enzyme genes. Furthermore, overexpression experiments in Arabidopsis and antisense oligonucleotide experiments in tea plants corroborated that CsHsf10 exerts a significant positive regulatory effect on drought resistance in tea plants. Yeast one-hybrid assays and dual luciferase reporter gene experiments demonstrated that CsHsf10 can directly target CsPOD17, significantly promoting its transcriptional expression. Additionally, we found that the expression of CsHsf10 contributes to the increased accumulation of catechin components in tea plants under drought stress. These findings suggest that, during the response of tea plants to drought stress, CsHsf10 not only enhances antioxidant capacity by regulating the activity of antioxidant enzymes but also optimizes the physiological state of tea plants by influencing the accumulation of secondary metabolites, thereby significantly improving their drought resistance.
1. Introduction
The regulatory network centered on heat shock transcription factors (Hsfs) plays a crucial role in plant responses to various stressors, including high temperatures, drought, low temperatures, and salt stress. Upon exposure to these stressors, Hsfs binds to the DNA binding domain (DBD) at the N-terminal, which consists of three helices (H1, H2, H3) and a four-chain anti-parallel β-fold structure. The heat shock element (HSE) is located in the promoter region (5′-nGAAn-3′), regulating the transcription of heat shock proteins (HSPs) [1]. Based on differences in their oligomerization domains, Hsfs are classified into three categories: A, B, and C [2,3]. Class A Hsfs primarily function as transcriptional activators, mediated by a short activation peptide (AHA) located in their C-terminal domain (CTD) [4]. In contrast, Class B Hsfs primarily act as inhibitors and contain a core LFGV tetrapeptide at their C-terminal [5]. The functions of Class C Hsfs still need to be further studied; however, some studies suggest that they may serve as transcriptional activation factors [6,7].
The Hsf gene can be induced by various stressors, including high temperature, low temperature, salinity, and drought, and plays a role in the corresponding stress response mechanisms. For instance, LlHsfC2 in the lily collaborates with Class A Hsf to balance the heat shock response, thereby enhancing the heat resistance of the lily [8]. Hsf21 in corn regulates cold resistance by maintaining lipid metabolism under low-temperature stress [9]. HsfB2 promotes the accumulation of flavonoids, thereby increasing the salt resistance of soybeans [10]. Studies have indicated that Tomato’s HsfA1a and HsfA1b, along with those in Arabidopsis mustard, play crucial roles in the drought tolerance of plants [11,12]. Additionally, the overexpression of soybean GmHsf34, carrot CarHsfB2, and pear PeHsfC1a has been shown to enhance the drought tolerance of Arabidopsis [13,14,15]. The OsHsfA3 gene in rice improves drought tolerance by regulating polyamine biosynthesis [16]. LmHsfA5 in ryegrass promotes drought tolerance by directly activating the expression of LmHSP18.2 and LmAPX2 [17]. MdHsfA8a in apples enhances drought tolerance by regulating flavonoid synthesis and ABA-mediated stomatal movement [18]. Furthermore, PbHsfC1a and PbERF3 synergistically enhance drought tolerance in pear through the ABA signaling pathway and the H2O2 signaling regulatory network [19].
Tea (Camellia sinensis) is an ancient commercial crop [20]. Due to its significant economic value, the area dedicated to tea plantations has rapidly increased in China and other tropical and subtropical countries [21]. Tea plants are susceptible to various environmental factors during their growth, including high-temperature stress [22,23], low-temperature stress [24], and drought stress, which is one of the most prevalent challenges [25,26]. Drought stress significantly impacts the growth and development of tea plants, affecting both yield and quality. Statistics indicate that drought can reduce tea yield by 14–33% and increase tea plant mortality by 6–19% [27]. In recent years, many provinces in China have experienced varying degrees of drought, adversely affecting the yield and quality of tea, which largely depend on favorable growing conditions. Drought stress severely hinders the growth and development of tea plants, ultimately impacting yield and quality [28]. As a sessile plant, the tea plant has evolved multiple defense mechanisms to withstand drought stress throughout its long-term adaptation process. Therefore, further research into the molecular mechanisms underlying drought tolerance in tea plants is essential. Hsf is a key regulator of plant responses to environmental stresses, including high temperature and drought conditions. However, research on tea plants has primarily focused on heat stress. Studies have demonstrated that the heterologous expression of CsHsfA2 enhances the heat tolerance of transgenic yeast, suggesting that it may play a role in regulating the heat stress response [29]. The inhibitory experiments mediated by the geldanamycin (GDA) inhibitors demonstrate that the tea plant’s high expression of the CsHsfA2 gene significantly enhances its heat shock response [30]. The expression of HsfB2b can be adjusted in the outer tea plant, which is associated with the heat-resistant mechanism [31]. Furthermore, under high-temperature conditions, CsHsfA1b and CsHsfA2 contribute to the accumulation of flavonoid substances in tea plants by influencing the negative regulation of jasmonic signaling [32].
To explore the response mechanism of tea plants under drought stress in-depth, this study conducted a systematic and comprehensive analysis of the Hsf gene family in tea plants, building on previous research [29,33]. Utilizing bioinformatics methods, we identified the tea plant Hsf gene family members, examining their genetic patterns, conserved motifs, and gene structures. Additionally, we determined the physiological changes in tea plants subjected to drought stress. We employed RT-qPCR technology to assess the relative expression levels of CsHsf gene family members across different tissues under drought conditions. Following identifying the candidate gene CsHsf10, we performed preliminary functional verification using Arabidopsis overexpression, yeast one-hybrid assays, and antisense oligonucleotides. The findings of this research not only enhance our understanding of the tea plant Hsf gene family but also establish a solid foundation for future in-depth studies on the functional mechanisms of these genes. More importantly, they provide highly valuable genetic resources for the molecular breeding of drought-resistant tea plants.
2. Materials and Methods
2.1. Identification and Physical and Chemical Analysis of Hsf Genes in Camellia sinensis
The reference genome data of C. sinensis were obtained from the TPIA database (http://tpia.teaplants.cn/download.html, accessed on 25 September 2024). At the same time, the conserved domain of the Hsf transcription factor was sourced from the Pfam database [34] (http://pfam.xfam.org/, accessed on 26 September 2024) using the Hidden Markov Model (HMM) associated with serial number PF00447. C. sinensis protein sequences containing Hsf domains were identified using TBtools (v2.122) [35] with default parameters (Evalue ≤ 1 × 10−5). Concurrently, the Arabidopsis thaliana Hsf protein sequence was retrieved from the TAIR database (https://www.arabidopsis.org/browse/gene_family/HSF, accessed on 26 September 2024), and the BLAST tool within TBtools (v2.122) was employed to perform comparisons with a cutoff E value of <1 × 10−20. Subsequently, the C. sinensis protein sequences were compared, and the two results were integrated to identify the final members of the CsHsf gene family. Additionally, the physical and chemical properties and the chromosomal locations of the Hsf protein sequences in C. sinensis were analyzed using TBtools (v2.122). The subcellular localization of the gene families was predicted using WOLF PSORT (https://wolfpsort.hgc.jp/, accessed on 27 September 2024) [36].
2.2. Phylogenetic Analysis of C. sinensis, A. thaliana, and Populus Trichocarpa Hsf Genes
Using MEGA (v11.0.13) software, the neighbor-joining method and 1000 bootstrap tests were employed to construct the phylogenetic tree of C. sinensis, A. thaliana, and P. trichocarpa. Subsequently, the tree was redrawn and annotated using the online platform iTOL (https://itol.embl.de/, accessed on 14 October 2024). Information regarding the Hsf family members of A. thaliana and P. trichocarpa was obtained from previous studies [37,38]. The protein sequence for poplar was sourced from the Phytozome database (https://phytozome-next.jgi.doe.gov/, accessed on 10 October 2024) [39].
2.3. Sequence, Conserved Motifs, and Gene Structure Analysis
Protein secondary structure predictions for DNA-binding domain (DBD) were conducted using JPred4 (https://www.compbio.dundee.ac.uk/jpred4/, accessed on 16 October 2024) [40]. MEGA (v11.0.13) was employed to edit multiple sequence alignment files, which were visualized with GeneDoc 2.7 [41]. The conserved motifs of C. sinensis Hsf proteins were predicted using the MEME tool (https://meme-suite.org/meme/, accessed on 6 November 2024), with the maximum number of motifs set to 10 and other parameters maintained at their default values. Annotation files for the C. sinensis genome were sourced from the TPIA database, from which the exons and UTR information of CsHsf were extracted, positioned, and visualized using TBtools (v2.122).
2.4. Gene Duplication and Collinearity Analysis of Tea Plant Hsf Gene
Genome data for A. thaliana, Populus nigra, Solanum lycopersicum, and Oryza sativa were obtained from the National Center for Biotechnology Information database (NCBI) (https://www.ncbi.nlm.nih.gov/, accessed on 8 November 2024). A systematic analysis of the Hsf gene family in C. sinensis was conducted using the MCScanX plug-in within TBtools (V2.122). This analysis included identifying tandem duplication events, segmental duplication events, and collinear relationships with the four species above. The results of this analysis were presented using visualization tools, which facilitated the calculation of genetic pairs involved in tandem duplication events and segmental duplication events within the Hsf gene family of C. sinensis, particularly under selective pressure.
2.5. Identification of Cis-Regulatory Elements in the Promoter Regions of CsHsf Genes
TBtools (v2.122) was used to extract the 2000 bp DNA sequence upstream of the Hsf promoter region in the C. sinensis genome. Subsequently, these sequences were submitted to the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plantcare/html/, accessed, accessed on 12 November 2024) [42] to identify cis-acting elements.
Focus on screening for elements related to stress response, plant growth and development, and hormone response.
2.6. Plant Materials and Drought Treatment
The experimental material for this study consisted of ‘Huangjincha 2’ annual tea seedlings sourced from the Gaoqiao Tea Garden of the Hunan Tea Research Institute (longitude: 113°08′, latitude: 28°20′). These seedlings had been cultivated in a greenhouse for approximately 1 year, during which they received regular irrigation and fertilization to promote healthy growth. For the experiment, seedlings exhibiting robust and consistent growth were selected as test materials and acclimatized in an artificial climate box for 4 weeks to prepare for the challenging environmental conditions. Cultivation was conducted in black square plastic pots measuring 10.2 cm in diameter, 13.8 cm in height, and 7.2 cm at the base. Before the onset of drought treatment, the tea seedlings were irrigated every 3 days with a volume of 100 mL per pot, supplemented with a slow-release compound fertilizer at a dosage of 5 g per pot. Following the initiation of drought treatment, all irrigation was halted to simulate a natural drought environment. The light conditions were set to 15,000 lux, with a photoperiod of 16 h of light and 8 h of darkness, maintaining day and night temperatures of 25 °C and 22 °C, respectively. After treatment, samples were collected from the second blade, which was fully expanded from top to bottom, on days 0, 5, 10, and 15. Meanwhile, the soil moisture content was measured using the JK-100 series soil moisture meter every 5:00 p.m. (Yoke, Taizhou, China). Additionally, a control group, consisting of the normal irrigation group, was established. Each treatment included at least three independent biological replicates. Samples were immediately quick-frozen in liquid nitrogen after collection to preserve their integrity and were stored at −80 °C in an ultra-low temperature freezer for subsequent experiments. A portion of the samples was utilized immediately to measure physiological and biochemical parameters.
2.7. Determination of Chlorophyll Fluorescence Parameters and Relative Conductivity
To prevent exposure to light, we placed the tested plant in the dark for 25 min at room temperature. Subsequently, we evaluated the leaves’ maximum actual photosynthetic efficiency (Fv/Fm) using the PAM fluorescence system (WALZ, Effeltrich, Germany) without detaching the plant. Subsequently, a sample of the leaf blade was placed in a 10 mL scale test tube filled with deionized water, covered with a glass plug, and allowed to soak at room temperature for 12 h. The conductivity meter was then employed to measure the conductance of the extracted liquid (R1). The test tube was placed in a boiling water bath for 30 min, allowed to cool to room temperature, and shaken thoroughly. The conductance of the leached liquid (R2) was measured again, and the relative conductivity was calculated using the formula R1/R2 × 100% [43].
2.8. Determination of Enzyme Activity, MDA Content, and Physiological Indices of Tea Plants
The enzymatic activities of SOD and POD in tea plants and Arabidopsis were quantitatively determined using specific activity detection kits (Item NO: BC0175, BC0095, Solarbio, Beijing, China, respectively). Similarly, the enzymatic activity of APX in Arabidopsis was quantitatively assessed using a specific activity detection kit (Item NO: BC0225, Solarbio, China). Tea plants’ malondialdehyde (MDA) content was measured using a detection kit (Item NO: BC0025, Solarbio, China), while the MDA content in Arabidopsis was determined using the CheKine detection kit (Item NO: KTB1050, Abbkine, Wuhan, China). A mixture of 3 g of ground sample was combined with 450 mL of distilled water and extracted at 100 °C for 45 min. After extraction, the mixture was filtered and cooled to room temperature. Distilled water was then added to the filtrate and mixed thoroughly to achieve a final 500 mL tea soup volume for subsequent measurements. A total of 1 mL of the solution, 0.5 mL of phosphate buffer, and 0.5 mL of chi-total were added to a 25 mL test tube. We heated the mixture at 100 °C for 15 min, then cooled it to room temperature, ensuring the final volume was 25 mL. A spectrophotometer measured the amino acid content at 570 nm [44]. Similarly, 1 mL of the solution was slowly dipped into a preprepared sulfate solution in a 25 mL capacity bottle, and the mixture was heated in boiling water for 3 min. After cooling to room temperature, the spectrophotometer assessed the soluble sugar content at a wavelength of 620 nm [43]. As the literature describes, total flavonoids were determined using aluminum chloride [45]. The tea polyphenol content was measured by the Folin–Ciocalteu method, as referenced in the literature [46].
2.9. RNA Extraction and Real-Time Fluorescence Quantitative PCR (RT-qPCR)
One-year-old potted tea seedlings were selected as the experimental materials in this study. Their leaf, stem, and root tissues were collected separately for subsequent experiments. For A. thaliana, leaves aged 2 months were chosen as the experimental materials. The same RNA extraction and q-PCR methods were employed for Arabidopsis and tea plants. Total RNA extraction was conducted using the Fastpure Universal Plant Total RNA Isolation Kit (Vazyme, Nanjing, China). The first strand cDNA for real-time fluorescence quantitative PCR (RT-qPCR) was synthesized from total RNA utilizing the Evo M-MLV reverse transcription reagent premix (Agbio, China). RT-qPCR was performed with the BYBR Green Premix Pro Taq HS qPCR kit (Agbio, China) on a QuantStudio 3 quantitative PCR instrument (Thermo Fisher Scientific, Waltham, MA, USA). The relative expression levels of genes were calculated using the 2−∆∆CT method, based on three biological replicates, with β-actin serving as the internal reference gene. Primers were designed utilizing the NCBI database (Table S1).
2.10. Homologous Sequence Alignment, Protein-Protein Interaction Network Prediction, and Subcellular Localization of CsHsf10
The amino acid sequences of CsHsf10 homologs from various species, such as Populus alba, Populus tomentose, P. trichocarpa, Populus simony, Populus euphratica, Manihot esculenta, Hevea brasiliensis, Tripterygium wilfordii, Forsythia ovata, Olea europaea, Cornus florida, Diospyros lotus, and Actinidia chinensis, were retrieved from the NCBI database (https://www.ncbi.nlm.nih.gov/, accessed on 10 December 2024). These sequences were visualized using GeneDoc 2.7 software, while MEGA (v11.0.13) software was employed to construct a phylogenetic tree based on 1000 bootstrap repeats utilizing the neighbor-joining method. The cDNA sequence of the CsHsf10 gene was ligated into the linearized vector pEAQ-GFP to create the pEAQ-CsHsf10-GFP construct. Following the transformation of Escherichia coli DH5α competent cells, the cloned plasmid was extracted and subsequently transformed into Agrobacterium tumefaciens GV3101. The Agrobacterium strain containing the pEAQ-CsHsf10-GFP construct, with an optical density at 600 nm (OD600) of 0.6–0.8, was injected into tobacco leaves’ epidermis. The cells were incubated under low light conditions at 25 °C for 12 h and then transferred to normal light conditions for 48 h. The GFP signal was detected using an Axio Scope A1 upright fluorescence microscope (Carl Zeiss AG, Oberkochen, Germany). The primers used in this study are listed in Table S1. Additionally, using tea plant protein as a model, the potential interaction relationships of the CsHsf10 protein were predicted using STRING (https://string-db.org/, accessed on 10 December 2024).
2.11. Arabidopsis thaliana Overexpression Experiment
The full-length coding region of CsHsf10 was inserted into the p1300 vector to construct the 35S::CsHsf10-EYFP recombinant vector. This recombinant vector was transformed into the Columbia ecotype of Arabidopsis using the A. tumefaciens strain GV3101 via the flower dipping method to obtain transgenic Arabidopsis plants. For further analysis, T3 generation homologous overexpression (OE) transgenic Arabidopsis lines were selected for experiments. The experimental conditions were established: light intensity was set at 10,000 lux, with a light cycle of 16 h of light and 8 h of darkness. The daytime temperature was maintained at 25 °C, while the nighttime temperature was controlled at 22 °C. A natural drought environment was simulated by ceasing irrigation, which was resumed 10 days after the drought treatment (Table S1).
2.12. Antisense Oligonucleotide (AsODNs) Experiment
The online software Soligo (https://sfold.wadsworth.org/cgi-bin/soligo.pl, accessed on 11 December 2024) was utilized to design and select four antisense oligonucleotides (AsODNs) targeting CsHsf10 (Table S1). Tea plant seedlings with one bud and two leaves were cultured in centrifuge tubes containing 20 μM of CsHsf10-related AsODNs. The samples were incubated under standard light conditions, while a control group was established using the sense strand solution. Samples were collected at 0, 24 h to measure the expression levels of CsHsf10. The group exhibiting the highest efficacy among the AsODNs was chosen for subsequent drought treatment and related assays. In the experimental group, the tea plant branches were placed in waterless centrifuge tubes for 24 h to simulate drought conditions, while those in the control group were immersed in centrifuge tubes containing sterile water to maintain normal growth conditions. Each group included a control group without CsHsf10 silencing (CK group) and a silencing group (AsODN-CsHsf10). After the 24 h treatment, mature leaves (the second leaves) were selected for subsequent measurements.
2.13. LC-MS/MS Chromatographic Conditions
We have employed liquid chromatography-tandem mass spectrometry (LC-MS/MS) technology to enhance our understanding of the changes in catechin components in tea plants under drought conditions. Analysis was conducted using a Nexera X2 LC-30AD high-performance liquid chromatography system (Shimadzu, Kyoto, Japan) equipped with an Acquity BEH C18 column (2.1 × 100 mm, 1.7 μm; Waters, Milford, MA, USA). The column temperature was maintained at 40 °C, and separation was achieved through gradient elution utilizing mobile phases A (0.1% formic acid in water) and B (0.1% formic acid in acetonitrile). An injection volume of 1 μL was employed, with a flow rate set at 0.3 mL/min.
2.14. Yeast One-Hybrid (Y1H) Assay
The full-length coding sequence of CsHsf10 was cloned into the pGADT7 vector to serve as the prey, while the promoter regions of CsPOD17 were cloned into the pHis2 vector to function as the bait. The prey and bait constructs were co-transformed into the Saccharomyces cerevisiae Y187 strain. To verify the interactions between CsHsf10 and CsPOD17, the co-transformed yeast cells were diluted and plated onto SD/-His/-Leu/-Trp media, supplemented with varying concentrations of 3-aminotriazole (3-AT) for selection (Table S1).
2.15. Dual-Luciferase Reporter Assay (LUC)
The full-length coding sequence of CsHsf10 was cloned into the pGreenII62-SK vector as an effector gene, while the promoter region of CsPOD17 was cloned into the pGreenII0800-LUC vector as a reporter gene. These plasmids were introduced into the A. tumefaciens strain GV3101 (pSoup-p19). Subsequently, Agrobacterium strains containing both the effector and reporter genes were co-injected into the lower epidermis of tobacco leaves. Following an incubation period of 12 h at 25 °C in low light, the plants were transferred to normal light conditions for 36 h. Finally, fluorescence imaging was conducted using the Viber Newton 7.0 bioimaging system (Vilber Bio Imaging, Paris, France) and potassium luciferase substrate. (Table S1)
2.16. Statistical Analysis
Correlation analysis, Student’s t-test, and Duncan’s multiple range comparisons were conducted using IBM SPSS Statistics 23.0 (IBM, Armonk, NY 10504, USA), with p < 0.05 indicating statistical significance. All data were presented as the mean ± standard deviation (SD) of at least three replicates. Graphs were generated using GraphPad Prism (v8.0.2) (GraphPad Software, USA). The Mantel test and Pearson correlation coefficient were employed for the analysis, while ChiPlot (https://www.chiplot.online/, accessed on 11 December 2024) was utilized to visualize the correlation results.
3. Results
3.1. Physiological Changes of Tea Plant Under Drought Stress
Tea seedlings were subjected to natural drought treatment to investigate the physiological changes in tea plants under drought conditions. The results indicated that the damage to the tea plants gradually intensified over time. On day 0, the leaves appeared glossy and erect. After 5 days of drought treatment, the leaves began to lose their luster and tended to curl and droop. After 10 days, the leaves showed signs of yellowing, with the edges turning reddish-brown. By day 15 of the drought treatment, the leaves were predominantly reddish-brown and completely drooped (Figure 1A). To gain deeper insights into the physiological responses of tea plants to drought stress, several parameters were assessed, including the levels of physiologically active substances, chlorophyll fluorescence, relative conductivity, and enzyme activity (Figure 1B). The research results indicate that, in contrast to the stability observed in the CK group, the physiological indicators of tea plants in the experimental group exhibited significant fluctuations due to drought. Tea polyphenol increased by 1.27 times at 10 days before beginning to decline. Amino acid levels reached their lowest point on the fifth day and subsequently increased. The phenol-ammonia ratio peaked at 9.25 on day 5 before commencing a decline. The soluble sugar content initially rose before decreasing, returning to baseline levels by day 15. Flavonoid and maximum photosynthetic efficiency (Fv/Fm) exhibited significant reductions during drought, decreasing by 56.69% and 72.00%, respectively, at 15 days. MDA content and relative conductivity also increased markedly, 3.15 times and 0.71 times, respectively. The activities of SOD and POD increased at 5 days of drought but then significantly declined by 20.28% and 44.49%, respectively. Meanwhile, the results of the soil moisture analysis indicated that, following the cessation of irrigation, the soil moisture content significantly decreased by 73.90% on the 15th day compared to the CK. This finding demonstrates that the tea plants were experiencing drought stress, which aligns with the previously mentioned results.
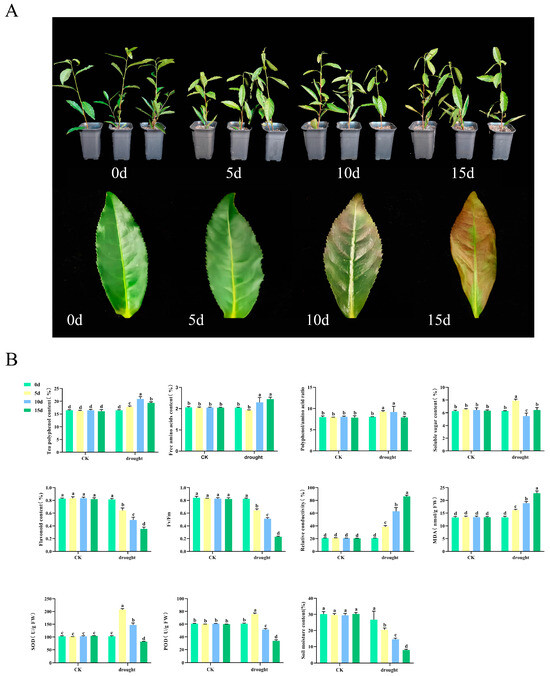
Figure 1.
Drought stress treatment and physiological changes of tea plants. (A) Drought stress treatment of tea plants. (B) Physiological changes of tea plants and soil moisture variations. CK is the normal irrigation group; drought is the drought treatment group. 0 d, the 0th day of the experiment; 5 d, the 5th day of the experiment; 10 d, the 10th day of the experiment; 15 d, the 15th day of the experiment. Fv/Fm, maximum photosynthetic efficiency; MDA, malondialdehyde; SOD, superoxide dismutase; POD, peroxidase; different letters indicate significant differences (p < 0.05); data are presented as the mean ± standard deviation (SD).
3.2. Relative Expression Levels and Correlation Analysis of the CsHsf Gene Family Under Drought Stress
We identified 31 CsHsf family members in the C.sinensis genome. Of these, 22 CsHsf genes are located on chromosomes, while nine are situated on unassembled scaffolds and have been named accordingly (Table S2). A systematic phylogenetic analysis (Figure S1; Table S3), gene structure analysis (Figure S2; Table S4), synteny analysis (Figure S3; Table S5), and promoter element analysis (Figure S4) were conducted. The results indicated that members of the CsHsf family play a significant role in the growth and development of tea plants and their response to stress. Following the investigation of physiological changes in tea leaves, we further explored the reaction of CsHsf family genes to drought stress by analyzing their expression levels in tea plants’ buds, leaves, stems, and roots. The expression levels of CsHsf family genes exhibit distinct characteristics across different tissues. Through clustering analysis of the expression patterns of these genes, we categorized them into four groups: Group I, Group II, Group III, and Group IV (Figure 2A). Group I genes showed high expression across all tissues. In Group II, the expression levels were notably higher in mature and old leaves. The expression levels of Group III genes in the tested samples are relatively low, which may be attributed to limitations in transcriptional activity. Conversely, Group IV genes demonstrated increased root expression levels while showing lower expression in buds and young leaves. These findings suggest that CsHsf family members may fulfill different roles in the growth of various tea plant tissues. Notably, ten genes—CsHsf 1-3, CsHsf 5, CsHsf 9-10, CsHsf 17, and CsHsf 28-30—were found to be highly expressed in mature leaves (second leaves), prompting further analysis through RT-qPCR (Figure 2B). The results indicated that all ten members of the CsHsf family responded to drought stress, exhibiting a consistent expression pattern characterized by an initial increase followed by a subsequent decrease. Specifically, CsHsf1-2, CsHsf9, and CsHsf29-30 peaked on day 10, with increases ranging from 1.27 to 2.53 times before beginning to decline. In contrast, CsHsf3, CsHsf5, CsHsf10, CsHsf17, and CsHsf28 reached their peak at day 5, followed by a gradual decrease. Notably, CsHsf10 exhibited the most pronounced response, with an increase of 11.23 times, while CsHsf17 showed a rise of 5.34 times; the remaining members increased by from 1.90 to 2.33 times. This variation in expression may be associated with the mechanisms underlying drought responses at different time points. It is worth noting that, compared to the CK, the expression of CsHsf10 was significantly induced following drought treatment, suggesting that it may play a crucial role in regulating drought tolerance in tea plants. To investigate the potential pathways of CsHsf10 in response to drought stress, we analyzed its correlation with physiological changes in tea plants (Figure 2C). The results indicate that CsHsf10 is significantly positively correlated with flavonoids, relative conductivity, MDA, and Fv/Fm. Conversely, it is significantly negatively correlated with tea polyphenols and SOD.
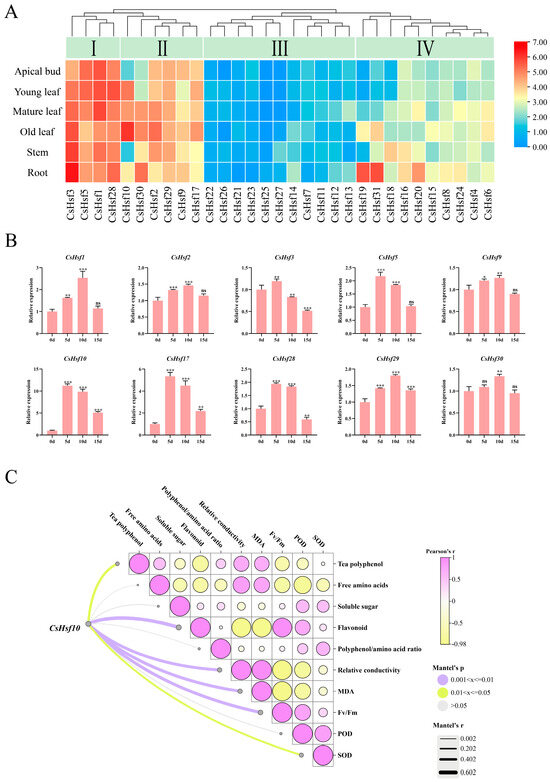
Figure 2.
Expression analysis of CsHsfs in different plant tissues and under drought conditions. (A) Heat map of CsHsf expression in different tissues. (B) Relative expression levels of 10 CsHsf genes in mature leaves (the second leaf) at different periods under drought stress. (C) Correlation analysis between CsHsf10 and physiological changes in mature leaves (the second leaf) of tea plants; 5 d, the 5th day of natural drought; 10 d, the 10th day of natural drought; 15 d, the 15th day of natural drought; Fv/Fm, maximum photosynthetic efficiency; MDA, malondialdehyde; SOD, superoxide dismutase; POD, peroxidase; significant differences from CK were determined by Student’s t-test (ns not significant; * p < 0.05; ** p < 0.01; *** p < 0.001); data are presented as the mean ± standard deviation (SD).
3.3. Bioinformatics Analysis and Subcellular Localization of CsHsf10
According to phylogenetic tree classification, CsHsf10 belongs to the A3 subfamily (Figure S1). Amino acid sequences from other species with high homology to CsHsf10 were retrieved from the NCBI database, compared, and utilized to construct a phylogenetic tree (Figure 3B, Table S6). Sequence analysis revealed that CsHsf10 and other homologous sequences contain a DBD, a coiled-coil domain, and two protein-disordered regions, with no significant differences in sequence length (Figure 3A). These species include Olea europaea, Populus alba, and Tripterygium wilfordii, among others. Particularly, CsHsf10 exhibits the highest homology with DIHsfA3 (Diospyros lotus) and AxHsfA3 (Actinidia chinensis). These three genes are positioned on the same branch of the phylogenetic tree, indicating a closer genetic relationship. The tea protein database was used as a reference to analyze the interaction relationship of the CsHsf10 protein (Figure 3C). The results indicated that TEA_016617 shares high homology with CsHsf10, showing a similarity of 94.60%. TEA_016617 interacts with several proteins, including TEA_028988, TEA_007283, TEA_006589, TEA_023371, TEA_020126, TEA_027305, TEA_024009, and TEA_017582. These genes may play a significant role in drought stress responses in tea plants.
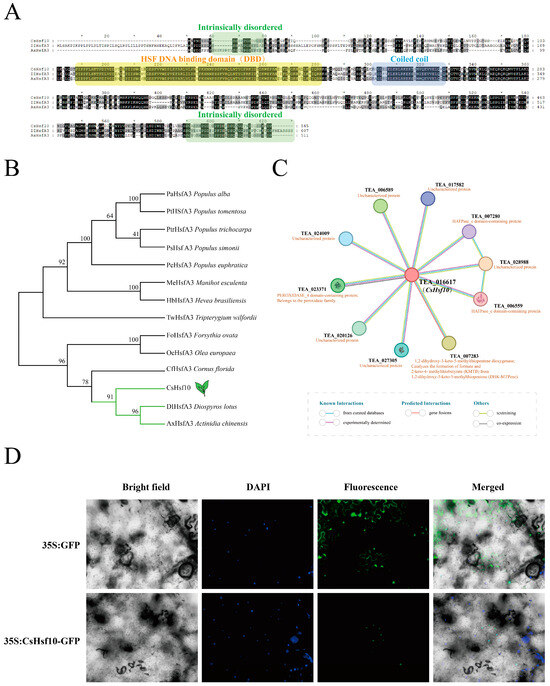
Figure 3.
Homologous sequence alignment, phylogeny, and subcellular localization analysis of CsHsf10. (A) Homologous sequence alignment. (B) Phylogenetic analysis of CsHsf10 and homologous sequences from other species. (C) CsHsf10 protein interaction network prediction. (D) Subcellular localization of CsHsf10.
The role of transcription factors in transcriptional regulation depends on their positioning within the nucleus. To ensure the subcellular localization of CsHsf10, the pEAQ-GFP construct was utilized as a control in the epidermal cells of tobacco leaves, while the pEAQ-CsHsf10-GFP construct was transiently expressed. Fluorescence signals were detected using a fluorescence microscope, revealing that green fluorescence was observed throughout the cells expressing 35S: GFP, whereas green fluorescence signals were exclusively detected in the nucleus of tobacco cells into which the pEAQ-CsHsf10-GFP fusion plasmid was introduced (Figure 3D). This indicates that CsHsf10 is localized in the nucleus and is classified as a nuclear protein, which aligns with the predicted results.
3.4. Effects of Overexpression of CsHsf10 Gene and Antisense Oligonucleotides on Drought Resistance
To investigate the role of CsHsf10 in drought resistance, we overexpressed the CsHsf10 gene in Arabidopsis and identified two transgenic lines in the T3 generation through RT-qPCR and DNA verification screening. The results of the analysis indicated that the transcript levels of these two lines (CsHsf10 OE1 and CsHsf10 OE2) were significantly higher than those of the Columbia ecotype, which serves as the wild type (WT) (Figure 4A). Before drought treatment, no significant differences were observed between the transgenic CsHsf10 seedlings and WT seedlings. After 10 days of natural drought, only 8.00% of WT seedlings survived, while most genetically modified seedlings exhibited only a slight water loss. The survival rates for OE1 and OE2 were 85.53% and 88.60%, respectively, significantly higher than that of WT (Figure 4B,C). In addition, we measured the chlorophyll fluorescence parameters of WT, OE1, and OE2 plants under normal growth conditions and on the 10th day of drought stress. Under normal conditions, there were no significant differences in the Fv/Fm values among WT, OE1, and OE2. However, on the 10th day of drought stress, the Fv/Fm values for OE1 and OE2 were significantly higher than that for WT, measuring 0.601 and 0.606, respectively, compared to 0.477 for WT (Figure 4D,E). Meanwhile, we measured the MDA content and the activities of SOD, POD, and APX (ascorbate peroxidase) enzymes in both WT and transgenic overexpression (OE) strain plants at day 10 of drought stress. The results indicate that there are no significant differences in MDA content or SOD, POD, and APX enzyme activities under normal growth conditions. However, the MDA content in OE strain plants is significantly lower under drought stress than in WT plants. At the same time, the activities of SOD, POD, and APX enzymes are considerably higher in the OE strains compared to in the WT (Figure 4F). These findings suggest that the overexpression of CsHsf10 enhances drought tolerance in Arabidopsis, potentially by improving antioxidant enzyme pathways.
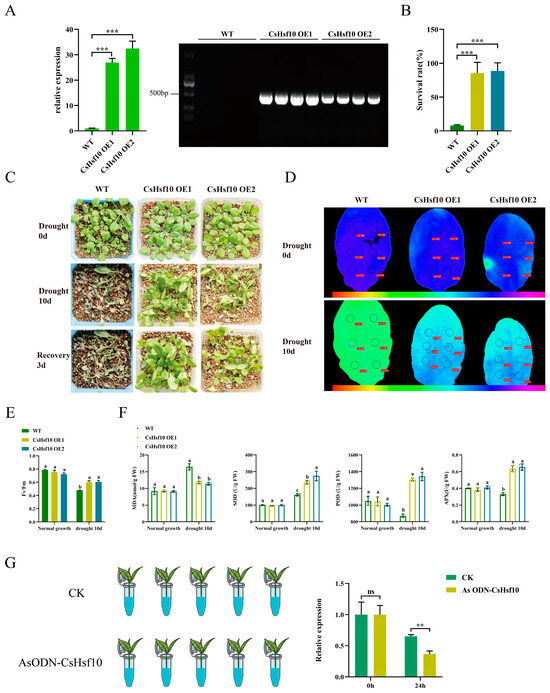
Figure 4.
Overexpression of CsHsf10 in Arabidopsis improves drought tolerance. (A) qRT-PCR detection and DNA verification results of CsHsf10 gene in transgenic lines (OE-1, OE-2) and WT plants. (B) The survival rate of Arabidopsis under drought stress. (C) The phenotype of soil planting plants before and after drought stress. (D) Chlorophyll fluorescence imaging before and after drought stress. (E) Changes in Fv/Fm values before and after drought stress. Fv/Fm, maximum photosynthetic efficiency. (F) Changes in MDA, SOD, POD, and APX content before and after drought stress. MDA, malondialdehyde; SOD, superoxide dismutase; POD, peroxidase; APX, ascorbate peroxidase. (G) Treatment of antisense oligonucleotides (AsODN) of CsHsf10 and relative expression level of CsHsf10 at 0 and 24 h after treatment. Different letters indicate significant differences (p < 0.05); significant differences from CK were determined by Student’s t-test (ns not significant; ** p < 0.01; *** p < 0.001); data are presented as the mean ± standard deviation (SD).
To further verify the drought resistance of CsHsf10, the oligodeoxynucleotide AsODN-CsHsf10 was designed to inhibit its expression in tea plants. Tea plant seedlings with one bud and two leaves were placed in a solution containing 20 μM AsODN-CsHsf10 (experimental group) alongside a sense strand solution containing CsHsf10 (CK group). After sampling at 0 and 24 h, qRT-PCR detected silencing of the CsHsf10 gene after 24 h. The expression of the CsHsf10 gene in the experimental group was significantly lower than that in the CK group (Figure 4G), leading to the selection of the tea plant that had been silenced for 24 h for subsequent drought experiments. The chlorophyll fluorescence detection results indicated that there was no significant difference in the Fv/Fm values under normal growth conditions between the two groups. However, after 24 h of drought, the Fv/Fm in the experimental group was significantly lower than in the CK group. Additionally, MDA content determination showed no significant difference in MDA content between the control and experimental groups under normal growth conditions. Under drought stress, however, MDA content increased significantly following the silencing of the CsHsf10 gene. This suggests that silencing the CsHsf10 gene enhances the sensitivity of tea plants to drought stress. In addition, we measured changes in the content of catechin components. Under normal growth conditions, no significant differences were observed in the levels of DL-C, EC, EGC, ECG, GCG, and EGCG between the CK and experimental groups. However, under drought stress, silencing the CsHsf10 gene significantly reduced the contents of DL-C, EC, EGC, ECG, GCG, and EGCG compared to the CK group (Table 1). It indicates that CsHsf10 may influence tea polyphenols, such as catechins, and other mechanisms related to drought resistance in tea plants.

Table 1.
Changes in physiological components of mature tea leaves (second leaf) before and after 24 h drought treatment.
3.5. CsHsf10 Positively Regulates CsPOD17 to Drought Response Mechanism
We performed a yeast one-hybrid assay to elucidate the potential regulatory relationship between CsHsf10 and antioxidant enzymes. The complete coding sequence of CsHsf10 was cloned into the pGADT7 vector as the prey, while the CsPOD17 promoter was inserted into the pHis2 vector as the bait. The experimental group consisted of a mixture of the pGADT7-CsHsf10 and pHis2-CsPOD17 plasmids, whereas the self-activated group was composed of pHis2-CsPOD17 and pGADT7 plasmids alone. The growth of the Y187 strain containing only the recombinant vector of the CsPOD17 promoter was inhibited on SD/-His/-Leu/-Trp medium supplemented with 50 mM 3-amino-1,2,4-triazole (3-AT). In contrast, when transformed with the prey vector pGADT7-CsHsf10, the colony growth of the experimental group was significantly enhanced compared to that of the self-activated group under the same dilution gradient (Figure 5A). A dual luciferase reporter gene assay revealed that fluorescence signals were detected in tobacco leaves co-injected with pGreenII62-SK-CsHsf10 and pGreenII0800-LUC-CsPOD17 in group IV. The relative LUC/REN values were also significantly higher than those in the other groups (Figure 5B). These results indicate that CsHsf10 can directly bind to CsPOD17 and enhance the transcriptional expression of CsPOD17.
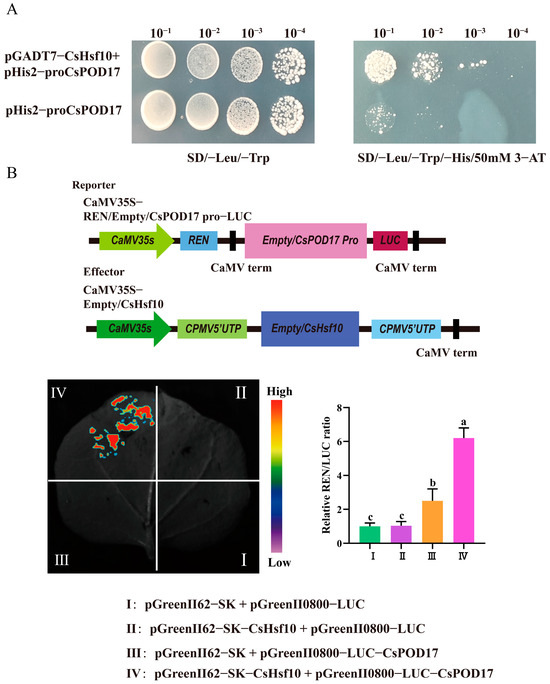
Figure 5.
Results of yeast monomer and double luciferase reporter gene. (A) Yeast monochrome experiment. (B) Results of the dual luciferase reporter gene experiment. Different letters indicate significant differences (p < 0.05); data are presented as the mean ± standard deviation (SD).
4. Discussion
Based on phylogenetic analysis, our study identified 31 Hsf family members in the tea plant ‘Shuchazao’ genome and categorized them into three subfamilies (A, B, C). This number is fewer than the 78 identified in wheat [47] but more significant than the counts found in grape (19) [48], plum (18) [49], garlic (22) [50], and loquat (30) [51]. This discrepancy may be attributed to the whole-genome duplication event that occurred during the genetic differentiation of the tea plant [52]. The tea plant Hsf gene family features highly conserved DBD, which consists of an α1-β1-β2-α2-α3-β3-β4 structure, consistent with those in other species [53]. Gene structure analysis indicates that motifs 1–5 collectively form the DBD domain of Hsf, which is crucial for recognizing and binding heat shock elements (HSEs) [1]. Motif 7, located at the C-terminus, is unique to class A heat shock factors (HsfAs) and is speculated to be an AHA motif involved in the transcriptional activity and stress response of CsHsf, aligning with previous research findings [4]. This study identified two pairs of tandem duplication genes and four pairs of segmental duplication genes, which indicates that these duplication events contribute significantly to expanding the tea plant Hsf gene family. Notably, the segmental duplication genes in the tea plant genome substantially impact the growth of the Hsf gene family. Furthermore, the Ka/Ks values of six pairs of homologous CsHsf genes were all found to be less than 1. This observation suggests that these gene pairs have undergone purifying selection, which helps to maintain the relative conservation and functionality of the Hsf gene family. Collinearity analysis with other species indicated that species with closer evolutionary relationships exhibit more collinear gene pairs. Additionally, we observed that the promoters of all CsHsf genes contain at least one stress-related cis-acting element, including the ARE (anaerobic induction), MBS (drought induction), LTR (low-temperature response), TC-rich repeats (defense and stress response), and GC-motifs (hypoxia-specific induction), among others related to abiotic stress, which suggests that Hsf plays a crucial role in regulating the abiotic stress adaptability of tea plants.
Current research on Hsfs primarily concentrates on their role in heat stress response. However, studies have demonstrated that Hsfs can also be induced under drought conditions and is responsive to drought stress [6,17,54,55]. Despite this, research on drought in tea plants remains relatively limited. To investigate the response mechanisms of the CsHsf family members under drought stress, ‘Huanghuangcha 2’ was subjected to natural drought treatment, revealing that its physiological components were affected to a certain extent. For instance, tea polyphenol levels increased by 1.27 times after 10 days but began declining. Flavonoid content decreased significantly with the onset of drought, showing reductions of 56.69%. SOD and POD activities increased at 5 days of drought but then significantly decreased by 20.28% and 44.49%. These results indicate that, as drought duration increased, the photosynthetic efficiency of tea plants significantly declined, leading to severe inhibition of photosynthesis, increased cell membrane permeability, and loss of selectivity. Following RT-QPCR analysis, the expression of Hsf genes in various tissues of tea plants was found to be specific. Then, ten family members exhibiting high expression levels in mature leaves were selected for relative expression analysis under drought stress. The results indicated that all ten CsHsfs responded to drought conditions, displaying a consistent expression trend of initially increasing followed by decreasing levels. In particular, CsHsf10 was significantly induced, showing an increase of 11.23 times on the 5th day compared to CK. As a promising candidate gene for drought resistance, the sequence and function of the CsHsf10 gene were analyzed. CsHsf10 features a typical Hsf DBD, a coiled-coil domain, and two protein-disordered regions. Notably, the coiled-coil domain is hypothesized to serve as a component of the oligomerization domain, which is essential for binding to HSE elements and inducing the expression of heat shock protein HSP [53,56]. Phylogenetic tree classification indicates that CsHsf10 belongs to the A3 subfamily. Sequence analysis reveals that CsHsf10 is closely related to DIHsfA3 (Diospyros lotus) and AxHsfA3 (Actinidia chinensis), suggesting they may share similar functions. Subcellular localization results show that CsHsf10 is localized in the nucleus.
Previous studies have demonstrated that HsfA3 is responsive to drought stress [16,57]. However, this aspect remains unexplored in tea plants. Correlation analyses conducted in conjunction with previously measured physiological indicators of tea leaves revealed that CsHsf10 may enhance drought resistance in tea plants by promoting the expression of tea polyphenols, including catechins, as well as antioxidant enzyme genes. To thoroughly investigate the role of CsHsf10 in the drought resistance response of tea plants, this study demonstrated that the survival rate and Fv/Fm ratio of Arabidopsis overexpressing CsHsf10 were significantly enhanced under drought stress. At the same time, the MDA content was notably reduced. These findings indicate that CsHsf10 can substantially improve the drought tolerance of Arabidopsis. Further analysis revealed that the activities of SOD, POD, and APX in Arabidopsis overexpressing CsHsf10 increased significantly, corroborating previous results from gene expression correlation analysis. Concurrently, this study also suppressed the expression of CsHsf10 in tea plants by designing oligodeoxynucleotides (ODN-CsHsf10). The results indicated that under drought stress, the Fv/Fm value of tea seedlings with silenced CsHsf10 decreased significantly, resulting in reduced photosynthetic efficiency, a marked increase in MDA content, and a substantially lower catechin component content compared to the control group. These findings further validate the positive regulatory role of CsHsf10 in the drought resistance response of tea plants. Subsequently, we conducted a verification experiment, where yeast monoamide and luciferase reporter gene assays demonstrated that CsHsf10 could directly bind to CsPOD17 and promote the transcriptional expression of CsPOD17. Although the current evidence indicates an interaction between CsHsf10 and CsPOD17, the specific mechanism of this interaction under drought conditions remains unclear. However, given that an increase in peroxidase (POD) activity was observed in plants overexpressing CsHsf10 under drought conditions, this interaction may potentially enhance plant drought tolerance, laying a foundation for our subsequent research. Furthermore, the expression of CsHsf10 significantly increases the content of catechin components in tea plants subjected to drought stress. These findings indicate that, in the context of drought stress response, CsHsf10 not only enhances antioxidant capacity by modulating the activity of antioxidant enzymes but also improves the physiological state of tea plants by influencing the accumulation of secondary metabolites, thereby enhancing their resistance. Consequently, this study offers new insights and a theoretical foundation for the molecular breeding of tea plants, highlighting its significant scientific relevance and practical application (Figure 6).
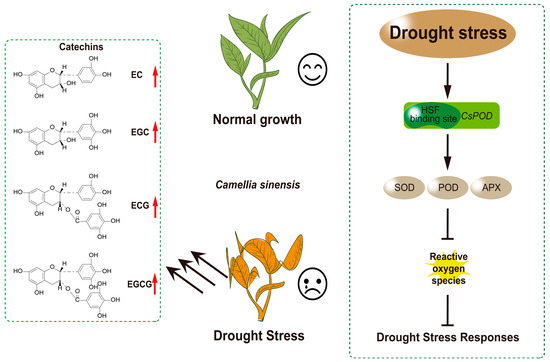
Figure 6.
Under drought stress conditions, the expression of CsHsf10 is closely associated with increased antioxidant enzyme activity, which may be linked to the interaction between CsHsf10 and CsPOD17. This interaction could enhance antioxidant enzyme activity, effectively reducing the damage caused by reactive oxygen species (ROS) to tea plant cells and significantly improving the drought resistance of tea plants. Furthermore, the expression of CsHsf10 can also lead to a notable increase in the content of catechin components in tea plants under drought stress. However, the specific regulatory mechanism through which CsHsf10 influences catechin content remains unclear and requires further investigation to elucidate its role in the secondary metabolism of tea plants.
5. Conclusions
This study identified the tea plant’s drought candidate gene CsHsf10 from 31 CsHsf genes. Experiments involving the overexpression of this gene in Arabidopsis and the use of antisense oligonucleotides in tea plants confirmed that CsHsf10 plays a positive role in the drought response of tea plants. Subsequently, the CsHsf10-CsPOD17 module was examined for its response to drought stress using yeast monoclonal assays and luciferase reporter gene experiments. However, the regulatory mechanism of the CsHsf10 gene about catechins under drought stress requires further investigation. This study provides strong evidence for identifying genes associated with drought resistance in tea plants and contributes to developing drought-resistant varieties.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/horticulturae11040373/s1, Figure S1: Phylogenetic analysis of Hsf proteins from Camellia sinensis (Cs), Arabidopsis thaliana (At) and Populus trichocarpa (Pt); Figure S2: Sequence alignment, conserved motif composition and gene structure of CsHsf gene; Figure S3: Collinear analysis; Figure S4: Distribution and statistical prediction of cis-acting elements in the 2000 bp upstream of the CsHsf gene promoter region; Table S1: All primer sequences in this study; Table S2: Physical and chemical properties of CsHsf gene family member proteins in tea plant; Table S3: Corresponding sequence numbers in Arabidopsis thaliana and Populus trichocarpa; Table S4: CsHsf protein sequence motifs; Table S5: Collinear analysis; Table S6: The other species homologous sequences of CsHsf10 correspond to sequence numbers.
Author Contributions
Conceptualization, investigation, methodology, data curation, writing—original draft, Y.L.; methodology, data curation, software, investigation, C.S. (Chenyu Shao) and R.X.; data curation, S.Q. and Q.H.; methodology, validation, Y.P. and J.G., and H.T., and Y.Z.; project administration, J.H. and Z.L.; writing—review and editing, funding acquisition, C.S (Chengwen Shen). All authors have read and agreed to the published version of the manuscript.
Funding
This study was financially supported by the project of the National Natural Science Foundation of China (32372765); the project of National Key Research and Development Plan (2022YFD1600801); the project of Chenzhou National Sustainable Development Agenda Innovation Demonstration Zone Construction Project (2022SFQ48); the project of Special Project for the Construction of Modern Agricultural Industrial Technology Systems in Hunan Province (HARS-10); the key projects from the Nature Science Foundation of Hunan Province (2021JC0007); the National Key Research and Development (R&D) Plan (2024YFD1200504-2).
Data Availability Statement
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Conflicts of Interest
The authors declare no conflicts of interest.
References
- Andrasi, N.; Pettko-Szandtner, A.; Szabados, L. Diversity of plant heat shock factors: Regulation, interactions, and functions. J. Exp. Bot. 2021, 72, 1558–1575. [Google Scholar] [PubMed]
- Von Koskull-Döring, P.; Scharf, K.D.; Nover, L. The diversity of plant heat stress transcription factors. Trends Plant Sci. 2007, 12, 452–457. [Google Scholar]
- Harrison, C.J.; Bohm, A.A.; Nelson, H.C. Crystal structure of the DNA binding domain of the heat shock transcription factor. Science 1994, 263, 224–227. [Google Scholar] [PubMed]
- Döring, P.; Treuter, E.; Kistner, C.; Lyck, R.; Chen, A.; Nover, L. The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell 2000, 12, 265–278. [Google Scholar] [CrossRef]
- Czarnecka-Verner, E.; Pan, S.; Salem, T.; Gurley, W.B. Plant class B HSFs inhibit transcription and exhibit affinity for TFIIB and TBP. Plant Mol. Biol. 2004, 56, 57–75. [Google Scholar] [PubMed]
- Xue, G.P.; Sadat, S.; Drenth, J.; McIntyre, C.L. The heat shock factor family from Triticum aestivum in response to heat and other major abiotic stresses and their role in regulation of heat shock protein genes. J. Exp. Bot. 2014, 65, 539–557. [Google Scholar]
- Zhuang, L.; Cao, W.; Wang, J.; Yu, J.; Yang, Z.; Huang, B. Characterization and functional analysis of FaHsfC1b from Festuca arundinacea conferring heat tolerance in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 2702. [Google Scholar] [CrossRef]
- Wu, Z.; Li, T.; Ding, L.; Wang, C.; Teng, R.; Xu, S.; Cao, X.; Teng, N. Lily LlHSFC2 coordinates with HSFAs to balance heat stress response and improve thermotolerance. New Phytol. 2024, 241, 2124–2142. [Google Scholar]
- Gao, L.; Pan, L.; Shi, Y.; Zeng, R.; Li, Z.; Zhang, X.; Zhao, X.; Gong, X.; Huang, W.; Yang, X.; et al. Genetic variation in a heat shock transcription factor modulates cold tolerance in maize. Mol. Plant. 2024, 17, 1423–1438. [Google Scholar]
- Bian, X.H.; Li, W.; Niu, C.F.; Wei, W.; Hu, Y.; Han, J.Q.; Lu, X.; Tao, J.J.; Jin, M.; Qin, H.; et al. A class B heat shock factor selected for during soybean domestication contributes to salt tolerance by promoting flavonoid biosynthesis. New Phytol. 2020, 225, 268–283. [Google Scholar]
- Wang, Y.; Cai, S.; Yin, L.; Shi, K.; Xia, X.; Zhou, Y.; Yu, J.; Zhou, J. Tomato HsfA1a plays a critical role in plant drought tolerance by activating ATG genes and inducing autophagy. Autophagy 2015, 11, 2033–2047. [Google Scholar] [PubMed]
- Bechtold, U.; Albihlal, W.S.; Lawson, T.; Fryer, M.J.; Sparrow, P.A.C.; Richard, F.; Persad, R.; Bowden, L.; Hickman, R.; Martin, C.; et al. Arabidopsis HEAT SHOCK TRANSCRIPTION FACTORA1b overexpression enhances water productivity, resistance to drought, and infection. J. Exp. Bot. 2013, 64, 3467–3481. [Google Scholar]
- Li, P.S.; Yu, T.F.; He, G.H.; Chen, M.; Zhou, Y.B.; Chai, S.C.; Xu, Z.S.; Ma, Y.Z. Genome-wide analysis of the Hsf family in soybean and functional identification of GmHsf-34 involvement in drought and heat stresses. BMC Genom. 2014, 15, 1009. [Google Scholar]
- Ma, H.; Wang, C.; Yang, B.; Cheng, H.; Wang, Z.; Mijiti, A.; Ren, C.; Qu, G.; Zhang, H.; Ma, L. CarHSFB2, a class B heat shock transcription factor, is involved in different developmental processes and various stress responses in chickpea (Cicer arietinum L.). Plant Mol. Biol. Rep. 2016, 34, 1–14. [Google Scholar] [CrossRef]
- Wang, L.; Liu, Y.; Chai, G.; Zhang, D.; Fang, Y.; Deng, K.; Aslam, M.; Niu, X.; Zhang, W.; Qin, Y.; et al. Identification of passion fruit HSF gene family and the functional analysis of PeHSF-C1a in response to heat and osmotic stress. Plant Physiol. Biochem. 2023, 200, 107800. [Google Scholar]
- Zhu, M.D.; Zhang, M.; Gao, D.J.; Zhou, K.; Tang, S.J.; Zhou, B.; Lv, Y.M. Rice OsHSFA3 gene improves drought tolerance by modulating polyamine biosynthesis depending on abscisic acid and ROS levels. Int. J. Mol. Sci. 2020, 21, 1857. [Google Scholar] [CrossRef] [PubMed]
- Ma, G.; Shen, J.; Yu, H.; Huang, X.; Deng, X.; Hu, Z.; Amee, M.; Chen, L.; Cao, L. Genome-wide identification and functional analyses of heat shock transcription factors involved in heat and drought stresses in ryegrass. Environ. Exp. Bot. 2022, 201, 104968. [Google Scholar]
- Wang, N.; Liu, W.; Yu, L.; Guo, Z.; Chen, Z.; Jiang, S.; Xu, H.; Fang, H.; Wang, Y.; Zhang, Z.; et al. HEAT SHOCK FACTOR A8a modulates flavonoid synthesis and drought tolerance. Plant Physiol. 2020, 184, 1273–1290. [Google Scholar]
- Zhang, F.; Pan, Z.; Han, C.; Dong, H.; Lin, L.; Qiao, Q.; Zhao, K.; Wu, J.; Tao, S.; Zhang, S.; et al. Pyrus betulaefolia ERF3 interacts with HsfC1a to coordinately regulate aquaporin PIP1;4 and NCED4 for drought tolerance. Hortic. Res. 2024, 11, uhae090. [Google Scholar]
- Ye, L.; Xue, H.; Li, N.; Ye, M.; Huang, J.; Wang, X.; Wu, J.; Ding, C. Genome-wide identification and expression analysis of pseudo-response regulators (PRRs) in the tea plant Camellia sinensis (L.) O. Kuntze. Horticulturae 2024, 10, 1294. [Google Scholar] [CrossRef]
- Xue, H.; Ren, X.; Li, S.; Wu, X.; Cheng, H.; Xu, B.; Gu, B.; Yang, G.; Peng, C.; Ge, Y.; et al. Assessment of private economic benefits and positive environmental externalities of tea plantation in China. Environ. Monit. Assess. 2013, 185, 8501–8516. [Google Scholar] [CrossRef] [PubMed]
- Huang, F.; Lei, Y.; Duan, J.; Kang, Y.; Luo, Y.; Ding, D.; Chen, Y.; Li, S. Investigation of heat stress responses and adaptation mechanisms by integrative metabolome and transcriptome analysis in tea plants (Camellia sinensis). Sci. Rep. 2024, 14, 10023. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Gao, T.; Wan, S.; Zhang, Y.; Yang, J.; Yu, Y.; Wang, W. Genome-wide identification, classification and expression analysis of the HSP gene superfamily in tea plant (Camellia sinensis). Int. J. Mol. Sci. 2018, 19, 2633. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Zhu, X.; Hou, M.; Luo, W.; Jiang, Y.; Yu, Y.; Wang, J.; Yuan, H.; Huang, X.; Hua, J. Effects of low-temperature stress on cold resistance biochemical characteristics of Dali and Siqiu tea seedlings. Horticulturae 2024, 10, 823. [Google Scholar] [CrossRef]
- Yang, L.; Zhao, P.; Song, X.; Ma, Y.; Fan, L.; Xie, M.; Song, Z.; Zhang, X.; Ma, H. Molecular and physiological responses of toona ciliata to simulated drought stress. Horticulturae 2024, 10, 1029. [Google Scholar] [CrossRef]
- Baek, S.G.; Shin, J.W.; Nam, J.I.; Seo, J.M.; Kim, J.M.; Woo, S.Y. Drought and salinity stresses response in three korean native herbaceous plants and their suitability as garden plants. Horticulturae 2024, 10, 1225. [Google Scholar] [CrossRef]
- Hasan, R.; Islam, A.F.M.S.; Maleque, M.A.; Islam, M.S.; Rahman, M.M. Effect of drought stress on leaf productivity and liquor quality of tea: A review. Asian J. Soil Sci. Plant Nutr. 2023, 9, 1–10. [Google Scholar] [CrossRef]
- Liu, S.C.; Jin, J.Q.; Ma, J.Q.; Yao, M.Z.; Ma, C.L.; Li, C.F.; Ding, Z.T.; Chen, L. Transcriptomic analysis of tea plant responding to drought stress and recovery. PLoS ONE. 2016, 11, e0147306. [Google Scholar] [CrossRef]
- Zhang, X.; Xu, W.; Ni, D.; Wang, M.; Guo, G. Genome-wide characterization of tea plant (Camellia sinensis) Hsf transcription factor family and role of CsHsfA2 in heat tolerance. BMC Plant Biol. 2020, 20, 224. [Google Scholar] [CrossRef]
- Seth, R.; Maritim, T.K.; Parmar, R.; Sharma, R.K. Underpinning the molecular programming attributing heat stress associated thermotolerance in tea (Camellia sinensis (L.) O. Kuntze). Hort. Res. 2021, 8, 99. [Google Scholar] [CrossRef]
- Zheng, S.; Liu, C.; Zhou, Z.; Xu, L.; Lai, Z. Physiological and transcriptome analyses reveal the protective effect of exogenous trehalose in response to heat stress in tea plant (Camellia sinensis). Plants 2024, 13, 1339. [Google Scholar] [CrossRef]
- Zhang, X.; Li, L.; He, Y.; Lang, Z.; Zhao, Y.; Tao, H.; Li, Q.; Hong, G. The CsHSFA-CsJAZ6 module-mediated high temperature regulates flavonoid metabolism in Camellia sinensis. Plant Cell Environ. 2023, 46, 2401–2418. [Google Scholar] [PubMed]
- Li, G.; Shi, X.; Lin, Q.; Lv, M.; Chen, J.; Wen, Y.; Feng, Z.; Azam, S.M.; Cheng, Y.; Wang, S.; et al. Genome-wide identification and expression analysis of heat shock transcription factors in Camellia sinensis under abiotic stress. Plants 2025, 14, 697. [Google Scholar] [CrossRef]
- Mistry, J.; Chuguransky, S.; Williams, L.; Qureshi, M.; Salazar, G.A.; Sonnhammer, E.L.L.; Tosatto, S.C.E.; Paladin, L.; Raj, S.; Richardson, L.J.; et al. Pfam: The protein families database in 2021. Nucleic Acids Res. 2021, 49, D412–D419. [Google Scholar] [PubMed]
- Chen, C.; Wu, Y.; Li, J.; Wang, X.; Zeng, Z.; Xu, J.; Liu, Y.; Feng, J.; Chen, H.; He, Y.; et al. TBtools-II: A “one for all, all for one” bioinformatics platform for biological big-data mining. Mol. Plant. 2023, 16, 1733–1742. [Google Scholar] [PubMed]
- Horton, P.; Park, K.J.; Obayashi, T.; Fujita, N.; Harada, H.; Adams-Collier, C.J.; Nakai, K. WoLF PSORT: Protein localization predictor. Nucleic Acids Res. 2007, 35, W585–W587. [Google Scholar]
- Hubel, A.; Lee, J.H.; Wu, C.; Schoffl, F. Arabidopsis heat shock factor is constitutively active in Drosophila and human cells. Mol. Gen. Genet. 1995, 248, 136–141. [Google Scholar] [CrossRef]
- Zhao, K.; Dang, H.; Zhou, L.; Hu, J.; Jin, X.; Han, Y.; Wang, S. Genome-wide identification and expression analysis of the HSF gene family in poplar. Forests 2023, 14, 510. [Google Scholar] [CrossRef]
- Goodstein, D.M.; Shu, S.; Howson, R.; Neupane, R.; Hayes, R.D.; Fazo, J.; Mitros, T.; Dirks, W.; Hellsten, U.; Putnam, N.; et al. Phytozome: A comparative platform for green plant genomics. Nucleic Acids Res. 2012, 40, D1178–D1186. [Google Scholar] [CrossRef]
- Drozdetskiy, A.; Cole, C.; Procter, J.; Barton, G.J. JPred4: A protein secondary structure prediction server. Nucleic Acids Res. 2015, 43, W389–W394. [Google Scholar]
- KB, N. GeneDoc: Analysis and visualization of genetic variation. EMBnet News. 1997, 4, 1–4. [Google Scholar]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [PubMed]
- Zhang, C.; He, Q.; Wang, M.; Gao, X.; Chen, J.; Shen, C. Exogenous indole acetic acid alleviates Cd toxicity in tea (Camellia sinensis). Ecotoxicol. Environ. Saf. 2020, 190, 110090. [Google Scholar] [PubMed]
- Shao, C.; Chen, J.; Lv, Z.; Gao, X.; Guo, S.; Xu, R.; Deng, Z.; Yao, S.; Chen, Z.; Kang, Y.; et al. Staged and repeated drought-induced regulation of phenylpropanoid synthesis confers tolerance to a water deficit environment in Camellia sinensis. Ind. Crops Prod. 2023, 201, 116843. [Google Scholar]
- Shraim, A.M.; Ahmed, T.A.; Rahman, M.M.; Hijji, Y.M. Determination of total flavonoid content by aluminum chloride assay: A critical evaluation. LWT-Food Sci. Technol. 2021, 150, 111932. [Google Scholar]
- Shao, C.; Deng, Z.; Liu, J.; Li, Y.; Zhang, C.; Yao, S.; Zuo, H.; Shi, Y.; Yuan, S.; Qin, L.; et al. Effects of preharvest shading on dynamic changes in metabolites, gene expression, and enzyme activity of three tea types during processing. J. Agric. Food Chem. 2022, 70, 14544–14558. [Google Scholar]
- Zhou, M.; Zheng, S.; Liu, R.; Lu, J.; Lu, L.; Zhang, C.; Liu, Z.; Luo, C.; Zhang, L.; Yant, L.; et al. Genome-wide identification, phylogenetic and expression analysis of the heat shock transcription factor family in bread wheat (Triticum aestivum L.). BMC Genom. 2019, 20, 505. [Google Scholar]
- Liu, G.; Chai, F.; Wang, Y.; Jiang, J.; Duan, W.; Wang, Y.; Wang, F.; Li, S.; Wang, L. Genome-wide Identification and classification of HSF family in grape, and their transcriptional analysis under heat acclimation and heat stress. Hortic. Plant J. 2018, 4, 133–143. [Google Scholar]
- Wan, X.; Yang, J.; Guo, C.; Bao, M.; Zhang, J. Genome-wide identification and classification of the Hsf and sHsp gene families in Prunus mume, and transcriptional analysis under heat stress. PeerJ 2019, 7, e7312. [Google Scholar] [CrossRef]
- Yang, Q.Q.; Yang, F.; Liu, C.Y.; Zhao, Y.Q.; Lu, X.J.; Ge, J.; Zhang, B.W.; Li, M.Q.; Yang, Y.; Fan, J.D. Genome-wide analysis of the HSF family in Allium sativum L. and AsHSFB1 overexpression in Arabidopsis under heat stress. BMC Genom. 2024, 25, 1072. [Google Scholar] [CrossRef]
- Deng, C.; Chen, Y.; Wei, W.; Chen, X.; Jiang, J. Genome-wide identification of heat shock transcription factor family and key members response analysis to heat stress in loquat. Horticulturae 2024, 10, 1195. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, F.; Ma, Y.; Zhang, T.; Sun, P.; Lan, M.; Li, F.; Fang, W. An ancient whole-genome duplication event and its contribution to flavor compounds in the tea plant (Camellia sinensis). Hortic. Res. 2021, 8, 176. [Google Scholar] [CrossRef] [PubMed]
- Scharf, K.D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. BBA-Gene Regul. Mech. 2012, 1819, 104–119. [Google Scholar] [CrossRef]
- Dossa, K.; Diouf, D.; Cisse, N. Genome-wide investigation of Hsf genes in sesame reveals their segmental duplication expansion and their active role in drought stress response. Front. Plant Sci. 2016, 7, 1522. [Google Scholar]
- Tashi, G.; Zhan, H.; Xing, G.; Chang, X.; Zhang, H.; Nie, X.; Ji, W. Genome-wide identification and expression analysis of heat shock transcription factor family in Chenopodium quinoa Willd. Agronomy 2018, 8, 103. [Google Scholar] [CrossRef]
- Peteranderl, R.; Rabenstein, M.; Shin, Y.K.; Liu, C.W.; Wemmer, D.E.; King, D.S.; Nelson, H.C. Biochemical and biophysical characterization of the trimerization domain from the heat shock transcription factor. Biochemistry 1999, 38, 3559–3569. [Google Scholar] [CrossRef]
- Schramm, F.; Larkindale, J.; Kiehlmann, E.; Ganguli, A.; Englich, G.; Vierling, E.; von Koskull-Doering, P. A cascade of transcription factor DREB2A and heat stress transcription factor HsfA3 regulates the heat stress response of Arabidopsis. Plant J. 2008, 53, 264–274. [Google Scholar]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).